Abstract
In the crystal structure of the title Schiff-base, C20H21N3O4, the amino group forms an N—H⋯O hydrogen bond to the acetyl group of an adjacent molecule, forming a zigzag chain. The 2-hydroxy group is internally hydrogen bonded to the amido group though an O—H⋯O hydrogen bond.
Related literature
For medicinal uses of the precursor Schiff base, see: Jin et al. (2006 ▶); Joshi et al. (2008 ▶); Szczepankiewicz et al. (2001 ▶).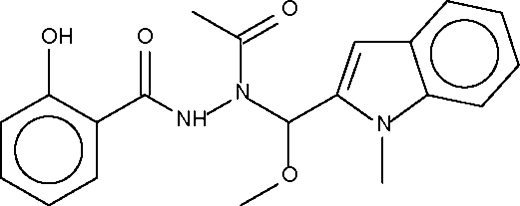
Experimental
Crystal data
C20H21N3O4
M r = 367.40
Monoclinic,
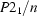
a = 11.0075 (3) Å
b = 10.5197 (3) Å
c = 15.4479 (4) Å
β = 93.967 (2)°
V = 1784.51 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 (2) K
0.20 × 0.15 × 0.10 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
16316 measured reflections
4072 independent reflections
2646 reflections with I > 2σ(I)
R int = 0.065
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.120
S = 1.02
4072 reflections
255 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.26 e Å−3
Δρmin = −0.26 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026846/tk2298sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026846/tk2298Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1o⋯O2 | 0.87 (1) | 1.81 (2) | 2.599 (2) | 150 (3) |
| N1—H1n⋯O3i | 0.85 (1) | 2.01 (1) | 2.812 (2) | 157 (2) |
Symmetry code: (i) 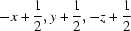 .
.
Acknowledgments
We thank the University of Malaya for supporting this study (grant No. FS338/2008 A).
supplementary crystallographic information
Comment
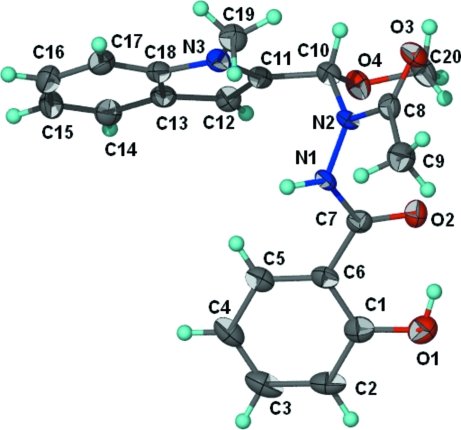
The Schiff base, N'-[(1-methyl-1H-indol-2-yl)methylene]-2-hydroxybenzohydrazide, exhibits useful medicinal properties (Jin et al., 2006; Joshi et al., 2008; Szczepankiewicz et al., 2001). When dissolved in acetic anhydride, the compound undergoes a reaction to yield the title compound, (I), Fig. 1. Essentially, a mole of methyl acetate has been added across the carbon-nitrogen double-bond. In the crystal structure of (I), the amino group forms an N–H···O hydrogen bond to the acetyl group of an adjacent molecule to result in a zigzag chain that runs along the b-axis of the orthorhombic unit cell, Table 1. The 2-hydroxy group is internally hydrogen bonded to the amido group though an O–H···O hydrogen bond, Table 1.
Experimental
2-Hydroxybenzohydrazide was condensed with 1-methylindole-3-carboxaldehyde to yield the corresponding Schiff base. To N'-[(1-methyl-1H-indol-2-yl)methylene]-2-hydroxybenzohydrazide (0.88 g, 3 mmol) was added acetic anhydride (10 ml). The mixture was heated to 398–403 K until the reactants dissolved completely. After 2 h of heating, the mixture was cooled and then treated with ethyl acetate and saturated aqueous sodium bicarbonate. The organic layer was separated and dried over anhydrous sodium sulfate. The solvent was evaporated and the resulting solid was recrystallized from methanol to give (I) as colorless crystals.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95 to 0.98 Å) and were included in the refinement in the riding model approximation, with U(H) set to 1.2–1.5U(C). The hydroxy- and ammonium H-atoms were located in a difference Fourier map, and were refined with a distance restraints O–H = N–H = 0.85±0.01 Å; their temperature factors were freely refined.
Figures
Fig. 1.
