Abstract
Human papillomavirus (HPV) type 16 E6 (16E6) is an oncogenic, multifunctional nuclear protein that induces p53 degradation and perturbs normal cell cycle control, leading to immortalization and transformation of infected keratinocytes and epithelial cells. Although it is unclear how 16E6 disrupts the epigenetic profile of host genes, its presence in the nucleus is a key feature. The present report describes intrinsic properties of 16E6 that influence its nuclear import in living cells. When the coding region of full-length 16E6 was inserted in frame into the C terminus of green fluorescent protein (GFP), it effectively prevented the 16E6 pre-mRNA from being spliced and led to the expression of a GFP-E6 fusion which localized predominantly to the nucleus. Further studies identified three novel nuclear localization signals (NLSs) in 16E6 that drive the protein to accumulate in the nucleus. We found that all three NLS sequences are rich in positively charged basic residues and that point mutations in these key residues could abolish the retention of 16E6 in the nucleus as well as the p53 degradation and cell immortalization activities of the protein. When inserted into corresponding regions of low-risk HPV type 6 E6, the three NLS sequences described for 16E6 functioned actively in converting the normally cytoplasmic HPV type 6 E6 into a nuclear protein. The separate NLS sequences, however, appear to play different roles in nuclear import and retention of HPV E6. The discovery of three unique NLS sequences in 16E6 provides new insights into the nuclear association of 16E6 which may reveal other novel activities of this important oncogenic protein.
Human papillomaviruses (HPVs) are small DNA tumor viruses that replicate and assemble exclusively in the nucleus. HPVs infect keratinocytes in the basal layers of stratified epithelia and replicate in infected keratinocytes in a differentiation-dependent manner. Genital HPV infection causes benign or sometimes malignant lesions. Certain types of HPVs, such as high-risk or oncogenic HPV type 16 (HPV16), HPV18, and HPV31, are frequently detected in cervical and other genital cancers. A characteristic of infection by these HPVs is that viral genomes are commonly found integrated into the cancer cell genome. Other types of HPVs, such as low-risk or nononcogenic HPV6 and HPV11, which induce benign genital warts, are very rarely found in genital malignancies (42).
High-risk HPV E6 and E7, the two viral oncoproteins, appear to be most important for malignant conversion, as demonstrated by their capacity to immortalize and transform keratinocytes and epithelial cells. Low-risk HPV E6 and E7, however, lack such biological activity (1, 26, 41, 47, 55, 60, 69). Biochemically, high-risk E6 but not low-risk E6 interacts with E6AP and tumor suppressor protein p53 to induce the ubiquitination-mediated degradation of p53 (27, 59, 74), while high-risk E7 but not low-risk E7 interacts with tumor suppressor protein pRb to promote cell cycle progression (5, 13, 18, 59, 74). Thus, interactions with cellular tumor suppressor proteins and perturbation of normal cell cycle control by high-risk E6 and E7 are believed to be the most important influences for malignant conversion (43, 45, 68). In this regard, HPV16 E6 (16E6) binds E6AP more strongly and drives the degradation of p53 more efficiently than HPV18 E6 (18E6) (59). In contrast, HPV11 E6 (11E6) has minimal binding affinity for E6AP (28) and influences the degradation of p53 in vivo only weakly (70).
Full-length 16E6 is translated from a single bicistronic E6E7 mRNA without splicing of an intron within the E6 coding region, although spliced 16E6*I and 16E6*II mRNAs frequently exist and encode 16E6*I and 16E6*II proteins having the same N-terminal 41- or 48-amino-acid (aa) residues as those seen in the full-length 16E6 protein. Full-length 16E6 is a basic protein (∼18 kDa) composed of either 151 or 158 aa residues, depending on which one of the two juxtaposed AUGs is used as a start codon (48). Among its 151 residues, 31 (21%) are basic in nature; this percentage is higher than those for most proteins, which contain only 14% basic residues (44). Like E6 proteins encoded by other papillomaviruses, 16E6 contains four zinc-binding motifs, Cys-X-X-Cys, and forms two Cys-Cys fingers that bind zinc directly (30). These features of 16E6 catagorize it as a nucleophilic protein, like nuclear proteins in general. Besides the ability to immortalize and transform cells and induce p53 degradation, 16E6 is known to be functionally involved in the regulation of gene transcription (10, 12). In addition to p53, 16E6 can interact with other transcription factors and coactivators, including p300/CBP (52, 78), IRF-3 (57), and c-Myc (22). The multifunctional activity of 16E6 is not restricted to the nucleus, as this protein can act as a regulator of signal transduction through interactions with cytoplasmic E6BP (Erc55) (7), E6TP (16, 17, 63), paxillin (72), tumor necrosis factor receptor 1 (14), and PSD-95, Dlg, Zona occludens-1 proteins, such as hDlg (32, 37). Therefore, 16E6 can be regarded as a multifaceted viral protein with characteristic and distinct activities in the nucleus and cytoplasm of HPV16-infected cells. In contrast, the biological functions of 16E6*I and 16E6*II remain to be defined, although 18E6*I can antagonize the activity of full-length 18E6 (53, 54).
Consistent with the known biological activities and structural features of 16E6, full-length 16E6 has been identified as a nuclear protein by immunofluorescence staining (30, 62). Attempts to discern how 16E6 is directed to the nucleus have provided uncertain evidence due to a lack of both extensive analysis and availability of reliable anti-16E6 antibodies. Cys-66, but not Cys-136, of 16E6 was initially identified as being essential for the presence of E6 in the nucleus; therefore, the N-terminal zinc finger was proposed to be involved in the nuclear localization of E6 (30). However, later studies demonstrated that the C terminus of 16E6, between residues 120 and 151, harbored a nuclear localization signal (NLS), as deletion of this region could abolish nuclear staining (62). More controversially, 16E6 also has been shown by immunofluorescence to colocalize with p53 in the cytoplasm in cervical cancer-derived CaSki and SiHa cells (40). This observation contrasts sharply with the finding that 18E6 colocalizes with p53 in the nucleus in transformed and nontransformed epithelial cells (36). Consequently, we believe that the subcellular distribution of 16E6 remains unresolved.
The lack of conclusive evidence for the cellular distribution of 16E6 and the fact that many uncertainties underlie previously published observations led us to study the molecular details of 16E6 in an attempt to determine whether a more defined NLS prompts 16E6 to enter the nucleus. We took advantage of green fluorescent protein (GFP) fusion technology to develop a strategy that would allow us to limit the splicing of HPV16 E6E7 (16E6E7) pre-mRNA and thereby to produce predominantly full-length E6 in living cells. GFP is a convenient, genetically encoded intrinsic fluorescent molecular label that has been widely and successfully used to study protein distribution in living cells (6, 73). In this report, we examined in detail the sequence of 16E6 and identified three novel NLS sequences (NLS1, NLS2, and NLS3) in 16E6 that are responsible for its nuclear localization. The presence of defined NLS sequences in 16E6 provides conclusive evidence that it localizes to the nucleus in living cells, where it may play important roles in the induction of cell immortalization and transformation during HPV16 infection.
MATERIALS AND METHODS
Construction of expression vectors.
Mammalian expression vectors pEGFP-C1 and pEGFP-N1 were purchased from Clontech (Palo Alto, Calif.). HPV6b E6E7 (6E6E7) (nucleotides [nt] 85 to 939), HPV11 E6E7 (11E6E7) (nt 85 to 924), HPV18 E6E7 (18E6E7) (nt 103 to 967), 16E6E7 (nt 81 to 880), 16E6 (nt 81 or 101 to 559), and HPV16 E7 (16E7) (nt 560 to 880) were amplified by PCR from individual HPV DNAs provided by E.-M. de Villiers and then cloned in frame in the C terminus of GFP at the XhoI and Asp718 sites of the polylinker region of expression vector pEGFP-C1, producing plasmids pZMZ66 (6E6E7), pZMZ67 (11E6E7), pZMZ69 (18E6E7), pZMZ70 (16E6E7), pZMZ73 (158-aa 16E6), pTMF36 (151-aa 16E6), and pZMZ74 (16E7). 16E6E7 (nt 81 to 880) or 16E6 (nt 81 to 559) with a mutation at the nt 226 5′ splice site (GU to GG), introduced by overlapping PCR, was constructed in frame in the C terminus of GFP and led to the generation of plasmid pZMZ81 or pZMZ82, respectively. The single nucleotide mutation at the nt 226 5′ splice site of 16E6E7 converts a GUA codon for valine in 16E6 to a GGA codon for glycine. The 16E6E7 or 16E6 coding region was also inserted in the N terminus of GFP, creating plasmids pWX1 (16E6E7, nt 81 to 880), pTMF1 (16E6E7, nt 81 to 880), and pZMZ79 (16E6, nt 81 to 559). Plasmid pTMF1 has a mutation at the nt 226 5′ splice site (GU to GG) in the E6 coding region. In addition, 16E6*I and 16E6*II cDNAs derived from CaSki cells were also inserted in the C terminus of GFP, resulting in plasmids pZMZ71 (16E6*I, nt 81 to 559) and pZMZ72 (16E6*II, nt 81 to 559), which lack coding sequences from nt 227 to nt 408 (16E6*I) and from nt 227 to nt 525 (16E6*II), respectively, in the E6 coding region due to alternative splicing of the pre-mRNAs.
