Abstract
The title compound, C7H15N3OP+·I3 −, is a derivative of the well known water-soluble aminophosphine 1,3,5-triaza-7-phosphaadamantane (PTA). The crystal structure is composed of a cage-like 1-methyl-1-azonia-3,5-diaza-7-phosphatricyclo[3.3.1.1]decane 7-oxide cation and a triiodide anion. The N-methylation of the PTA cage results in a slight elongation of the corresponding C—N bonds, while the oxidation of the P atom leads to a slight shortening of the C—P bonds in comparison with those of PTA. In general, most of the bonding parameters are comparable with those reported for related compounds bearing the PTA core. Two intermolecular C—H⋯O hydrogen bonds between methylene groups and the P=O group are responsible for the linkage of neighbouring cations into linear one-dimensional hydrogen-bonded chains.
Related literature
For a comprehensive review of PTA chemistry, see: Phillips et al. (2004 ▶). For general background, see: Kirillov et al. (2007 ▶); Smoleński & Pombeiro (2008 ▶). For synthesis of PTA and its N-methylated derivative, see: Daigle et al. (1974 ▶); Daigle (1998 ▶). For related structures, see: Forward et al. (1996a
▶,b
▶); Otto et al. (2005 ▶); Frost et al. (2006 ▶); Marsh et al. (2002 ▶).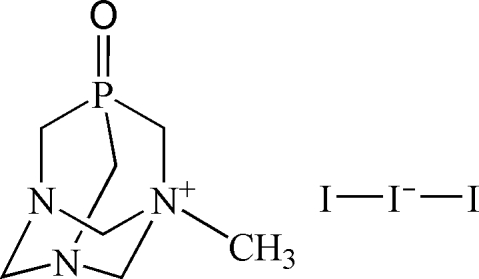
Experimental
Crystal data
C7H15N3OP+·I3 −
M r = 568.89
Monoclinic,
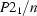
a = 7.1570 (8) Å
b = 8.2257 (8) Å
c = 25.903 (3) Å
β = 92.472 (7)°
V = 1523.5 (3) Å3
Z = 4
Mo Kα radiation
μ = 6.24 mm−1
T = 150 (2) K
0.13 × 0.10 × 0.10 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.497, T max = 0.574 (expected range = 0.464–0.536)
11526 measured reflections
2789 independent reflections
2214 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.102
S = 1.12
2789 reflections
172 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 2.44 e Å−3
Δρmin = −1.03 e Å−3
Data collection: SMART (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: WinGX (Version 1.70.01; Farrugia, 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808001426/kp2159sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808001426/kp2159Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| C1—N1 | 1.479 (8) |
| C1—P1 | 1.821 (8) |
| C2—N2 | 1.486 (8) |
| C2—P1 | 1.799 (7) |
| C3—N3 | 1.495 (9) |
| C3—P1 | 1.825 (8) |
| C4—N3 | 1.496 (9) |
| C12—N1 | 1.462 (9) |
| C12—N2 | 1.467 (10) |
| C23—N2 | 1.440 (9) |
| C23—N3 | 1.550 (9) |
| C31—N1 | 1.441 (9) |
| C31—N3 | 1.551 (9) |
| O1—P1 | 1.483 (5) |
| I1—I3 | 2.9067 (8) |
| I1—I2 | 2.9127 (7) |
| I3—I1—I2 | 172.41 (2) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C23—H23A⋯O1i | 0.99 (10) | 2.26 (11) | 3.161 (9) | 150 (9) |
| C31—H31A⋯O1i | 0.97 (10) | 2.23 (10) | 3.160 (9) | 161 (8) |
Symmetry code: (i) 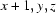 .
.
Acknowledgments
This work was supported by the Foundation for Science and Technology (FCT), Portugal, and its POCI 2010 programme (FEDER funded).
supplementary crystallographic information
Comment
Within our ongoing research (Kirillov et al., 2007; Smoleński & Pombeiro, 2008) on the synthesis of transition metal complexes with PTA or derived ligands, we have attempted the reaction of a copper(II) salt with N-methyl-1,3,5-triaza-7-phospha-adamantane iodide, which resulted in the formation of the title compound, (I), as a by-product. Its crystal structure is reported herein.