Thermal ellipsoid plot (Barbour, 2001) of (I) drawn at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C20H21N3O4 | F000 = 776 |
| Mr = 367.40 | Dx = 1.367 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1739 reflections |
| a = 11.0075 (3) Å | θ = 2.2–21.4º |
| b = 10.5197 (3) Å | µ = 0.10 mm−1 |
| c = 15.4479 (4) Å | T = 100 (2) K |
| β = 93.967 (2)º | Block, colorless |
| V = 1784.51 (8) Å3 | 0.20 × 0.15 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 2646 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.065 |
| Monochromator: graphite | θmax = 27.5º |
| T = 100(2) K | θmin = 2.2º |
| ω scans | h = −14→14 |
| Absorption correction: None | k = −13→13 |
| 16316 measured reflections | l = −20→20 |
| 4072 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.120 | w = 1/[σ2(Fo2) + (0.0466P)2 + 0.3679P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.001 |
| 4072 reflections | Δρmax = 0.26 e Å−3 |
| 255 parameters | Δρmin = −0.26 e Å−3 |
| 2 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.75833 (13) | 0.62701 (15) | 0.28349 (10) | 0.0328 (4) | |
| H1O | 0.717 (2) | 0.5560 (16) | 0.2811 (18) | 0.069 (10)* | |
| O2 | 0.57533 (11) | 0.47150 (13) | 0.28757 (9) | 0.0266 (3) | |
| O3 | 0.27682 (12) | 0.25872 (13) | 0.22300 (9) | 0.0279 (3) | |
| O4 | 0.41144 (11) | 0.35118 (13) | 0.43051 (8) | 0.0257 (3) | |
| N1 | 0.38731 (14) | 0.55433 (15) | 0.29735 (10) | 0.0197 (4) | |
| H1N | 0.3430 (17) | 0.6189 (15) | 0.3061 (14) | 0.037 (6)* | |
| N2 | 0.33109 (13) | 0.43653 (14) | 0.29605 (10) | 0.0188 (3) | |
| N3 | 0.12618 (13) | 0.52539 (15) | 0.40494 (10) | 0.0205 (4) | |
| C1 | 0.68446 (17) | 0.7144 (2) | 0.31706 (12) | 0.0262 (5) | |
| C2 | 0.73287 (19) | 0.8337 (2) | 0.33795 (14) | 0.0315 (5) | |
| H2 | 0.8144 | 0.8530 | 0.3262 | 0.038* | |
| C3 | 0.6634 (2) | 0.9230 (2) | 0.37533 (14) | 0.0354 (5) | |
| H3 | 0.6972 | 1.0042 | 0.3892 | 0.042* | |
| C4 | 0.54370 (19) | 0.8971 (2) | 0.39356 (13) | 0.0308 (5) | |
| H4 | 0.4966 | 0.9592 | 0.4209 | 0.037* | |
| C5 | 0.49478 (18) | 0.78036 (18) | 0.37132 (12) | 0.0249 (4) | |
| H5 | 0.4132 | 0.7621 | 0.3837 | 0.030* | |
| C6 | 0.56213 (17) | 0.68846 (18) | 0.33118 (12) | 0.0225 (4) | |
| C7 | 0.51091 (17) | 0.56257 (18) | 0.30497 (11) | 0.0207 (4) | |
| C8 | 0.31298 (16) | 0.36884 (19) | 0.22067 (12) | 0.0221 (4) | |
| C9 | 0.33655 (19) | 0.4356 (2) | 0.13833 (13) | 0.0294 (5) | |
| H9A | 0.3008 | 0.3867 | 0.0889 | 0.044* | |
| H9B | 0.4246 | 0.4435 | 0.1337 | 0.044* | |
| H9C | 0.2997 | 0.5204 | 0.1383 | 0.044* | |
| C10 | 0.30451 (16) | 0.38509 (18) | 0.38166 (12) | 0.0212 (4) | |
| H10 | 0.2504 | 0.3091 | 0.3736 | 0.025* | |
| C11 | 0.24263 (16) | 0.48432 (18) | 0.43178 (12) | 0.0208 (4) | |
| C12 | 0.28551 (17) | 0.55066 (18) | 0.50327 (12) | 0.0227 (4) | |