All plasmids expressing N-terminal or C-terminal deletions of 16E6 (see Fig. 5) were also prepared by PCR from plasmid pZMZ81 and inserted in frame in the same polylinker region of vector pEGFP-C1. Plasmid pTMF15 (151 aa with V42G), derived from plasmid pZMZ81, has a 16E6 coding region (nt 101 to 559) for encoding 16E6 with 151 aa residues using the second AUG as the only start codon. Plasmid pTMF13 (ΔN6) has a deletion of aa 1 to 6 in 151-aa 16E6, pTMF14 (ΔN13) has a deletion of aa 1 to 13, pTMF16 (ΔN30) has a deletion of aa 1 to 30, pTMF19 (ΔN43) has a deletion of aa 1 to 43, pTMF17 (ΔN56) has a deletion of aa 1 to 56, pTMF20 (ΔN74) has a deletion of aa 1 to 74, pTMF21 (ΔN30+ΔC130) has deletions of aa 1 to 30 and aa 130 to 151, and pTMF22 (ΔN30+ΔC112) has deletions of aa 1 to 30 and aa 112 to 151.
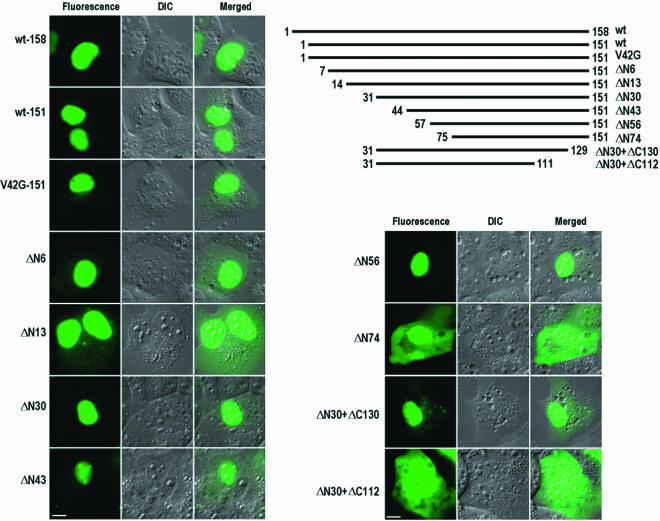
FIG. 5.
Mapping of NLSs in 16E6 in COS-1 cells. Three versions (158 aa, 151 aa, and 151 aa with V42G) of full-length 16E6 were compared for their subcellular localizations. The 151-aa version with V42G was similar to wild-type 151-aa 16E6 except for a V-to-G mutation at residue 42 due to an nt 226 5′ splice site mutation (GU to GG). 16E6 was truncated either from the N terminus to the C terminus or from the C terminus to the N terminus. The truncated E6-GFP fusions were expressed in COS-1 cells transfected with plasmids and imaged at 24 h after transfection. Numbers at the ends of the lines are positions of the first and last residues of the protein. DIC, differential interference contrast; wt, wild type. Scale bars, ∼8 μm.
To create point mutations in 16E6 NLS1, NLS2, and NLS3 or to introduce 16E6 NLS sequences into HPV6b E6 (6E6), various overlapping PCRs were conducted and the resultant amplified PCR fragments containing the expected mutations were cloned into vector pEGFP-C1 as described above. All of the desired mutations in the plasmids were then confirmed by sequencing. In this regard, plasmids pTMF34 and pTMF35 (with mutated NLS1) were derived, respectively, from pZMZ71 and pZMZ72, and plasmids pTMF23 (with mutated NLS2) and pTMF24 (with mutated NLS3) were derived, respectively, from pTMF17 and pTMF21. Plasmid pTMF26 with both mutated NLS2 and mutated NLS3 in truncated 16E6 was derived from pTMF23.
Plasmids pTMF30 (158 aa) and pTMF32 (151 aa) had mutations in both NLS2 and NLS3 of full-length 16E6. Plasmids pTMF44 (151 aa), pTMF45 (151 aa), and pTMF33 (151 aa) encoded full-length 16E6 with mutant NLS1 plus mutant NLS2, mutant NLS1 plus mutant NLS3, and all three mutant NLSs, respectively.
Various plasmids were constructed to contain an insert of 16E6 NLS1 (pTMF38), NLS2 (pTMF27), or NLS3 (pTMF29) only or combined inserts of NLS1 plus NLS2 (pTMF39), NLS1 plus NLS3 (pTMF40), or NLS2 plus NLS3 (pTMF28) in 6E6. All of the plasmids were derived from plasmid pZMZ66 and were designed to express chimeric 6E6-16E6 NLS protein.
Cells and transfection.
293 (human), HaCaT (human), and COS-1 (monkey) cells were seeded on glass coverslips in 60-mm-diameter dishes and grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) was used to transfect 2 μg of vector DNA into cells by following the recommendations of the manufacturer.
Reverse transcription (RT)-PCR analysis of spliced 16E6E7 mRNAs.
Total cell RNA was extracted from 293 cells at 24 h after transfection by using TRIzol(Invitrogen) as recommended by the manufacturer. Following RNase-free DNase I digestion, 500 ng of total cell RNA was reverse transcribed at 42°C with random hexamers as primers and then amplified for 35 cycles with primers Pr106 (′-GTTTCAGGACCCACAGGAGC-3′) and Pr559 (′-CACTGAGGTACC/TTACAGCTGGGTTTCTCTACG-3′).
Western blotting analysis.
COS-1 cells at 24 h after transfection with various plasmids (4 μg) were lysed directly with sodium dodecyl sulfate (SDS) sample buffer. Western blot analysis was performed as described previously (76) with an enhanced chemiluminescence (ECL) Western blotting detection system (Amersham) and monoclonal anti-GFP antibody (Zymed Laboratories, Inc., South San Francisco, Calif.).
Cell immortalization and p53 degradation.
16E6 or chimeric GFP-16E6-6E6, including the mutant forms described above, was introduced into human mammary epithelial cell line 76N (MECs) by either transient transfection with retroviral vector pLXSN or infection with high-titer viruses obtained from transiently transfected packaging cell line LinX-A (25). Immortal MECs were selected in D2 medium containing G418 (2) for at least 1 month. For studies of E6-mediated p53 degradation, plasmids expressing 16E6 with various NLS mutants or 6E6 with the 16E6 NLS replacement (see Fig. 8) were cloned into pcDNA3.1 or pSP65. E6 proteins and p53 were expressed by using in vitro transcription-translation conducted with a rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine. Degradation of 35S-labeled p53 by E6 proteins was monitored by immunoprecipitation of p53 products with polyclonal anti-p53 antibody as described previously (41).
FIG. 8.
Conversion of a cytoplasmic 6E6 protein into a nuclear protein by 16E6 NLS motifs. The corresponding 16E6 NLS regions in 6E6 were replaced with individual 16E6 NLS motifs. The cellular distribution of the chimeric E6 protein with one or two 16E6 NLS motifs in COS-1 cells was imaged at 6 h after transfection. Arrows are as described in the legends to Fig. 4 and 6. DIC, differential interference contrast; WT, wild type. Scale bar, ∼8 μm.
Live-cell imaging by fluorescence microscopy.
After transfection, the expression of GFP fusions was monitored first with an Olympus IX70 microscope equipped with a fluorescence observation attachment. To capture high-resolution images, coverslips were recovered after 6 or 24 h of transfection, mounted on modified glass slides with 10% fetal calf serum-containing cell culture medium, and used immediately for imaging. Cell images were collected with a Zeiss Axioplan 2 wide-field epifluorescence microscope equipped with a Plan-Apochromat ×63 (N.A. 1.40) oil immersion objective lens, a fluorescence filter set suitable for GFP fluorescence, a Hamamatsu ORCA ER cooled charge-coupled device camera (Hamamatsu Photonics K.K., Hamamatsu, Japan), and MetaMorph version 6.0 image acquisition and processing software (Universal Imaging Inc., Downingtown, Pa.). Fluorescence images were collected at various exposure times suited to the individual protein expression levels displayed by cells within a particular sample. Images were saved in TIFF format. Adobe Photoshop version 6.0. (Adobe Systems, Inc., San Jose, Calif.) was used to arrange processed images into composite figures. To count cell fluorescence distribution, 100 cells were examined for each plasmid (pZMZ66, pZMZ67, pZMZ69,and pZMZ70) from ∼20 random fields viewed through the Zeiss Axioplan 2 wide-field epifluorescence microscope.
RESULTS
Prevention of 16E6 pre-mRNA from splicing in vivo to express full-length E6 protein.
