Abstract
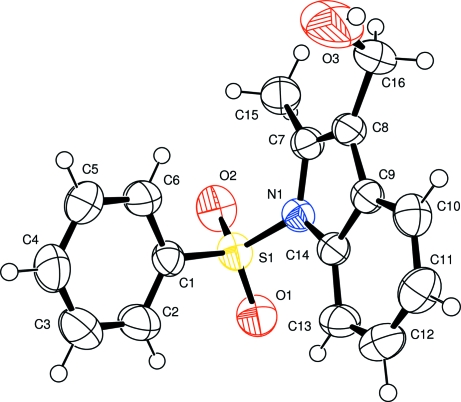
In the title compound, C16H15NO3S, the plane of the phenyl ring forms a dihedral angle of 80.37 (8)° with the indole ring system. The crystal packing is stabilized by weak O—H⋯O hydrogen bonds which link the molecules into infinite chains along the a axis of the crystal.
Related literature
For biological activity, see: Nieto et al. (2005 ▶); Pomarnacka & Kozlarska-Kedra (2003 ▶). For the structure of closely related compounds, see: Chakkaravarthi et al. (2007 ▶); Liu et al. (2007 ▶).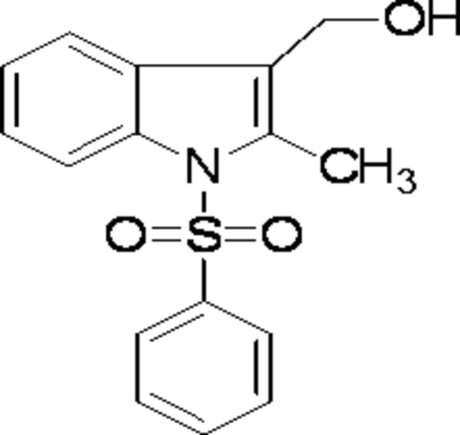
Experimental
Crystal data
C16H15NO3S
M r = 301.35
Triclinic,
a = 8.3780 (4) Å
b = 9.6969 (5) Å
c = 9.9630 (4) Å
α = 78.718 (2)°
β = 65.347 (3)°
γ = 78.884 (2)°
V = 715.77 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.23 mm−1
T = 295 (2) K
0.22 × 0.18 × 0.16 mm
Data collection
Bruker Kappa APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.920, T max = 0.963
14857 measured reflections
4088 independent reflections
3267 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.058
wR(F 2) = 0.241
S = 1.05
4088 reflections
191 parameters
H-atom parameters constrained
Δρmax = 0.43 e Å−3
Δρmin = −0.48 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808003024/ya2066sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808003024/ya2066Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O2i | 0.82 | 2.59 | 3.276 (5) | 142 |
Symmetry code: (i) 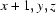 .
.
Acknowledgments
The authors acknowledge the Sophisticated Analytical Instrument Facility, Indian Institute of Technology, Madras, for the data collection.
supplementary crystallographic information
Comment
In continuation of our studies of benzenesulfonamide derivatives, which are known to exhibit antibacterial (Nieto et al., 2005), anticancer and anti - HIV (Pomarnacka & Kozlarska-Kedra, 2003) activities, we determined the crystal structure of the title compound, (I). The geometric parameters of the molecule of (I) (Fig. 1) agree well with those reported for similar structures (Chakkaravarthi et al., 2007; Liu et al., 2007).
The plane of the phenyl ring forms the dihedral angle of 80.37 (8)° with the indole ring system. The N1—S1—C1 plane is also approximately orthogonal to indole (dihedral angle 79.21 (6)°) and makes an angle of 57.86 (11)° with the phenyl plane.
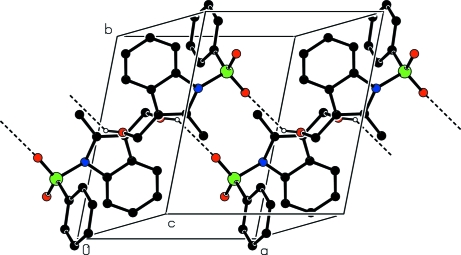
The crystal packing of (I) is stabilized by a rather weak O—H···O bonds which link the molecules into the infinite chains along the x-axis of the crystal (Fig. 2, Table 2).