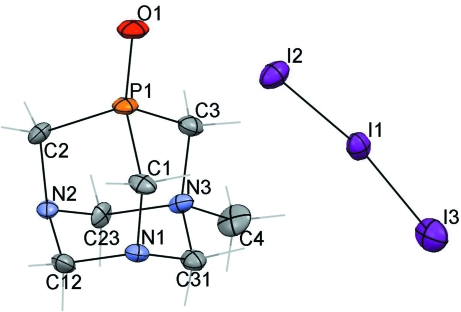
The molecular structure of (I) (Fig. 1) bears a cage-like cation [C7H15N3OP]+ and a tri-iodide anion, with the shortest cation···anion separation of ca 4.0 Å. The N-methylation of the PTA cage results in a slight elongation of the C—N bonds around N3 atom [avg. 1.53 (1) Å] in comparison with the C—N bonds around N1 and N2 atoms [avg. 1.46 (1) Å] (Table 1). The oxidation of P1 atom also slightly affects the C—P bonds [avg. 1.82 (1) Å] which are somewhat shorter than those in PTA [avg. 1.86 (1) Å]. The tri-iodide anion with the I2—I1—I3 angle of 172.41 (2)° deviates from the linear geometry. In general, most of the bonding parameters of (I) agree within values reported for the related compound, [C7H15N3OP][BPh4] (Forward et al., 1996a,b), possessing similar cation, as well as for other N-alkylated (Otto et al., 2005; Forward et al., 1996a,b) or P-oxidized (Frost et al., 2006; Marsh et al., 2002) PTA derivatives.
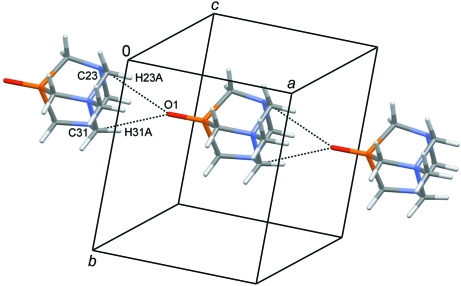
In (I), the neighbouring cationic units are combined into the linear one-dimensional H-bonded chains (Fig. 2) by means of two intermolecular C—H···O hydrogen bonds [C23—H23A···O1i 1.00 (11) Å, 2.26 (11) Å, 3.161 (9) Å, 150 (9)°; C31—-H31A···O1i 0.97 (10) Å, 2.23 (10) Å, 3.160 (9) Å, 161 (8)°; symmetry code: 1 + x, y, z], which link the methylene groups (C23, C31) with the O1 atom of the P=O moiety.
Experimental
The aqueous solutions (5 ml each) of Cu(NO3)2.2.5 H2O (116 mg, 0.50 mmol) and N-methyl-1,3,5-triaza-7-phospha-adamantane iodide, [C7H15N3P]I (299 mg, 1.00 mmol) [for the synthesis of this compound, see: Daigle et al. (1974); Daigle (1998)], were combined and left stirring in air at ambient temperature for 1 h. The resulting white suspension containing mainly a CuI aminophosphine compound was filtered off. The colourless filtrate was left to evaporate in a beaker in air for two weeks, leading to the formation of a small crop of red X-ray quality crystals of compound (I) as a by-product (it is typically contaminated by a colourless crystalline material). FT–IR (KBr pellet), cm-1: 2967 w, 2939 w, 1449 m, 1384 s, 1304 m, 1279 w, 1246 w, 1195 s [ν(P=O)], 1108 w, 1091 w, 1066 w, 1019 m, 983 m, 934 m, 900 w, 876 w, 816 m, 792 w, 752 m, 544 w, 441 w, 408 w. FAB-MS+ (m-nitrobenzylicalcohol), m/z: 188 [C7H15N3OP]+.
Refinement
All hydrogen atoms were located except from H4A, H4B and H4C which were inserted in calculated positions.
Figures
Fig. 1.
The molecular structure of the title compound with the atom labelling scheme. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented as grey sticks. C, grey; N, blue; P, orange; O, red; I, purple.
Fig. 2.
Fragment of the crystal packing diagram of (I) showing the generation of a one-dimensional linear chain from the neighbouring cations via intermolecular C—H···O hydrogen bonds (dotted lines). Tri-iodide anions are omitted for clarity. C, grey; N, blue; P, orange; O, red; H, pale grey.