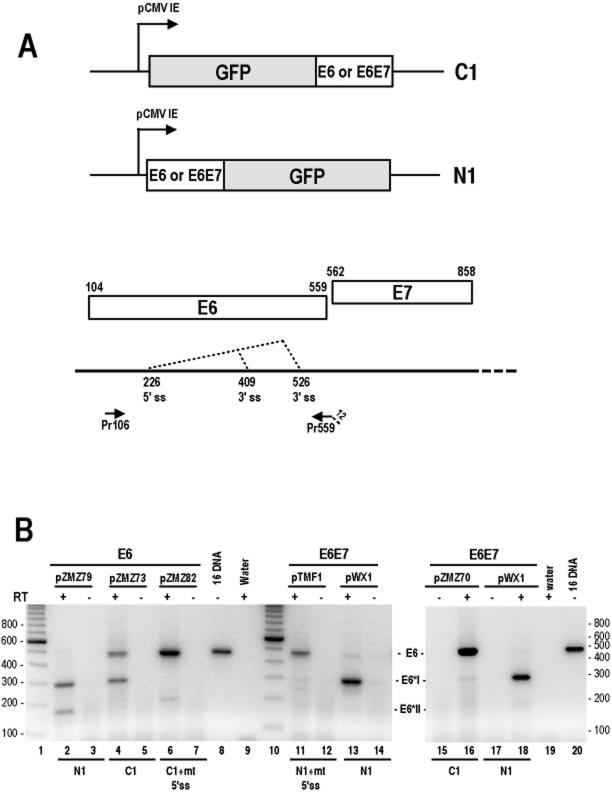
16E6 and 16E7 in cervical cancer tissues or in cervical cancer-derived cell lines were expressed as a single bicistronic pre-mRNA transcript from the same promoter, p97 (67). The bicistronic 16E6E7 pre-mRNA has an intron in the E6 coding region with one 5′ splice site and two 3′ splice sites at nt 409 and 526, respectively, in the virus genome, producing the 16E6*I and 16E6*II mRNAs (66). Full-length unspliced E6 mRNA is considered to be a result of intron escape and is responsible for translation of the oncogenic E6 protein. Thus, it is necessary to prevent 16E6E7 pre-mRNA from splicing in order to study the cellular localization and function of the full-length E6 protein. To satisfy this criterion, the 16E6E7 coding regions were inserted in the C terminus of GFP (pZMZ70) in mammalian expression vector pEGFP-C1 (the 16E6E7-C1 vector). This approach provided full-length unspliced E6E7 mRNA to be adequately expressed in vector-transfected cells, as shown in Fig. 1B, lane 16. Compared to the 16E6E7-C1 vector, the 16E6E7-N1 vector, containing the 16E6E7 coding regions (pWX1) inserted in the N terminus of GFP, produced mainly 16E6*I and few 16E6*II mRNAs (Fig. 1B, lanes 13 and 18). Similarly, an expression vector containing the E6 open reading frame (ORF) inserted alone (pZMZ79) in the N terminus of GFP (the 16E6-N1 vector) produced predominantly 16E6*I mRNA (compare lane 2 with lane 4 in Fig. 1B). Although the E6 pre-mRNA was partially spliced when expressed from the corresponding 16E6-C1 vector (Fig. 1B, lane 4), it was distinguishable from the pre-mRNA that came from its counterpart, the 16E6-N1 vector (Fig. 1B, lane 2).When a mutant nt 226 5′ splice site (GU to GG) was introduced into those same vectors, none of the mRNAs expressed was spliced (Fig. 1B, lanes 6 and 11). Collectively, these data indicate that the 16E6E7-C1 or 16E6-C1 construct provided GFP-tagged E6 from unspliced, full-length mRNA.
FIG. 1.
Strategy for preventing E6 pre-mRNA from splicing for the production of full-length 16E6-GFP fusions. (A) Diagram of the 16E6E7 (nt 81 to 880) or 16E6 (nt 81 to 559) coding region inserted either upstream (N1) or downstream (C1) of the GFP gene in corresponding pEGFP expression vectors. The E6 and E7 ORFs, the splicing directions of the 16E6 pre-mRNA, and a primer pair used for RT-PCR analysis are depicted below the diagram. The numbers above the E6 and E7 ORFs are start and stop codons for each ORF. ss, splice site. Drawings are not to scale. (B) Expression of 16E6 in 293 cells by transient transfection. Total cell RNA isolated from transfected cells after the removal of plasmid DNA by RNase-free DNase I treatment was examined by RT-PCR analysis with primers Pr106 and Pr559 for 16E6 RNA detection. Both plasmids pZMZ82 and pTMF1, having an mutant nt 226 5′ splice site (GU to GG), were used to transcribe unspliced E6 or E6E7 RNA as a control. The identities of the spliced and unspliced E6 RNA products are indicated between the gels. Size markers (lanes 1 and 10) from a 100-bp ladder are indicated at the left and right of the gels.
Cellular distributions of low-risk and high-risk HPV E6 proteins.
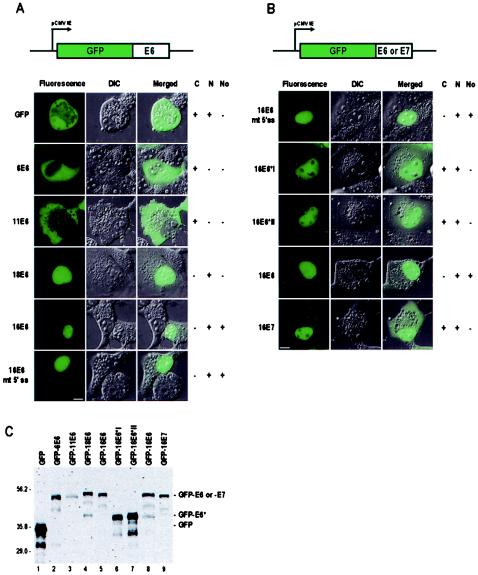
Next, we wished to compare the cellular distributions of high-risk and low-risk HPV E6 proteins. Viral E6E7 coding regions were inserted in the C terminus of vector pEGFP-C1, allowing E6 proteins from different HPVs to be expressed as GFP-E6 fusion proteins. GFP alone, when expressed in COS-1 cells, exhibited a diffuse signal, being present in both the nucleus and the cytoplasm (Fig. 2A). GFP-E6 from low-risk HPV6 and HPV11 expressed as a full-length fusion protein (Fig. 2C) at low and moderate levels (empirically determined) exhibited a predominantly cytoplasmic distribution, with a weak signal in the nucleus (Fig. 2A), in 76% of the cells for 6E6 and in 93% for 11E6. At higher expression levels, low-risk 6E6 again localized predominantly in the cytoplasm; however, variably sized fluorescent aggregates had a tendency to form in about 24% of the cells (data not shown). It is unclear what the basis is for the aggregate formation by 6E6, but since the aggregates were present in cells with higher levels of expression, we chose to focus on cells with low to moderate levels of expression, which we believe are more representative of E6 protein levels that would be expressed in an endogenous system. Nonetheless, both low-risk 6E6 and 11E6 proteins localized to the cytoplasm in mammalian cells. In contrast, high-risk 16E6 and 18E6 proteins, regardless of whether they were expressed from vectors encoding E6E7 or E6 alone (16E6 only), were found in the nucleus in 99% (16E6) or 98% (18E6) of the cells, with very little fluorescence signal in the cytoplasm (Fig. 2A and B). Interestingly, 16E6, but not 18E6, was also found in the cell nucleolus. The subcellular distribution pattern of 16E6 truly represented full-length 16E6, since localization to the nucleus was seen when 16E6 was expressed from either the 16E6E7 or the 16E6 coding region containing a mutant 5′ splice site and the 16E6 protein was distinguishable from the 16E6*I and 16E*II proteins expressed from the spliced E6 mRNA. Full-length 16E6 and 18E6 expressed from unspliced mRNAs were also verified by Western blotting analysis (Fig. 2C, lanes 4, 5, and 8).
FIG. 2.
Cellular localizations of low-risk and high-risk HPV E6 in COS-1 cells. COS-1 cells were transfected with plasmids pZMZ66 (6E6E7, nt 85 to 939), pZMZ67 (11E6E7, nt 85 to 924), pZMZ69 (18E6E7, nt 103 to 967), pZMZ70 (16E6E7, nt 81 to 880), and pZMZ81 (16E6E7, nt 81 to 880 with a mutant nt 226 5′ splice site [GU to GG]) (A) and pZMZ82 (16E6, nt 81 to 559 with a mutant nt 226 5′ splice site [GU to GG]), pZMZ71 (16E6*I cDNA, nt 81 to 559), pZMZ72 (16E6*II cDNA, nt 81 to 559), pZMZ73 (16E6, nt 81 to 559), and pZMZ74 (16E7, nt 560 to 880) (B). 16E6*I and 16E6*II cDNA constructs have no coding sequences in the E6 intron region because of pre-mRNA splicing. Diagrams at the top of each panel show the insertion and position of the “E6” or “E7” coding region in mammalian expression vector pEGFP-C1. The individual images were collected at 6 (6E6 and 11E6) or 24 h after transfection. Plasmid pEGFP-C1 was used as a GFP control in each transfection. A fluorescence image, a differential interference contrast (DIC) image, and a merged fluorescence-DIC image for each fusion protein arepresented. (A) Localization of 6E6, 11E6, 16E6, and 18E6 translated from the inserted E6E7 coding region in COS-1 cells. 16E6 mt 5′ ss indicates the plasmid containing the 16E6E7 coding region but having a mutant 5′ splice site at nt 226 position (GU to GG), used as a transfection control for full-length 16E6, since this mutation blocks the splicing of the E6E7 pre-mRNA. The single nucleotide mutation in the 5′ splice site of 16E6E7 converts a GUA codon for valine in 16E6 to a GGA codon for glycine. (B) Localization of various 16E6 proteins translated from the inserted E6 coding region and E6*I or E6*II cDNA. 16E7 protein translated from plasmid pZMZ74 was used for comparison. + or − indicates the presence or absence of cytoplasmic (C), nuclear (N), or nucleolar (No) localization of GFP fusions. Scale bars, ∼8 μm. (C) Western blotting analysis of GFP-E6 or GFP-E7 fusions expressed from pEGFP-C1 vectors. As shown in panels A and B, all GFP fusions except for mutant E6 were included for comparison of the expected full-length E6 or E7 protein, even through the 6E6 and 11E6 coding regions do not have an intron. GFP-16E6 in lane 5, encoded from a plasmid with the 16E6E7 coding region, was included for comparison with GFP-16E6 in lane 8, encoded from a plasmid with the 16E6 coding region. Protein samples prepared from COS-1 cells transfected with the various plasmids were resolved on an SDS-12% polyacrylamide gel and blotted with anti-GFP antibody. GFP and its derived fusions are indicated above the lanes. Prestained protein markers (Bio-Rad) in kilodaltons are shown at the left.