Experimental
Benzenesulfonyl chloride (7.75 ml, 43.9 mmol), 60% NaOH solution (60 g in 100 ml), along with tetrabutylammonium hydrogensulfate (1.5 g) were added to the solution of 2-methylindole-3-carboxaldehyde (8.0 g, 50.3 mmol) in distilled benzene (200 ml). The two-phase system thus formed was stirred at room temperature for 2 h. It was then diluted with water (200 ml) and the organic layer was separated. The aqueous layer was extracted with benzene (two times by 30 ml). The combined organic layer was dried (Na2SO4). The benzene was then completely removed and the crude product was recrystallized from methanol to get 1-phenylsulfonyl-2-methylindole-3-carboxaldehyde.
NaBH4 (2.97 g, 78.60 mmol) was added slowly to a solution of 1-phenylsulfonyl-2-methylindole-3-carboxaldehyde (3 g, 13.10 mmol) in THF (30 ml). The reaction mixture was stirred at room temperature for 3 hrs. Dilute HCl (10%) was cautiously added until the solution became acidic. After most of the THF was removed in vacuo, the solution was extracted with dichloromethane and dried (MgSO4); the solvent was then removed under vacuo. The crude product thus obtained was recrystallized from 10% ethyl acetate/n-hexane.
Refinement
H atoms were positioned geometrically and refined using riding model approximation with C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C) for aromatic C—H, C—H = 0.97 Å and Uiso(H) = 1.2Ueq(C) for CH2, C—H = 0.96 Å and Uiso(H) = 1.5Ueq(C) for CH3 and O—H = 0.82 Å and Uiso(H) = 1.5Ueq(O) for OH. The hydroxyl O3 atom showed rather high thermal displacement parameters, however the attempts to introduce an alternative position and refine disordered model proved unsuccessful.
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 50% probability displacement ellipsoids; H atoms are shown as small spheres of arbitrary radius.
Fig. 2.
The packing of (I), viewed down the c axis. H-bonds are shown as dashed lines; H atoms not involved in hydrogen bonding have been omitted.
Crystal data
| C16H15NO3S | Z = 2 |
| Mr = 301.35 | F000 = 316 |
| Triclinic, P1 | Dx = 1.398 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 8.3780 (4) Å | Cell parameters from 8443 reflections |
| b = 9.6969 (5) Å | θ = 2.7–26.2º |
| c = 9.9630 (4) Å | µ = 0.23 mm−1 |
| α = 78.718 (2)º | T = 295 (2) K |
| β = 65.347 (3)º | Block, colourless |
| γ = 78.884 (2)º | 0.22 × 0.18 × 0.16 mm |
| V = 715.77 (6) Å3 |
Data collection
| Bruker Kappa APEXII diffractometer | 4088 independent reflections |
| Radiation source: fine-focus sealed tube | 3267 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.021 |
| T = 295(2) K | θmax = 30.0º |
| ω and φ scans | θmin = 2.2º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −11→11 |
| Tmin = 0.920, Tmax = 0.963 | k = −13→13 |
| 14857 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.058 | H-atom parameters constrained |