| H12 | 0.3624 | 0.5400 | 0.5343 | 0.027* | |
| C13 | 0.19310 (17) | 0.63891 (18) | 0.52267 (12) | 0.0222 (4) | |
| C14 | 0.18300 (18) | 0.73351 (19) | 0.58605 (12) | 0.0261 (5) | |
| H14 | 0.2474 | 0.7477 | 0.6291 | 0.031* | |
| C15 | 0.07752 (18) | 0.8057 (2) | 0.58468 (13) | 0.0282 (5) | |
| H15 | 0.0704 | 0.8708 | 0.6267 | 0.034* | |
| C16 | −0.01854 (18) | 0.7841 (2) | 0.52232 (13) | 0.0295 (5) | |
| H16 | −0.0899 | 0.8348 | 0.5230 | 0.035* | |
| C17 | −0.01235 (17) | 0.6909 (2) | 0.45972 (13) | 0.0260 (5) | |
| H17 | −0.0785 | 0.6754 | 0.4182 | 0.031* | |
| C18 | 0.09490 (16) | 0.62076 (18) | 0.46006 (12) | 0.0220 (4) | |
| C19 | 0.05096 (17) | 0.4838 (2) | 0.32897 (12) | 0.0269 (5) | |
| H19A | −0.0351 | 0.4872 | 0.3415 | 0.040* | |
| H19B | 0.0726 | 0.3963 | 0.3144 | 0.040* | |
| H19C | 0.0647 | 0.5397 | 0.2799 | 0.040* | |
| C20 | 0.4677 (2) | 0.24041 (19) | 0.39780 (14) | 0.0313 (5) | |
| H20A | 0.5395 | 0.2183 | 0.4358 | 0.047* | |
| H20B | 0.4924 | 0.2574 | 0.3392 | 0.047* | |
| H20C | 0.4097 | 0.1696 | 0.3960 | 0.047* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0245 (7) | 0.0335 (9) | 0.0408 (9) | −0.0008 (7) | 0.0048 (6) | 0.0009 (7) |
| O2 | 0.0224 (7) | 0.0240 (8) | 0.0339 (8) | 0.0036 (6) | 0.0054 (6) | −0.0027 (6) |
| O3 | 0.0311 (8) | 0.0184 (8) | 0.0342 (8) | −0.0058 (6) | 0.0016 (6) | −0.0067 (6) |
| O4 | 0.0282 (7) | 0.0211 (8) | 0.0272 (7) | 0.0051 (6) | −0.0028 (6) | −0.0001 (6) |
| N1 | 0.0201 (8) | 0.0107 (8) | 0.0284 (9) | −0.0007 (7) | 0.0024 (7) | −0.0012 (7) |
| N2 | 0.0219 (8) | 0.0112 (8) | 0.0235 (8) | −0.0031 (6) | 0.0023 (6) | −0.0011 (7) |
| N3 | 0.0213 (8) | 0.0190 (9) | 0.0213 (8) | −0.0025 (7) | 0.0010 (6) | −0.0028 (7) |
| C1 | 0.0270 (10) | 0.0277 (12) | 0.0236 (10) | −0.0010 (9) | −0.0011 (8) | 0.0057 (9) |
| C2 | 0.0291 (11) | 0.0302 (12) | 0.0345 (12) | −0.0109 (10) | −0.0027 (9) | 0.0081 (10) |
| C3 | 0.0452 (13) | 0.0230 (12) | 0.0364 (12) | −0.0101 (10) | −0.0088 (10) | 0.0048 (10) |
| C4 | 0.0420 (13) | 0.0197 (11) | 0.0297 (11) | −0.0021 (9) | −0.0028 (10) | −0.0017 (9) |
| C5 | 0.0296 (10) | 0.0209 (11) | 0.0239 (10) | −0.0027 (9) | −0.0014 (8) | 0.0013 (9) |
| C6 | 0.0252 (10) | 0.0184 (10) | 0.0236 (10) | −0.0025 (8) | 0.0000 (8) | 0.0049 (8) |
| C7 | 0.0243 (10) | 0.0205 (10) | 0.0172 (9) | −0.0013 (8) | 0.0020 (7) | 0.0030 (8) |
| C8 | 0.0194 (9) | 0.0207 (11) | 0.0259 (10) | 0.0033 (8) | 0.0002 (8) | −0.0026 (8) |
| C9 | 0.0338 (11) | 0.0306 (12) | 0.0241 (10) | −0.0007 (10) | 0.0028 (9) | −0.0032 (9) |
| C10 | 0.0228 (9) | 0.0176 (10) | 0.0232 (10) | −0.0010 (8) | 0.0007 (8) | 0.0005 (8) |
| C11 | 0.0222 (9) | 0.0175 (10) | 0.0228 (9) | −0.0016 (8) | 0.0022 (8) | 0.0017 (8) |