Crystal data
| C7H15N3OP+·I3– | F000 = 1040 |
| Mr = 568.89 | Dx = 2.480 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation λ = 0.71069 Å |
| Hall symbol: -P2yn | Cell parameters from 2835 reflections |
| a = 7.1570 (8) Å | θ = 2.6–27.9º |
| b = 8.2257 (8) Å | µ = 6.24 mm−1 |
| c = 25.903 (3) Å | T = 150 (2) K |
| β = 92.472 (7)º | Plate, red |
| V = 1523.5 (3) Å3 | 0.13 × 0.10 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2789 independent reflections |
| Radiation source: fine-focus sealed tube | 2214 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.040 |
| T = 150(2) K | θmax = 25.4º |
| φ and ω scans | θmin = 2.9º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −8→8 |
| Tmin = 0.497, Tmax = 0.574 | k = −9→9 |
| 11526 measured reflections | l = −29→31 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.102 | w = 1/[σ2(Fo2) + (0.045P)2 + 6.2243P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max = 0.017 |
| 2789 reflections | Δρmax = 2.44 e Å−3 |
| 172 parameters | Δρmin = −1.03 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.5522 (9) | 0.4541 (10) | 0.1979 (3) | 0.0186 (16) | |
| C2 | 0.5510 (9) | 0.1132 (8) | 0.1974 (3) | 0.0156 (15) | |
| C3 | 0.5535 (10) | 0.2823 (10) | 0.1042 (3) | 0.0188 (15) | |
| C4 | 0.8586 (11) | 0.2823 (11) | 0.0631 (3) | 0.0294 (18) | |
| H4A | 0.9942 | 0.2819 | 0.0701 | 0.044* | |
| H4B | 0.8230 | 0.3796 | 0.0432 | 0.044* | |
| H4C | 0.8222 | 0.1851 | 0.0432 | 0.044* | |
| C12 | 0.8197 (10) | 0.2815 (10) | 0.2227 (3) | 0.0192 (15) | |
| C23 | 0.8255 (10) | 0.1301 (8) | 0.1442 (3) | 0.0162 (15) | |
| C31 | 0.8255 (10) | 0.4347 (10) | 0.1446 (3) | 0.0192 (16) | |
| N1 | 0.7565 (7) | 0.4292 (7) | 0.1960 (2) | 0.0169 (13) | |
| N2 | 0.7569 (7) | 0.1334 (7) | 0.1956 (2) | 0.0141 (12) | |
| N3 | 0.7614 (8) | 0.2828 (7) | 0.1131 (2) | 0.0168 (12) | |
| O1 | 0.2300 (7) | 0.2825 (7) | 0.1582 (2) | 0.0268 (12) | |
| P1 | 0.4367 (2) | 0.2827 (2) | 0.16547 (7) | 0.0168 (4) | |
| I1 | 0.42016 (7) | 0.78153 (6) | 0.088953 (18) | 0.02270 (15) | |
| I2 | 0.14227 (7) | 0.78170 (6) | 0.16786 (2) | 0.02691 (16) | |
| I3 | 0.73414 (8) | 0.78543 (8) | 0.02143 (2) | 0.03606 (18) | |
| H1A | 0.517 (12) | 0.452 (11) | 0.232 (4) | 0.043* | |
| H1B | 0.498 (12) | 0.558 (11) | 0.183 (3) | 0.043* | |
| H2A | 0.501 (12) | 0.108 (11) | 0.232 (4) | 0.043* | |
| H2B | 0.521 (12) | 0.008 (12) | 0.182 (3) | 0.050* | |
| H3A | 0.512 (14) | 0.190 (12) | 0.084 (4) | 0.060* | |
| H3B | 0.522 (14) | 0.380 (13) | 0.085 (4) | 0.060* | |
| H12A | 0.948 (16) | 0.286 (12) | 0.225 (4) | 0.060* | |
| H12B | 0.793 (14) | 0.282 (11) | 0.256 (4) | 0.050* | |
| H23A | 0.964 (15) | 0.135 (13) | 0.147 (4) | 0.060* | |
| H23B | 0.769 (13) | 0.034 (13) | 0.127 (4) | 0.060* | |
| H31A | 0.959 (14) | 0.414 (12) | 0.147 (4) | 0.060* | |
| H31B | 0.768 (13) | 0.538 (13) | 0.126 (4) | 0.060* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.010 (3) | 0.026 (5) | 0.020 (4) | 0.005 (3) | 0.001 (3) | −0.003 (3) |