| wR(F2) = 0.241 | w = 1/[σ2(Fo2) + (0.0931P)2 + 0.2366P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 4088 reflections | Δρmax = 0.43 e Å−3 |
| 191 parameters | Δρmin = −0.48 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.70206 (7) | 0.26671 (6) | 0.24015 (6) | 0.0505 (2) | |
| O1 | 0.7232 (3) | 0.1941 (2) | 0.1217 (2) | 0.0682 (6) | |
| O2 | 0.5477 (2) | 0.3641 (2) | 0.2984 (3) | 0.0705 (6) | |
| O3 | 1.1581 (6) | 0.5512 (4) | 0.3706 (4) | 0.1316 (14) | |
| H3 | 1.2640 | 0.5450 | 0.3519 | 0.197* | |
| N1 | 0.8742 (2) | 0.35733 (18) | 0.17689 (19) | 0.0434 (4) | |
| C1 | 0.7297 (3) | 0.1424 (2) | 0.3853 (2) | 0.0464 (4) | |
| C2 | 0.7397 (5) | 0.0001 (3) | 0.3817 (4) | 0.0705 (8) | |
| H2 | 0.7350 | −0.0317 | 0.3013 | 0.085* | |
| C3 | 0.7567 (6) | −0.0952 (3) | 0.4989 (4) | 0.0831 (10) | |
| H3A | 0.7620 | −0.1917 | 0.4982 | 0.100* | |
| C4 | 0.7657 (4) | −0.0484 (3) | 0.6162 (3) | 0.0709 (8) | |
| H4 | 0.7779 | −0.1133 | 0.6946 | 0.085* | |
| C5 | 0.7569 (5) | 0.0929 (3) | 0.6185 (3) | 0.0705 (8) | |
| H5 | 0.7647 | 0.1236 | 0.6981 | 0.085* | |
| C6 | 0.7364 (4) | 0.1917 (3) | 0.5037 (3) | 0.0594 (6) | |
| H6 | 0.7275 | 0.2883 | 0.5064 | 0.071* | |
| C7 | 0.8842 (3) | 0.4756 (2) | 0.2371 (2) | 0.0449 (4) | |
| C8 | 1.0560 (3) | 0.4853 (2) | 0.2018 (2) | 0.0453 (4) | |
| C9 | 1.1626 (3) | 0.3694 (2) | 0.1214 (2) | 0.0413 (4) | |
| C10 | 1.3442 (3) | 0.3292 (3) | 0.0597 (3) | 0.0537 (5) | |
| H10 | 1.4219 | 0.3825 | 0.0652 | 0.064* | |
| C11 | 1.4065 (4) | 0.2092 (3) | −0.0093 (3) | 0.0594 (6) | |
| H11 | 1.5275 | 0.1794 | −0.0481 | 0.071* | |
| C12 | 1.2929 (4) | 0.1321 (3) | −0.0224 (3) | 0.0615 (6) | |
| H12 | 1.3393 | 0.0523 | −0.0718 | 0.074* | |
| C13 | 1.1125 (4) | 0.1701 (2) | 0.0358 (3) | 0.0525 (5) | |
| H13 | 1.0363 | 0.1171 | 0.0277 | 0.063* | |
| C14 | 1.0485 (3) | 0.2911 (2) | 0.1073 (2) | 0.0400 (4) | |
| C15 | 0.7267 (4) | 0.5750 (3) | 0.3152 (3) | 0.0650 (7) | |
| H15A | 0.6612 | 0.5303 | 0.4133 | 0.097* | |
| H15B | 0.6525 | 0.5993 | 0.2601 | 0.097* | |
| H15C | 0.7648 | 0.6594 | 0.3226 | 0.097* | |
| C16 | 1.1300 (4) | 0.5945 (3) | 0.2393 (3) | 0.0649 (7) | |
| H16A | 1.2413 | 0.6143 | 0.1580 | 0.078* | |
| H16B | 1.0488 | 0.6816 | 0.2494 | 0.078* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0467 (3) | 0.0609 (4) | 0.0548 (4) | −0.0134 (2) | −0.0299 (3) | −0.0024 (2) |
| O1 | 0.0776 (13) | 0.0891 (14) | 0.0633 (11) | −0.0306 (11) | −0.0455 (10) | −0.0057 (10) |
| O2 | 0.0440 (9) | 0.0813 (13) | 0.0859 (14) | −0.0033 (9) | −0.0311 (9) | −0.0021 (11) |
| O3 | 0.224 (4) | 0.128 (3) | 0.104 (2) | −0.087 (3) | −0.104 (2) | 0.0019 (19) |
| N1 | 0.0453 (9) | 0.0437 (8) | 0.0451 (9) | −0.0082 (7) | −0.0214 (7) | −0.0037 (6) |
| C1 | 0.0469 (10) | 0.0499 (10) | 0.0458 (10) | −0.0151 (8) | −0.0192 (8) | −0.0027 (8) |
| C2 | 0.100 (2) | 0.0570 (14) | 0.0674 (16) | −0.0225 (14) | −0.0387 (16) | −0.0107 (12) |
| C3 | 0.122 (3) | 0.0477 (13) | 0.081 (2) | −0.0179 (16) | −0.043 (2) | 0.0014 (13) |