Analysis of relative fluorescence signal intensities, measured from line profiles drawn across individual cells, including the nucleoplasm and nucleoli, showed that 16E6 not only was uniformly distributed in the nucleus but also accumulated in the nucleolus to a greater degree than 16E6*I and 16E6*II, which did not accumulate in nucleoli (Fig. 2B and data not shown). Both 16E6*I and 16E*II were distributed predominantly within the nucleus, with perhaps a higher cytoplasmic signal than that observed for 16E6. Although this difference in subcellular distributions is subtle, the difference in subnuclear distributions is more striking, with 16E6*I and 16E6*II clearly being excluded from nucleoli (Fig. 2B). The cellular distributions of 16E6*I and 16E6*II also resembled that of 16E7, with 16E7 displaying a higher signal in the cytoplasm than in the nucleus (Fig. 2B). These findings are consistent with 16E7 being reported as a nuclear protein (20, 65). Overall, in COS-1 cells, high-risk, full-length E6 proteins are distributed predominantly in the nucleus, whereas low-risk, full-length E6 proteins are localized in the cytoplasm.
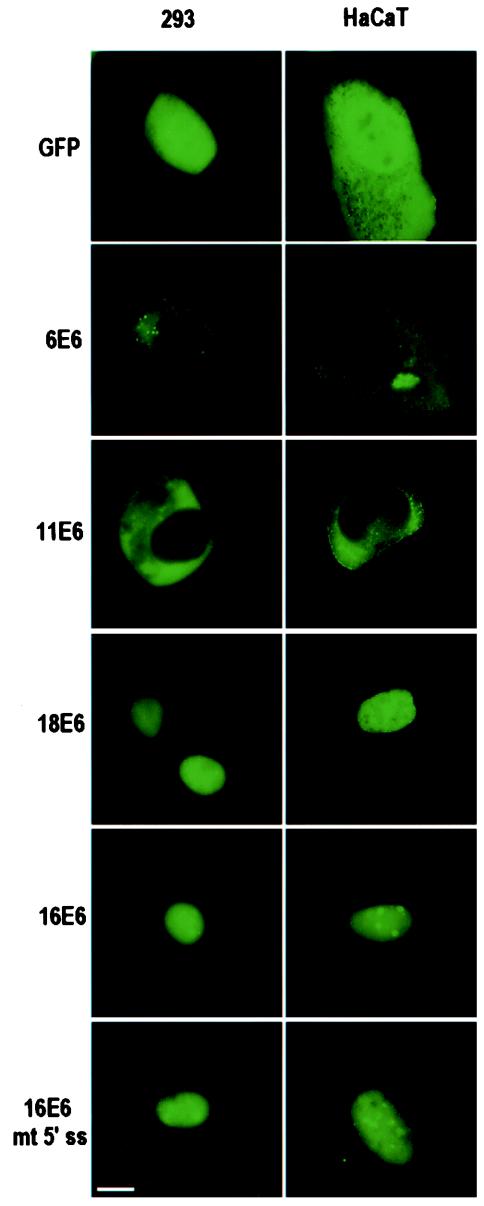
Several additional cell lines, including 293, HaCaT, and 3T3, were used to study E6 expression. The cellular distributions of high-risk and low-risk E6 proteins in 293 and HaCaT cells were similar to those observed in COS-1 cells (Fig. 3). HaCaT cells displayed very low transfection and protein expression efficiencies compared to COS-1 and 293 cells; consequently, COS-1 and 293 cells were used predominantly in our studies. The subcellular distributions of 16E6 and 18E6 in mouse 3T3 cells also appeared to be similar to those observed in the other three cell types, except that 16E6 did not accumulate in the nucleoli of 3T3 cells (data not shown). Thus, the difference in the cellular distributions of high-risk and low-risk E6 proteins indicates that there are differences in protein sequence that may be associated with cellular compartmentalization.
FIG. 3.
Cellular localizations of low-risk and high-risk HPV E6 in 293 and HaCaT cells. Each fluorescence image was captured at 6 (6E6 and 11E6) or 24 h after transfection of plasmids as described in the legend to Fig. 2. 16E6 mt 5′ ss indicates the plasmid (pZMZ81) containing the 16E6E7 coding region but having a mutant 5′ splice site at nt 226 position (GU to GG), used as a transfection control for full-length 16E6 as described in the legend to Fig. 2A. Scale bar, ∼8 μm.
The N-terminal region of 16E6 contains an NLS.
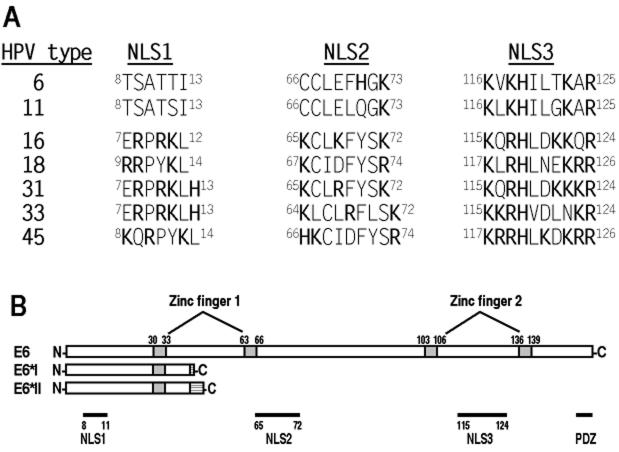
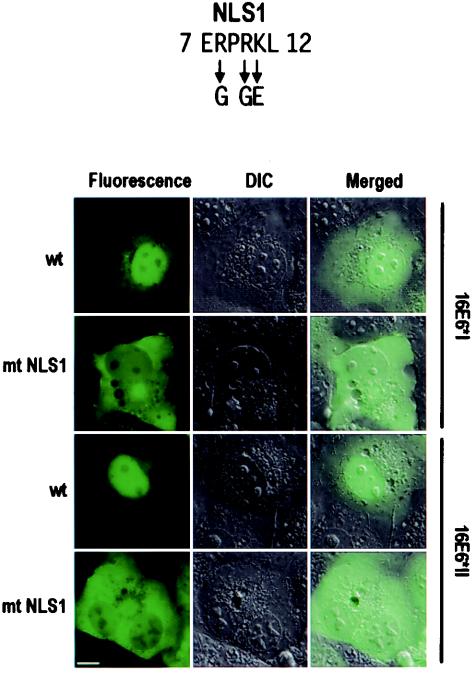
Considering that both 16E6*I and 16E6*II accumulate within the nucleus (Fig. 2B) and both have the same N-terminal 41 aa residues as the full-length 16E6 protein, we suspected that an NLS sequence resided in this region. Analysis of amino acid residues in this region of the 16E6 protein revealed a putative NLS motif, RPRK (aa 8 to 11), which contains several basic residues (in bold type). The putative NLS motif appears to exist only in oncogenic E6 proteins from high-risk HPVs and not low-risk HPV6 and HPV11 (see Fig. 10). Functional characterization of this putative NLS was approached by introducing point mutations. Conversion of R to G or K to E in the putative NLS effectively disrupted the nuclear localization of the E6* proteins (Fig. 4). Thus, the basic residues in this N-terminal NLS are responsible for the accumulation of 16E6*I and 16E6*II in the nucleus. We designate this sequence NLS1.
FIG. 10.
HPV E6 proteins and their functional motifs. (A) Corresponding amino acid residues of putative NLS motifs in representative HPV E6 proteins aligned with individual NLS motifs identified in 16E6. Bold letters indicate positively charged residues in each NLS. The superscript numbers are amino acid residue positions in each E6 protein. (B) Schematic diagram of 16E6 domains. Drawn for comparison are 16E6*I (43 aa) and 16E6*II (46 aa). A hatched box in the C terminus of each protein indicates the addition of 2 aa (E6*I) or 5 aa (E6*II) to the N-terminal 41 aa from an in-frame shift resulting from alternative splicing of 3′ splice sites (Fig. 1A). There are four zinc-binding motifs (grey boxes), Cys-X-X-Cys, that form two hypothetical fingers involving zinc binding (3, 30). Domains are depicted below the 16E6 diagram. The PSD-95, Dlg, Zona occludens-1 (PDZ)-binding site has the sequence X-(S/T)-X-(V/I/L)-COOH (43, 46).