| C2 | 0.014 (3) | 0.011 (4) | 0.022 (4) | −0.002 (3) | 0.006 (3) | 0.000 (3) |
| C3 | 0.013 (3) | 0.021 (4) | 0.023 (4) | 0.002 (3) | −0.003 (3) | 0.000 (3) |
| C4 | 0.027 (4) | 0.041 (5) | 0.022 (4) | −0.001 (4) | 0.017 (3) | −0.007 (4) |
| C12 | 0.016 (4) | 0.021 (4) | 0.020 (4) | −0.001 (3) | −0.006 (3) | 0.000 (3) |
| C23 | 0.018 (4) | 0.006 (4) | 0.026 (4) | 0.004 (3) | 0.005 (3) | 0.002 (3) |
| C31 | 0.012 (4) | 0.027 (5) | 0.019 (4) | 0.001 (3) | 0.002 (3) | 0.001 (3) |
| N1 | 0.011 (3) | 0.020 (3) | 0.019 (3) | −0.001 (2) | −0.002 (2) | 0.000 (3) |
| N2 | 0.009 (3) | 0.017 (3) | 0.017 (3) | 0.001 (2) | 0.002 (2) | 0.002 (2) |
| N3 | 0.014 (3) | 0.020 (3) | 0.016 (3) | −0.005 (3) | 0.002 (2) | −0.004 (3) |
| O1 | 0.009 (2) | 0.029 (3) | 0.042 (3) | 0.000 (2) | −0.002 (2) | 0.002 (3) |
| P1 | 0.0075 (8) | 0.0187 (9) | 0.0244 (10) | 0.0007 (7) | 0.0003 (7) | −0.0004 (8) |
| I1 | 0.0282 (3) | 0.0183 (3) | 0.0215 (3) | 0.0009 (2) | −0.00114 (19) | −0.0002 (2) |
| I2 | 0.0230 (3) | 0.0207 (3) | 0.0376 (3) | 0.0000 (2) | 0.0081 (2) | 0.0000 (2) |
| I3 | 0.0341 (3) | 0.0527 (4) | 0.0218 (3) | 0.0017 (3) | 0.0054 (2) | −0.0023 (3) |
Geometric parameters (Å, °)
| C1—N1 | 1.479 (8) | C12—N1 | 1.462 (9) |
| C1—P1 | 1.821 (8) | C12—N2 | 1.467 (10) |
| C1—H1A | 0.94 (9) | C12—H12A | 0.92 (11) |
| C1—H1B | 1.01 (9) | C12—H12B | 0.90 (11) |
| C2—N2 | 1.486 (8) | C23—N2 | 1.440 (9) |
| C2—P1 | 1.799 (7) | C23—N3 | 1.550 (9) |
| C2—H2A | 0.98 (9) | C23—H23A | 0.99 (10) |
| C2—H2B | 0.97 (10) | C23—H23B | 0.99 (10) |
| C3—N3 | 1.495 (9) | C31—N1 | 1.441 (9) |
| C3—P1 | 1.825 (8) | C31—N3 | 1.551 (9) |
| C3—H3A | 0.96 (10) | C31—H31A | 0.97 (10) |
| C3—H3B | 0.96 (11) | C31—H31B | 1.06 (10) |
| C4—N3 | 1.496 (9) | O1—P1 | 1.483 (5) |
| C4—H4A | 0.9800 | I1—I3 | 2.9067 (8) |
| C4—H4B | 0.9800 | I1—I2 | 2.9127 (7) |
| C4—H4C | 0.9800 | ||
| N1—C1—P1 | 107.9 (5) | N2—C23—H23A | 108 (6) |
| N1—C1—H1A | 109 (5) | N3—C23—H23A | 106 (6) |
| P1—C1—H1A | 107 (6) | N2—C23—H23B | 107 (6) |
| N1—C1—H1B | 118 (5) | N3—C23—H23B | 107 (6) |
| P1—C1—H1B | 109 (5) | H23A—C23—H23B | 117 (8) |
| H1A—C1—H1B | 105 (7) | N1—C31—N3 | 110.8 (6) |
| N2—C2—P1 | 109.3 (5) | N1—C31—H31A | 109 (6) |
| N2—C2—H2A | 116 (5) | N3—C31—H31A | 99 (6) |
| P1—C2—H2A | 106 (5) | N1—C31—H31B | 108 (5) |
| N2—C2—H2B | 107 (5) | N3—C31—H31B | 108 (5) |
| P1—C2—H2B | 114 (5) | H31A—C31—H31B | 122 (8) |
| H2A—C2—H2B | 104 (7) | C31—N1—C12 | 110.7 (6) |
| N3—C3—P1 | 110.8 (5) | C31—N1—C1 | 114.0 (5) |
| N3—C3—H3A | 112 (6) | C12—N1—C1 | 112.6 (6) |
| P1—C3—H3A | 109 (6) | C23—N2—C12 | 110.4 (6) |
| N3—C3—H3B | 106 (6) | C23—N2—C2 | 113.9 (5) |
| P1—C3—H3B | 110 (6) | C12—N2—C2 | 111.2 (6) |
| H3A—C3—H3B | 109 (8) | C4—N3—C3 | 111.3 (6) |
| N3—C4—H4A | 109.5 | C4—N3—C31 | 108.6 (5) |
| N3—C4—H4B | 109.5 | C3—N3—C31 | 110.7 (6) |
| H4A—C4—H4B | 109.5 | C4—N3—C23 | 108.0 (6) |