| C4 | 0.085 (2) | 0.0636 (15) | 0.0549 (14) | −0.0142 (14) | −0.0242 (13) | 0.0098 (11) |
| C5 | 0.094 (2) | 0.0730 (17) | 0.0455 (12) | −0.0156 (15) | −0.0283 (13) | −0.0044 (11) |
| C6 | 0.0810 (17) | 0.0508 (12) | 0.0519 (12) | −0.0122 (11) | −0.0300 (12) | −0.0066 (9) |
| C7 | 0.0538 (11) | 0.0378 (9) | 0.0425 (9) | −0.0039 (8) | −0.0198 (8) | −0.0041 (7) |
| C8 | 0.0566 (12) | 0.0404 (9) | 0.0430 (9) | −0.0100 (8) | −0.0218 (9) | −0.0055 (7) |
| C9 | 0.0468 (10) | 0.0430 (9) | 0.0384 (8) | −0.0086 (7) | −0.0207 (8) | −0.0031 (7) |
| C10 | 0.0458 (11) | 0.0631 (13) | 0.0558 (12) | −0.0105 (10) | −0.0229 (10) | −0.0058 (10) |
| C11 | 0.0522 (13) | 0.0661 (14) | 0.0540 (12) | 0.0030 (11) | −0.0202 (10) | −0.0070 (11) |
| C12 | 0.0697 (16) | 0.0582 (13) | 0.0569 (13) | 0.0096 (11) | −0.0272 (12) | −0.0205 (10) |
| C13 | 0.0667 (14) | 0.0486 (11) | 0.0529 (11) | −0.0072 (10) | −0.0307 (10) | −0.0137 (9) |
| C14 | 0.0472 (10) | 0.0410 (9) | 0.0363 (8) | −0.0073 (7) | −0.0210 (7) | −0.0023 (7) |
| C15 | 0.0632 (15) | 0.0511 (12) | 0.0709 (16) | 0.0048 (11) | −0.0186 (12) | −0.0157 (11) |
| C16 | 0.0821 (18) | 0.0538 (13) | 0.0680 (15) | −0.0242 (13) | −0.0294 (13) | −0.0134 (11) |
Geometric parameters (Å, °)
| S1—O2 | 1.421 (2) | C7—C8 | 1.350 (3) |
| S1—O1 | 1.422 (2) | C7—C15 | 1.491 (3) |
| S1—N1 | 1.6619 (18) | C8—C9 | 1.439 (3) |
| S1—C1 | 1.757 (2) | C8—C16 | 1.499 (3) |
| O3—C16 | 1.396 (4) | C9—C10 | 1.389 (3) |
| O3—H3 | 0.8200 | C9—C14 | 1.394 (3) |
| N1—C14 | 1.412 (3) | C10—C11 | 1.371 (4) |
| N1—C7 | 1.425 (3) | C10—H10 | 0.9300 |
| C1—C2 | 1.373 (4) | C11—C12 | 1.377 (4) |
| C1—C6 | 1.382 (3) | C11—H11 | 0.9300 |
| C2—C3 | 1.380 (4) | C12—C13 | 1.377 (4) |
| C2—H2 | 0.9300 | C12—H12 | 0.9300 |
| C3—C4 | 1.368 (5) | C13—C14 | 1.391 (3) |
| C3—H3A | 0.9300 | C13—H13 | 0.9300 |
| C4—C5 | 1.362 (5) | C15—H15A | 0.9600 |
| C4—H4 | 0.9300 | C15—H15B | 0.9600 |
| C5—C6 | 1.388 (4) | C15—H15C | 0.9600 |
| C5—H5 | 0.9300 | C16—H16A | 0.9700 |
| C6—H6 | 0.9300 | C16—H16B | 0.9700 |
| O2—S1—O1 | 119.32 (13) | C7—C8—C16 | 127.6 (2) |
| O2—S1—N1 | 106.85 (11) | C9—C8—C16 | 123.9 (2) |
| O1—S1—N1 | 106.16 (11) | C10—C9—C14 | 119.9 (2) |
| O2—S1—C1 | 109.90 (12) | C10—C9—C8 | 132.5 (2) |
| O1—S1—C1 | 109.10 (12) | C14—C9—C8 | 107.59 (18) |
| N1—S1—C1 | 104.41 (9) | C11—C10—C9 | 118.6 (2) |
| C16—O3—H3 | 109.5 | C11—C10—H10 | 120.7 |
| C14—N1—C7 | 107.57 (17) | C9—C10—H10 | 120.7 |
| C14—N1—S1 | 120.92 (14) | C10—C11—C12 | 121.1 (2) |
| C7—N1—S1 | 125.77 (15) | C10—C11—H11 | 119.4 |
| C2—C1—C6 | 121.3 (2) | C12—C11—H11 | 119.4 |
| C2—C1—S1 | 120.2 (2) | C13—C12—C11 | 121.6 (2) |
| C6—C1—S1 | 118.42 (18) | C13—C12—H12 | 119.2 |
| C1—C2—C3 | 119.1 (3) | C11—C12—H12 | 119.2 |
| C1—C2—H2 | 120.4 | C12—C13—C14 | 117.4 (2) |
| C3—C2—H2 | 120.4 | C12—C13—H13 | 121.3 |
| C4—C3—C2 | 120.4 (3) | C14—C13—H13 | 121.3 |