FIG. 4.
Point mutational analysis of a putative N-terminal NLS in 16E6 protein. 16E6*I and 16E6*II proteins were chosen for the assay because they have the same N-terminal 41 aa residues as the full-length 16E6 protein and could simplify the assay. The basic residues in the putative NLS were mutated to G or E residues, as indicated by arrows at the top. DIC, differential interference contrast; wt, wild type; mt, mutant. Scale bar, ∼8 μm.
Mapping of two other NLSs in 16E6 by deletion analysis.
The coding region of 16E6 has tandem AUG sequences, the second of which is presumably the usual start codon, resulting in a 16E6 protein with 151 residues (48). To determine whether particular motifs in other parts of 16E6 are responsible for the nuclear localization of the protein, we constructed a 16E6 sequence with 151 aa residues and a series of 16E6 deletion mutants that were linked in frame to the C terminus of GFP. As a result of the lack of a precisely defined NLS in earlier literature, we made successive deletions in 151-aa 16E6, beginning at the N terminus and progressing to the C terminus. Additional mutants that contained both N-terminal and C-terminal deletions were constructed. All mutants with deletions were transfected into COS-1 cells; the expression of these mutant proteins was examined, and their intracellular distributions were documented. As shown in Fig. 5, both 158- and 151-aa forms of full-length 16E6 localized predominantly in the nucleoplasm and even in nucleoli, with little fluorescence signal observed in the cytoplasm. The same was true for full-length 151-aa 16E6 having a V-to-G mutation at residue 42. Mutant ΔN6 and even mutant ΔN13, which lacked the N-terminal NLS, as described above, exhibited some E6 signal in the cytoplasm but, like full-length E6, displayed predominantly a nuclear localization pattern, indicating that other NLSs must exist in downstream regions. The distributions of three shorter truncation mutants, ΔN30, ΔN43, and ΔN56, were almost completely restricted to the nucleoplasm, with some accumulation in nucleoli and, to a much lower extent, in the cytoplasm. Interestingly, further truncation to residue 75, as represented by ΔN74, resulted in the protein being distributed equally throughout the cells, indicating that the region of residues 57 to 74 most likely contains an NLS sequence.
Deletion of the first 30 N-terminal residues from 16E6 (ΔN30) had no effect on the distribution of 16E6 in the nucleus; therefore, this mutant was used for the C-terminal truncation analysis. As shown in Fig. 5, mutant ΔN30+ΔC130 has an additional deletion of 21 C-terminal residues. When expressed in COS-1 cells, the truncated E6 protein localized predominantly to the nucleus, with some signal in the cytoplasm, indicating that the region between residues 130 and 151 might include weak nuclear localization activity (62). Interestingly, further truncation of the C terminus to residue 111, as represented by ΔN30+ΔC112, caused this form of E6 to be distributed equally throughout the cells. The difference in cellular distributions between the ΔN30+ΔC130 and ΔN30+ΔC112 forms of 16E6 indicates that the region between residues 112 and 129 in the C terminus harbors another NLS.
The same sets of plasmids, containing various truncations, were also expressed in parallel in 293 cells, and the mutant forms of 16E6 showed the same distribution patterns as those described for COS-1 cells (data not shown). Overall, the expression of the different truncation mutants indicates that there are at least two other NLS sequences in the full-length high-risk 16E6 protein.
Nuclear retention of truncated 16E6 depends on intact NLS sequences.
Having demonstrated that two regions, aa 57 to 74 and aa 112 to 129, are involved in the accumulation of truncated 16E6 within the nucleus, we wished to determine whether the two regions represent sufficient and active NLS sequences. We also sought to better define the residues responsible for nuclear localization. Analysis of the amino acid compositions within the two regions revealed that the region from aa 57 to 74 has a motif composed of 8 residues, KCLKFYSK (aa 65 to 72), with three lysine (K) residues separated by weak or nonpolar residues, whereas the region from aa 112 to 129 has a motif consisting of 10 residues, KQRHLDKKQR (aa 115 to 124), enriched with 6 basic residues. Collectively, the features of these two motifs, containing multiple basic residues, and the fact that their deletion influences the subcellular distribution of 16E6 make the two motifs reminiscent of NLS sequences present in several other papillomavirus proteins (Table 1) and other, nonpapillomavirus proteins (4, 49). Thus, we designate these two 16E6 putative NLS motifs NLS2 (KCLKFYSK) and NLS3 (KQRHLDKKQR).
TABLE 1.
NLSs characterized in papillomaviruses
| Virus | Viral protein | NLS sequence(s)a | Reference or source |
|---|---|---|---|
| HPV5 | E2 | 246RGRRYGRRPSSKSRRSQTQQRRSRSRHRSRSRSRSRSKSQTHTTWSTTRSRSTSVGKTRALTSRSRSRGRSPSTCRRGGGRSPRRRSR333 | 35 |
| HPV6b | L2 | 5RARRRKR11 | 71 |
| 444RKRRKR449 | |||
| HPV11 | E2 | 238RKRAR242 | 79 |
| L1 | 481KRPAVSKPSTAPKRKRTKTKK501 | 50 | |
| HPV16 | E6 | 8RPRK11 | This study |
| 65KCLKFYSK72 | |||
| 115KQRHLDKKQR124 | |||
| E7b | 17PETTDLYCYEQLNDSSEEEDEIDGP41 | 15 | |
| L1 | 510KRKATPTTSSTSTTAKRKKRK530 | 77 | |
| HPV45 | L1 | 505RRRPTIGPRKRPAASTSTASTASRPAKRVRIRSKK539 | 50 |
| Bovine papillomavirus type 1 | E1 | 84KRKVLGSSQNSSGSEASETPVKRRK108 | 38 |
| E2 | 106PKRCFKKGAR115 | 64 | |
| 340KCYRFRVKKNHRHR353 |
Bold type indicates basic residues.
HPV16 E7 has no obvious NLS, and the mechanism by which wild-type E7 is targeted to the nucleus is unknown. The sequence presented here was suggested by deletion mutation analysis (15).
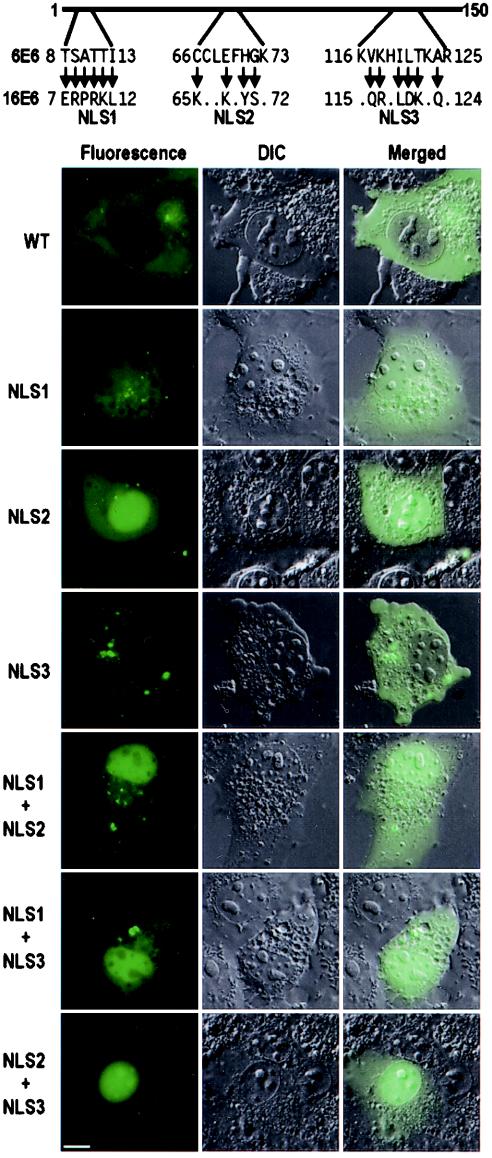
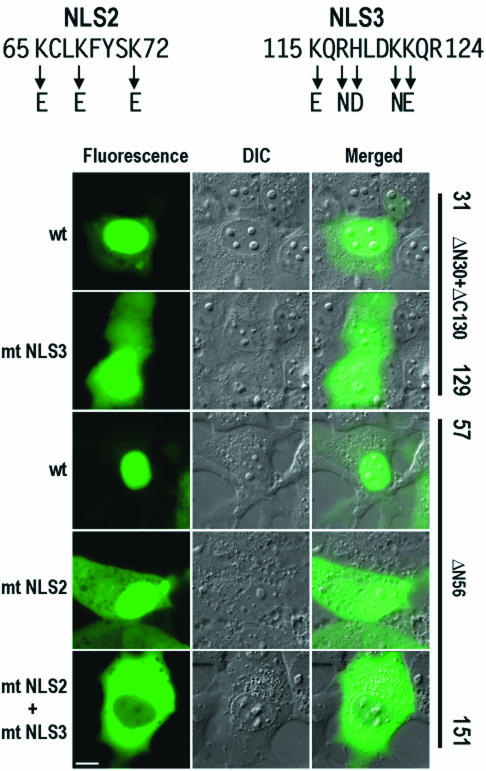
To precisely define the functions of the two NLS motifs, several point mutations were introduced into each NLS. Acidic amino acid residues were substituted for the endogenous basic amino acid residues as shown in Fig. 6. Two mutants, ΔN56 and ΔN30+ΔC130, were chosen for introducing point mutations because they were the shortest E6 truncated proteins containing both putative NLS motifs (Fig. 5) and localized mainly in the nucleus. As shown in Fig. 6, point mutations of NLS2 or NLS3 alone in truncated 16E6 impaired but did not eliminate the ability of the fusion proteins to accumulate within the nucleus of COS-1 cells. Fusion proteins containing point mutations of either NLS still displayed a substantial presence in the nucleus, although with a more noticeable fluorescence signal present in the cytoplasm. Subsequent mutations of both NLS motifs completely abolished the ability of the truncated protein to accumulate in the nucleus, as observed by the exclusively cytoplasmic distribution of truncated 16E6-GFP. A parallel experiment was also conducted with 293 cells, and the results were consistent with the observations made with COS-1 cells (data not shown). Based on these results, we conclude that the two putative NLS motifs, NLS2 and NLS3, in truncated 16E6 are functional NLS sequences responsible for the nuclear localization of the protein in a complementary fashion.