| N3—C4—H4C | 109.5 | C3—N3—C23 | 110.4 (6) |
| H4A—C4—H4C | 109.5 | C31—N3—C23 | 107.8 (5) |
| H4B—C4—H4C | 109.5 | O1—P1—C2 | 119.2 (3) |
| N1—C12—N2 | 112.4 (5) | O1—P1—C1 | 119.3 (3) |
| N1—C12—H12A | 107 (6) | C2—P1—C1 | 101.5 (3) |
| N2—C12—H12A | 111 (6) | O1—P1—C3 | 112.4 (3) |
| N1—C12—H12B | 112 (6) | C2—P1—C3 | 100.5 (4) |
| N2—C12—H12B | 113 (6) | C1—P1—C3 | 100.8 (4) |
| H12A—C12—H12B | 100 (9) | I3—I1—I2 | 172.41 (2) |
| N2—C23—N3 | 111.1 (5) | ||
| N3—C31—N1—C12 | 57.7 (7) | N1—C31—N3—C4 | −172.0 (6) |
| N3—C31—N1—C1 | −70.5 (8) | N1—C31—N3—C3 | 65.6 (7) |
| N2—C12—N1—C31 | −59.3 (8) | N1—C31—N3—C23 | −55.2 (7) |
| N2—C12—N1—C1 | 69.6 (8) | N2—C23—N3—C4 | 172.4 (6) |
| P1—C1—N1—C31 | 65.5 (7) | N2—C23—N3—C3 | −65.7 (7) |
| P1—C1—N1—C12 | −61.6 (7) | N2—C23—N3—C31 | 55.3 (7) |
| N3—C23—N2—C12 | −57.4 (7) | N2—C2—P1—O1 | 174.7 (4) |
| N3—C23—N2—C2 | 68.6 (7) | N2—C2—P1—C1 | −52.0 (5) |
| N1—C12—N2—C23 | 59.1 (7) | N2—C2—P1—C3 | 51.4 (5) |
| N1—C12—N2—C2 | −68.4 (7) | N1—C1—P1—O1 | −175.4 (4) |
| P1—C2—N2—C23 | −64.0 (7) | N1—C1—P1—C2 | 51.3 (6) |
| P1—C2—N2—C12 | 61.6 (6) | N1—C1—P1—C3 | −51.8 (6) |
| P1—C3—N3—C4 | 179.9 (5) | N3—C3—P1—O1 | 179.9 (5) |
| P1—C3—N3—C31 | −59.2 (7) | N3—C3—P1—C2 | −52.2 (6) |
| P1—C3—N3—C23 | 60.0 (7) | N3—C3—P1—C1 | 51.8 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C23—H23A···O1i | 0.99 (10) | 2.26 (11) | 3.161 (9) | 150 (9) |
| C31—H31A···O1i | 0.97 (10) | 2.23 (10) | 3.160 (9) | 161 (8) |
Symmetry codes: (i) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: KP2159).
References
- Bruker (2004). APEX2, SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Daigle, D. J. (1998). Inorg. Synth.32, 40–45.
- Daigle, D. J., Pepperman, A. B. Jr & Vail, S. L. (1974). J. Heterocycl. Chem.11, 407–408.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Forward, J. M., Staples, R. J. & Fackler, J. P. Jr (1996a). Z. Kristallogr.211, 129–130.
- Forward, J. M., Staples, R. J. & Fackler, J. P. Jr (1996b). Z. Kristallogr.211, 131–132.
- Frost, B. J., Mebi, C. A. & Gingrich, P. W. (2006). Eur. J. Inorg. Chem. pp. 1182–1189.
- Kirillov, A. M., Smoleński, P., Guedes da Silva, M. F. C. & Pombeiro, A. J. L. (2007). Eur. J. Inorg. Chem. pp. 2686–2692.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Marsh, R. E., Kapon, M., Hu, S. & Herbstein, F. H. (2002). Acta Cryst. B58, 62–77. [DOI] [PubMed]
- Otto, S., Ionescu, A. & Roodt, A. (2005). J. Organomet. Chem.690, 4337–4342.
- Phillips, A. D., Gonsalvi, L., Romerosa, A., Vizza, F. & Peruzzini, M. (2004). Coord. Chem. Rev.248, 955–993.
- Sheldrick, G. M. (1996). SADABS. University of Gottingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Smoleński, P. & Pombeiro, A. J. L. (2008). Dalton Trans. pp. 87–91. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808001426/kp2159sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808001426/kp2159Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report