| C4—C3—H3A | 119.8 | C13—C14—C9 | 121.3 (2) |
| C2—C3—H3A | 119.8 | C13—C14—N1 | 131.2 (2) |
| C5—C4—C3 | 120.1 (3) | C9—C14—N1 | 107.57 (17) |
| C5—C4—H4 | 119.9 | C7—C15—H15A | 109.5 |
| C3—C4—H4 | 119.9 | C7—C15—H15B | 109.5 |
| C4—C5—C6 | 120.9 (3) | H15A—C15—H15B | 109.5 |
| C4—C5—H5 | 119.5 | C7—C15—H15C | 109.5 |
| C6—C5—H5 | 119.5 | H15A—C15—H15C | 109.5 |
| C1—C6—C5 | 118.1 (2) | H15B—C15—H15C | 109.5 |
| C1—C6—H6 | 121.0 | O3—C16—C8 | 112.6 (2) |
| C5—C6—H6 | 121.0 | O3—C16—H16A | 109.1 |
| C8—C7—N1 | 108.74 (19) | C8—C16—H16A | 109.1 |
| C8—C7—C15 | 127.7 (2) | O3—C16—H16B | 109.1 |
| N1—C7—C15 | 123.4 (2) | C8—C16—H16B | 109.1 |
| C7—C8—C9 | 108.49 (19) | H16A—C16—H16B | 107.8 |
| O2—S1—N1—C14 | −178.08 (15) | C15—C7—C8—C9 | −177.5 (2) |
| O1—S1—N1—C14 | −49.73 (17) | N1—C7—C8—C16 | 178.9 (2) |
| C1—S1—N1—C14 | 65.49 (17) | C15—C7—C8—C16 | 3.4 (4) |
| O2—S1—N1—C7 | 32.0 (2) | C7—C8—C9—C10 | 180.0 (2) |
| O1—S1—N1—C7 | 160.38 (18) | C16—C8—C9—C10 | −0.9 (4) |
| C1—S1—N1—C7 | −84.39 (18) | C7—C8—C9—C14 | 1.1 (2) |
| O2—S1—C1—C2 | 123.0 (3) | C16—C8—C9—C14 | −179.8 (2) |
| O1—S1—C1—C2 | −9.6 (3) | C14—C9—C10—C11 | −2.3 (3) |
| N1—S1—C1—C2 | −122.7 (2) | C8—C9—C10—C11 | 178.9 (2) |
| O2—S1—C1—C6 | −55.4 (2) | C9—C10—C11—C12 | 2.2 (4) |
| O1—S1—C1—C6 | 172.0 (2) | C10—C11—C12—C13 | −1.4 (4) |
| N1—S1—C1—C6 | 58.9 (2) | C11—C12—C13—C14 | 0.7 (4) |
| C6—C1—C2—C3 | 0.0 (5) | C12—C13—C14—C9 | −0.9 (3) |
| S1—C1—C2—C3 | −178.3 (3) | C12—C13—C14—N1 | 179.7 (2) |
| C1—C2—C3—C4 | −0.8 (6) | C10—C9—C14—C13 | 1.8 (3) |
| C2—C3—C4—C5 | 0.4 (6) | C8—C9—C14—C13 | −179.15 (18) |
| C3—C4—C5—C6 | 0.8 (5) | C10—C9—C14—N1 | −178.74 (18) |
| C2—C1—C6—C5 | 1.1 (4) | C8—C9—C14—N1 | 0.3 (2) |
| S1—C1—C6—C5 | 179.5 (2) | C7—N1—C14—C13 | 177.9 (2) |
| C4—C5—C6—C1 | −1.6 (5) | S1—N1—C14—C13 | 23.2 (3) |
| C14—N1—C7—C8 | 2.2 (2) | C7—N1—C14—C9 | −1.5 (2) |
| S1—N1—C7—C8 | 155.38 (16) | S1—N1—C14—C9 | −156.26 (14) |
| C14—N1—C7—C15 | 178.0 (2) | C7—C8—C16—O3 | 93.7 (4) |
| S1—N1—C7—C15 | −28.9 (3) | C9—C8—C16—O3 | −85.2 (3) |
| N1—C7—C8—C9 | −2.0 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O2i | 0.82 | 2.59 | 3.276 (5) | 142 |
Symmetry codes: (i) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: YA2066).
References
- Bruker (2004). APEX2 Version 1.0–27. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chakkaravarthi, G., Ramesh, N., Mohanakrishnan, A. K. & Manivannan, V. (2007). Acta Cryst. E63, o3564.
- Liu, Y., Gribble, G. W. & Jasinski, J. P. (2007). Acta Cryst. E63, o738–o740.
- Nieto, M. J., Alovero, F. L., Manzo, R. H. & Mazzieri, M. R. (2005). Eur. J. Med. Chem.40, 361–369. [DOI] [PubMed]
- Pomarnacka, E. & Kozlarska-Kedra, I. (2003). Il Farmaco, 58, 423–429. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808003024/ya2066sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808003024/ya2066Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report