FIG. 6.
Mutational analysis of 16E6 NLS2 and NLS3 motifs in truncated 16E6 in COS-1 cells. The images were captured from cells expressing truncated 16E6, ΔN30+ΔC130, and ΔN56, with or without mutations in NLS2, NLS3, or both, at 24 h after transfection. The basic residues in the two NLS motifs were mutated to acidic residues, as indicated by arrows at the top. DIC, differential interference contrast; wt, wild type; mt, mutant. Scale bar, ∼8 μm.
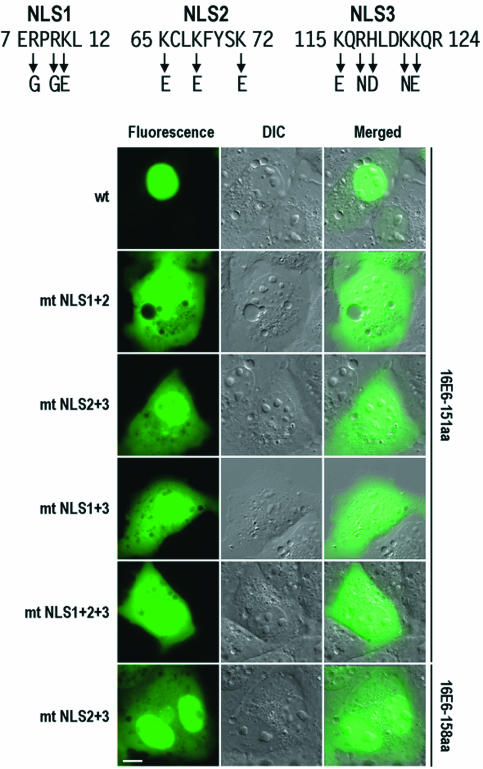
Nuclear localization of full-length 16E6 can be altered by combinational NLS mutations.
Having identified three NLS sequences separately in truncated 16E6 and in 16E6*I and 16E6*II, we next wished to examine the potential of these NLS sequences to localize full-length 16E6 to the nucleus. Combinational mutations of the NLS sequences were chosen because individual NLS mutations in truncated E6 proteins did not exclude the protein completely from the nucleus and thus would not provide a distinguishable cytoplasmic distribution of mutant full-length 16E6. As shown in Fig. 7, mutations of all basic residues in the two NLS sequences (mutant NLS1 plus mutant NLS2 or mutant NLS1 plus mutant NLS3) or in the three NLS sequences effectively made the subcellular distribution of the full-length 16E6-GFP fusion indistinguishable from that of GFP alone (cf. Fig. 7 and Fig. 2A). However, the distributions of both 151- and 158-aa forms of full-length 16E6 with wild-type NLS1 but mutant NLS2 and mutant NLS3 resembled those of 16E6*I and 16E6*II (cf. Fig. 7 and Fig. 2B and 4). These results demonstrated that the three NLS sequences identified in truncated 16E6 function in localizing the full-length 16E6 protein to the nucleus.
FIG. 7.
Mutational analysis of 16E6 NLS motifs in full-length 16E6 in COS-1 cells. Point mutations in the individual NLS motifs were the same as those described in the legends to Fig. 4 and 6. Cell images were captured at 24 h after transfection. DIC, differential interference contrast; wt, wild type; mt, mutant. Scale bar, ∼8 μm.
16E6 NLS sequences promote the nuclear localization of normally cytoplasmic 6E6.
When the sequence of 16E6 is compared to that of 6E6, it becomes apparent that the two proteins are highly divergent in the regions corresponding to 16E6 NLS1 and NLS2, in particular, the basic amino acid content. Previous results outlined in Fig. 2 and 3 demonstrated that low-risk 6E6 and 11E6 are cytoplasmic proteins. We wished to determine whether the 16E6 NLS motifs could also function in a cytoplasmic protein and lead to the accumulation of a normally cytoplasmic protein within the nucleus. The 6E6 protein was chosen for this assay, and the corresponding regions in the 6E6 sequence were replaced with 16E6 NLS1, NLS2, and NLS3. When expressed in COS-1 cells, the chimeric 6E6 protein containing either 16E6 NLS1 or 16E6 NLS2 alone was directed to the nucleus, but only partially (Fig. 8). Notably, 16E6 NLS2 provided 6E6 with a much stronger nuclear signal than did 16E6 NLS1. In contrast, 16E6 NLS3 by itself could not enhance the accumulation of 6E6 in the nucleus. However, when combined with NLS1 or NLS2, NLS3 could promote the conversion of 6E6 by NLS1 or NLS2 into a protein with a stronger nuclear signal, as did NLS1 when combined with NLS2. In this regard, NLS2 combined with NLS3 could convert 6E6 into a predominantly nuclear, even nucleolar, protein. All chimeric 6E6-16E6 proteins expressed in 293 cells (data not shown) showed the same distribution patterns as those seen in COS-1 cells. We conclude that the newly identified 16E6 NLS motifs promote the nuclear localization of a cytoplasmic protein.
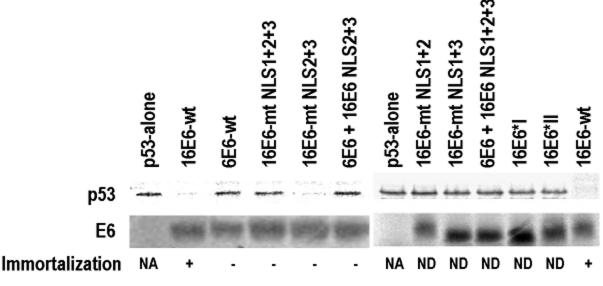
16E6 NLS sequences are important but not sufficient for cell immortalization and p53 degradation.
We next examined whether the 16E6 NLS sequences are necessary for human MEC immortalization. Because wild-type16E6 with the GFP tag was unable to immortalize these cells, we tested mutants with and without the GFP moiety in animmortalization assay (Fig. 9). Only wild-type 16E6 was immortalization competent. In addition, the 6E6 mutant with 16E6 NLS2 and NLS3 in the corresponding regions was also unable to induce immortalization. We also assayed these mutants for the ability to degrade p53 in vitro, an activity which would bypass the reliance on a subcellular localization. Introduction of mutations into the 16E6 NLS sequences was found to abolish the degradation of p53 by 16E6, but the influence of individual NLS sequences was not directly related to the degradation of p53 in vitro. 16E6 with intact NLS1 but defective NLS2 and NLS3 retained the ability to stimulate the degradation of p53. All other 16E6 mutants and the 6E6 mutant with NLS2 and NLS3 or all three NLS sequences from 16E6 were unable to induce p53 degradation (Fig. 9). While all mutations of NLS1 failed to lead to p53 degradation, the finding that 16E6*I and 6E6*II, bearing only NLS1, were unable to induce the degradation of p53 in vitro demonstrates that the NLS1 region is not sufficient for p53 degradation. Taken together, these results indicatethat 16E6 NLS sequences are important but not sufficient for cell immortalization or p53 degradation.
FIG. 9.
16E6 NLS sequences are important in the degradation of p53 in vitro and in the immortalization of human MECs. E6 and p53 proteins were prepared with a rabbit reticulocyte lysate transcription-translation system in the presence of [35S]methionine. The in vitro p53 degradation assay was described by Liu et al. (41). To minimize the background, p53 polyclonal antibody was used to immunoprecipitate p53 from the degradation reaction mixture. Briefly, 10 μl of p53 degradation solution, 25 μl of protein G-Sepharose beads, and 100 μl of p53 antibody were mixed with 400 μl of binding buffer (100 mM NaCl, 100 mM Tris-HCl [pH 8.0], 0.5% EDTA, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 0.5% protease inhibitor cocktail [Roche]). The mixture was rotated at 4°C overnight and washed three times with binding buffer. The immunoprecipitated proteins were then resolved on an SDS-12% polyacrylamide gel and visualized by autoradiography. Individual proteins used for p53 degradation are indicated above the lanes. The 16E6 NLS sequences with mutations are shown in Fig. 7, and their substitutions for the corresponding regions in 6E6 are shown in Fig. 8. The 16E6 mutant (16E6-mt) NLS lanes indicate combined mutations of the individual NLS sequences. The chimeric 6E6-16E6 (6E6 + 16E6) NLS lane indicates the corresponding regions of 6E6 being replaced by 16E6 NLS sequences. Shown below the p53 gel are the corresponding E6 proteins with (+) or without (−) immortalization competency, as examined in human MECs. wt, wild type; NA, not applicable; ND, not done.
DISCUSSION
In this report, we used a mammalian GFP expression system to express full-length 16E6 protein with the minimal production of other, truncated 16E6*I and 16E6*II proteins arising from 16E6 mRNA splicing. Using this strategy, we identified three distinct NLS motifs in 16E6 by three approaches. First, the expression of GFP-16E6 demonstrated a predominantly nuclear localization of the fusion protein which was directed by three NLS coding regions present in the 16E6 gene. Second, point mutations in the identified NLS regions of 16E6 resulted in cytoplasmic retention of the fusion protein. Third, the newly identified 16E6 NLS motifs, when inserted into the corresponding regions of 6E6, could promote the nuclear accumulation of the otherwise cytoplasmic 6E6 protein. Importantly, we found that two of the three NLS motifs in 16E6 are required to work together to promote nearly complete nuclear localization of the protein. None of the NLS motifs alone is sufficient for the protein to be retained exclusively in the nucleus, although NLS1 and NLS2 seem to have a stronger influence than NLS3 when present independently of each other in 6E6. As determined by sequence analysis, the three NLS motifs identified appear to be conserved in the corresponding regions of other high-risk E6 proteins, but only a putative NLS3 motif can be found in low-risk E6 proteins (Fig. 10).
Intracellular communication between the nucleus and the cytoplasm occurs through nuclear pore complexes (NPCs) present in the nuclear envelope. NPCs allow passive diffusion of molecules smaller than 40 to 60 kDa (up to 9 nm in diameter) between the nucleus and the cytoplasm (58, 75). The uniform cellular distribution, both cytoplasmic and nuclear, of GFP (approximately 27 kDa by calculation and approximately 35 kDa in our Western blotting studies) can be accounted for as a consequence of passive diffusion and has been well documented in the literature (6, 73). However, larger macromolecules, and even some proteins or RNAs smaller than 20 to 30 kDa, such as histones and tRNA (29), must be transported actively in a multistep manner through NPCs, usually involving proteins containing at least one NLS, via NLS-mediated import machinery. All of our chimeric GFP-E6 or GFP-E7 fusions (∼50 kDa) are at the threshold of the NPC transport limit and may or may not enter the nucleus by passive diffusion, depending on the features of the individual protein. 6E6 and 11E6 demonstrated a preference for distribution to the cytoplasm, whereas 16E6, 16E7, and 18E6 localized predominantly to the nucleus, even nucleoli (16E6). Our demonstration that 16E6 contains three NLS sequences provides conclusive evidence that an active import machinery mediates the nuclear import of 16E6. While we were preparing this article, Le Roux and Moroianu reported that the nuclear entry of high-risk 16E6, mediated by part of NLS3, as described in this study, could occur through several pathways (39).
To examine the subcellular localizations of E6 proteins, our experiments included the use of vectors that are prone to overexpressing GFP-tagged proteins. Being aware of potential overexpression artifacts, we examined the cellular distributions of E6 proteins with low and high expression levels at either 6 or 24 h posttransfection in the same and different cell lines. Except for 6E6, all other E6 fusion proteins exhibited similar distribution patterns at both low and high expression levels. The 6E6 protein was found to localize predominantly in the cytoplasm, with some nuclear signal, in cells with low to moderate levels of expression but shifted to an exclusively cytoplasmic localization, with the presence of aggregates, in cells with higher levels of expression. Larger proportions of cells with low to moderate levels of expression were observed at 6 h after transfection, while at 24 h after transfection, most cells were expressing 6E6 at high levels. At present, we do not know what causes 6E6 aggregation in the cytoplasm. Nonetheless, the low-risk 6E6 and 11E6 proteins displayed a predominantly cytoplasmic distribution in all cell lines tested. This finding is in contrast with a report on the nuclear localization of hemagglutinin (HA)-tagged 11E6 expressed in U2OS and HaCaT cells (23). We do not have an explanation for this discrepancy. Our results indicate that the lack of efficient NLS1 and NLS2 in the 6E6 and 11E6 proteins is probably the main reason for the cytoplasmic retention of these proteins.
The cellular distribution of 18E6 is very similar to that of 16E6 with regard to predominant localization in the nucleus but differs from that of 16E6 with regard to a much lower protein signal in the nucleolus. 18E6 has been described as a nuclear protein (36), consistent with our observation of a nuclear localization in living cells (Fig. 2A). However, the finding by us and others that 18E6 is a nuclear protein is in contrast to the results of a study with HA-tagged 18E6, in which 18E6 was observed to be cytoplasmic (23). It is unclear whether the cytoplasmic HA-tagged 18E6 protein described in that study (23) was truely representative of full-length 18E6 from unspliced 18E6 RNA. We have observed that 18E6 RNA is expressed mainly as 18E6*I (Z.-M. Zheng, et al., unpublished data).
It remains unknown why 16E6 should have more than two NLS motifs for specifying its nuclear localization and what cellular factors of the nucleocytoplasmic transport machinery, especially members of the importin family (33), are involved in the transport of 16E6 to the nucleus. For a classical, single-NLS-containing protein such as simian virus 40 large-T antigen, the NLS sequence (PKKKRKV) binds to importin α interacting with importin β. The trimeric NLS protein-importin α-importin β complex then docks to the cytoplasmic side of the NPC via importin β and follows via translocation to the nuclear side of the NPC (19). There are many reports of imported nuclear proteins having at least two NLSs. These include polyomavirus large-T antigen (56), influenza virus NS1 (21), bovine papillomavirus E2 (64), and herpes simplex virus type 1 γ1 34.5 (8). Further, it has been proposed that two signals may be required for different steps in nuclear import. One signal may mediate an importin-dependent association with the NPC, and the other may subsequently engage the transport machinery to foster translocation (24). For 16E6, three signals may carry out different functions. One signal may mediate nuclear import, with assistance from another, and the third one may prefer to interact with DNA for nuclear retention of the protein, as the NLS and DNA- or RNA-binding domain in many proteins have been shown to overlap (9, 34). This assumption is supported by evidence that two NLS sequences work better than a single NLS sequence in E6 nuclear localization (Fig. 5 and 8), that a motif similar to 16E6 NLS3 exists in low-risk E6 but does not provide nuclear localization of the protein, and that 16E6 NLS3 by itself in the corresponding region of 6E6 does not convert 6E6 into a nuclear protein but enhances the protein signal in the nucleus when combined with 16E6 NLS1 and/or NLS2.
We used p53 degradation in vitro as a measure of the activities of E6 mutant proteins. Several studies have shown that p53 degradation by E6 is not necessary for MEC immortalization (41). The mutants constructed in this report were intended to disrupt the NLS and change several amino acids in the peptides. However, this approach may alter other properties of E6. Only one mutant was able to degrade p53 in vitro, and all failed to immortalize MECs. The 16E6 mutant that retained NLS1 and lacked NLS2 and NLS3 retained the ability to induce p53 degradation but did not immortalize MECs. These results suggest that NLS1 is not necessary for epithelial cell immortalization and that the NLS2 and NLS3 mutations inactivate another required function of E6. Introduction of the 16E6 NLS sequences into the corresponding regions of 6E6 did not confer on 6E6 the ability to degrade p53 or to immortalize MECs, indicating that the NLS sequences identified in 16E6 are important but not sufficient for p53 degradation and cell immortalization. In addition, the inability of the GFP-16E6 chimera to immortalize MECs implies that this additional peptide blocked an activity of E6 necessary for immortalization. Further studies with limited mutations of the NLS sequences to test their effects on E6 biological activities are under way.
Several studies have consistently demonstrated that the region of the newly identified NLS3 is also biologically important for 16E6 to function as an oncoprotein. Deletion mutant 16E6Δ118-122 has a partial deletion (118HLDKK122) of NLS3 and leads to a low binding affinity in vitro for p53, E6AP, and E6BP (11, 41, 61), to impotent induction of telomerase activity and cell immortalization (31), and to a low capacity to maintain viral episomal DNA replication (51). 16E6 with an H118D or H118N mutation also showed reduced binding in vitro to E6AP, E6BP, and p53, but 16E6 with an L119R, D120A, or D120T mutation showed no changes in binding (41). The negative results obtained with the latter three mutants could be a result of such mutations being neutral and thus not disrupting the positively charged residues in NLS3.
In summary, we have identified and characterized three NLS motifs in the 16E6 protein that are responsible for the localization of 16E6 to the nucleus. Our results indicate that NLS1 and NLS2 have a stronger influence than NLS3 for retaining 16E6 in the nucleus. Although the three 16E6 NLS sequences identified play important roles in p53 degradation and cell immortalization, other sequences participating in those activities seem necessary. Given the complexity of the functional motifs present in E6 proteins, the relationship between nuclear localization and viral oncogenesis appears to be attributed to multiple protein motifs rather than NLS sequences alone. Nevertheless, identification of the three NLS motifs in 16E6 provides new insight into the novel activities of this oncoprotein.
Acknowledgments
We gratefully acknowledge E.-M. de Villiers, German Cancer Institute (Heidelberg, Germany), for providing HPV6, HPV11, HPV16, and HPV18 plasmids and Alison McBride, NIAID, for providing HaCaT cells. We also thank Douglas Lowy at NCI for critical comments on the manuscript.
REFERENCES
- 1.Band, V., S. Dalal, L. Delmolino, and E. J. Androphy. 1993. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6-immortalized human mammary epithelial cells. EMBO J. 12:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Band, V., D. Zajchowski, V. Kulesa, and R. Sager. 1990. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc. Natl. Acad. Sci. USA 87:463-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerheide, W., H. U. Bernard, Y. J. Tan, A. Ganesan, W. G. Rice, and A. E. Ting. 1999. Potential drugs against cervical cancer: zinc-ejecting inhibitors of the human papillomavirus type 16 E6 oncoprotein. J. Natl. Cancer Inst. 91:1211-1220. [DOI] [PubMed] [Google Scholar]
- 4.Boulikas, T. 1993. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3:193-227. [PubMed] [Google Scholar]
- 5.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 6.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, G., M. E. Brett, and B. He. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the γ1 34.5 protein of herpes simplex virus type 1. J. Virol. 76:9434-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547-556. [DOI] [PubMed] [Google Scholar]
- 11.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desaintes, C., S. Hallez, P. Van Alphen, and A. Burny. 1992. Transcriptional activation of several heterologous promoters by the E6 protein of human papillomavirus type 16. J. Virol. 66:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 14.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 277:21730-21739. [DOI] [PubMed] [Google Scholar]
- 15.Fujikawa, K., M. Furuse, K. Uwabe, H. Maki, and O. Yoshie. 1994. Nuclear localization and transforming activity of human papillomavirus type 16 E7-beta-galactosidase fusion protein: characterization of the nuclear localization sequence. Virology 204:789-793. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Q., L. Singh, A. Kumar, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 75:4459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 20.Greenfield, I., J. Nickerson, S. Penman, and M. Stanley. 1991. Human papillomavirus 16 E7 protein is associated with the nuclear matrix. Proc. Natl. Acad. Sci. USA 88:11217-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guccione, E., P. Massimi, A. Bernat, and L. Banks. 2002. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology 293:20-25. [DOI] [PubMed] [Google Scholar]
- 24.Hall, M. N., C. Craik, and Y. Hiraoka. 1990. Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc. Natl. Acad. Sci. USA 87:6954-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannon, G. J., P. Sun, A. Carnero, L. Y. Xie, R. Maestro, D. S. Conklin, and D. Beach. 1999. MaRX: an approach to genetics in mammalian cells. Science 283:1129-1130. [DOI] [PubMed] [Google Scholar]
- 26.Hawley-Nelson, P., K. H. Vousden, N. L. Hubbert, D. R. Lowy, and J. T. Schiller. 1989. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 8:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 13:4918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakel, S., W. Albig, U. Kutay, F. R. Bischoff, K. Schwamborn, D. Doenecke, and D. Gorlich. 1999. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda, T., S. Watanabe, S. Zanma, H. Sato, A. Furuno, and K. Yoshiike. 1991. Human papillomavirus type 16 E6 proteins with glycine substitution for cysteine in the metal-binding motif. Virology 185:536-543. [DOI] [PubMed] [Google Scholar]
- 31.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 32.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaCasse, E. C., and Y. A. Lefebvre. 1995. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 23:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, M. C., R. I. Lin, S. Y. Huang, C. W. Tsai, and W. Y. Tarn. 2000. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275:7950-7957. [DOI] [PubMed] [Google Scholar]
- 36.Lechner, M. S., D. H. Mack, A. B. Finicle, T. Crook, K. H. Vousden, and L. A. Laimins. 1992. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 11:3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lentz, M. R., D. Pak, I. Mohr, and M. R. Botchan. 1993. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J. Virol. 67:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Roux, L. G., and J. Moroianu. 2003. Nuclear entry of high-risk human papillomavirus type 16 E6 oncoprotein occurs via several pathways. J. Virol. 77:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang, X. H., M. Volkmann, R. Klein, B. Herman, and S. J. Lockett. 1993. Co-localization of the tumor-suppressor protein p53 and human papillomavirus E6 protein in human cervical carcinoma cell lines. Oncogene 8:2645-2652. [PubMed] [Google Scholar]
- 41.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 44.Mattews, C. A., and K. E. van Holde. 1990. Introduction to proteins: the primary level of protein structure, p. 133-156. In C. A. Mattews and K. E. van Holde (ed.), Biochemistry. The Benjamin/Cummings Publishing Co., Redwood City, Calif.
- 45.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 46.Munger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 47.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers, G., and E. Androphy. 1995. The E6 protein, p. III47-III57. In G. Myers (ed.), Human papillomavirus compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 49.Nair, R., P. Carter, and B. Rost. 2003. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 31:397-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson, L. M., R. C. Rose, L. LeRoux, C. Lane, K. Bruya, and J. Moroianu. 2000. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell. Biochem. 79:225-238. [PubMed] [Google Scholar]
- 51.Park, R. B., and E. J. Androphy. 2002. Genetic analysis of high-risk E6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 76:11359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pim, D., and L. Banks. 1999. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene 18:7403-7408. [DOI] [PubMed] [Google Scholar]
- 54.Pim, D., P. Massimi, and L. Banks. 1997. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257-264. [DOI] [PubMed] [Google Scholar]
- 55.Pim, D., A. Storey, M. Thomas, P. Massimi, and L. Banks. 1994. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene 9:1869-1876. [PubMed] [Google Scholar]
- 56.Richardson, W. D., B. L. Roberts, and A. E. Smith. 1986. Nuclear location signals in polyoma virus large-T. Cell 44:77-85. [DOI] [PubMed] [Google Scholar]
- 57.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rout, M. P., and J. D. Aitchison. 2000. Pore relations: nuclear pore complexes and nucleocytoplasmic exchange. Essays Biochem. 36:75-88. [DOI] [PubMed] [Google Scholar]
- 59.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 60.Sedman, S. A., M. S. Barbosa, W. C. Vass, N. L. Hubbert, J. A. Haas, D. R. Lowy, and J. T. Schiller. 1991. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 65:4860-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman, L., H. Itzhaki, A. Jackman, J. J. Chen, D. Koval, and R. Schlegel. 2002. Inhibition of serum- and calcium-induced terminal differentiation of human keratinocytes by HPV 16 E6: study of the association with p53 degradation, inhibition of p53 transactivation, and binding to E6BP. Virology 292:309-320. [DOI] [PubMed] [Google Scholar]
- 62.Sherman, L., and R. Schlegel. 1996. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J. Virol. 70:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh, L., Q. Gao, A. Kumar, T. Gotoh, D. E. Wazer, H. Band, L. A. Feig, and V. Band. 2003. The high-risk human papillomavirus type 16 E6 counters the GAP function of E6TP1 toward small Rap G proteins. J. Virol. 77:1614-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skiadopoulos, M. H., and A. A. McBride. 1996. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 70:1117-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith-McCune, K., D. Kalman, C. Robbins, S. Shivakumar, L. Yuschenkoff, and J. M. Bishop. 1999. Intranuclear localization of human papillomavirus 16 E7 during transformation and preferential binding of E7 to the Rb family member p130. Proc. Natl. Acad. Sci. USA 96:6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smotkin, D., H. Prokoph, and F. O. Wettstein. 1989. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J. Virol. 63:1441-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song, S., A. Liem, J. A. Miller, and P. F. Lambert. 2000. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology 267:141-150. [DOI] [PubMed] [Google Scholar]
- 69.Storey, A., and L. Banks. 1993. Human papillomavirus type 16 E6 gene cooperates with EJ-ras to immortalize primary mouse cells. Oncogene 8:919-924. [PubMed] [Google Scholar]
- 70.Storey, A., M. Thomas, A. Kalita, C. Harwood, D. Gardiol, F. Mantovani, J. Breuer, I. M. Leigh, G. Matlashewski, and L. Banks. 1998. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393:229-234. [DOI] [PubMed] [Google Scholar]
- 71.Sun, X. Y., I. Frazer, M. Muller, L. Gissmann, and J. Zhou. 1995. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology 213:321-327. [DOI] [PubMed] [Google Scholar]
- 72.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 74.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 75.Yoneda, Y., M. Hieda, E. Nagoshi, and Y. Miyamoto. 1999. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct. Funct. 24:425-433. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, Z. M., and S. Specter. 1996. Dynamic production of tumour necrosis factor-alpha (TNF-alpha) messenger RNA, intracellular and extracellular TNF-alpha by murine macrophages and possible association with protein tyrosine phosphorylation of STAT1 alpha and ERK2 as an early signal. Immunology 87:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou, J., J. Doorbar, X. Y. Sun, L. V. Crawford, C. S. McLean, and I. H. Frazer. 1991. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology 185:625-632. [DOI] [PubMed] [Google Scholar]
- 78.Zimmermann, H., R. Degenkolbe, H. U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zou, N., B. Y. Lin, F. Duan, K. Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]