| C12 | 0.0238 (10) | 0.0231 (11) | 0.0213 (9) | 0.0003 (8) | 0.0008 (8) | 0.0007 (8) |
| C13 | 0.0251 (10) | 0.0199 (10) | 0.0218 (10) | −0.0024 (8) | 0.0036 (8) | 0.0030 (8) |
| C14 | 0.0304 (11) | 0.0268 (12) | 0.0216 (10) | −0.0033 (9) | 0.0043 (8) | −0.0024 (9) |
| C15 | 0.0356 (11) | 0.0238 (11) | 0.0263 (10) | 0.0006 (9) | 0.0090 (9) | −0.0047 (9) |
| C16 | 0.0282 (11) | 0.0289 (12) | 0.0326 (11) | 0.0042 (9) | 0.0101 (9) | 0.0009 (10) |
| C17 | 0.0229 (10) | 0.0266 (11) | 0.0286 (11) | −0.0011 (8) | 0.0036 (8) | 0.0012 (9) |
| C18 | 0.0236 (9) | 0.0193 (10) | 0.0236 (10) | −0.0040 (8) | 0.0059 (8) | 0.0000 (8) |
| C19 | 0.0247 (10) | 0.0279 (12) | 0.0274 (11) | −0.0023 (9) | −0.0028 (8) | −0.0050 (9) |
| C20 | 0.0367 (12) | 0.0185 (11) | 0.0378 (12) | 0.0085 (9) | −0.0026 (10) | 0.0007 (9) |
Geometric parameters (Å, °)
| O1—C1 | 1.354 (2) | C8—C9 | 1.491 (3) |
| O1—H1O | 0.871 (10) | C9—H9A | 0.9800 |
| O2—C7 | 1.233 (2) | C9—H9B | 0.9800 |
| O3—C8 | 1.226 (2) | C9—H9C | 0.9800 |
| O4—C10 | 1.400 (2) | C10—C11 | 1.492 (3) |
| O4—C20 | 1.428 (2) | C10—H10 | 1.0000 |
| N1—C7 | 1.360 (2) | C11—C12 | 1.363 (3) |
| N1—N2 | 1.385 (2) | C12—C13 | 1.424 (3) |
| N1—H1N | 0.852 (9) | C12—H12 | 0.9500 |
| N2—C8 | 1.368 (2) | C13—C14 | 1.406 (3) |
| N2—C10 | 1.477 (2) | C13—C18 | 1.413 (3) |
| N3—C18 | 1.375 (2) | C14—C15 | 1.387 (3) |
| N3—C11 | 1.389 (2) | C14—H14 | 0.9500 |
| N3—C19 | 1.456 (2) | C15—C16 | 1.399 (3) |
| C1—C2 | 1.393 (3) | C15—H15 | 0.9500 |
| C1—C6 | 1.406 (3) | C16—C17 | 1.382 (3) |
| C2—C3 | 1.365 (3) | C16—H16 | 0.9500 |
| C2—H2 | 0.9500 | C17—C18 | 1.392 (3) |
| C3—C4 | 1.393 (3) | C17—H17 | 0.9500 |
| C3—H3 | 0.9500 | C19—H19A | 0.9800 |
| C4—C5 | 1.375 (3) | C19—H19B | 0.9800 |
| C4—H4 | 0.9500 | C19—H19C | 0.9800 |
| C5—C6 | 1.390 (3) | C20—H20A | 0.9800 |
| C5—H5 | 0.9500 | C20—H20B | 0.9800 |
| C6—C7 | 1.485 (3) | C20—H20C | 0.9800 |
| C1—O1—H1O | 106.0 (19) | O4—C10—C11 | 107.19 (14) |
| C10—O4—C20 | 112.71 (14) | N2—C10—C11 | 109.52 (15) |
| C7—N1—N2 | 120.11 (16) | O4—C10—H10 | 109.6 |
| C7—N1—H1N | 121.0 (15) | N2—C10—H10 | 109.6 |
| N2—N1—H1N | 117.1 (15) | C11—C10—H10 | 109.6 |
| C8—N2—N1 | 121.13 (15) | C12—C11—N3 | 110.09 (17) |
| C8—N2—C10 | 123.05 (15) | C12—C11—C10 | 129.33 (17) |
| N1—N2—C10 | 115.52 (14) | N3—C11—C10 | 120.52 (16) |
| C18—N3—C11 | 107.92 (15) | C11—C12—C13 | 106.97 (16) |
| C18—N3—C19 | 124.45 (16) | C11—C12—H12 | 126.5 |
| C11—N3—C19 | 127.50 (16) | C13—C12—H12 | 126.5 |
| O1—C1—C2 | 118.06 (18) | C14—C13—C18 | 118.61 (18) |
| O1—C1—C6 | 122.32 (18) | C14—C13—C12 | 134.43 (18) |
| C2—C1—C6 | 119.6 (2) | C18—C13—C12 | 106.95 (17) |
| C3—C2—C1 | 120.1 (2) | C15—C14—C13 | 118.93 (18) |
| C3—C2—H2 | 119.9 | C15—C14—H14 | 120.5 |
| C1—C2—H2 | 119.9 | C13—C14—H14 | 120.5 |
| C2—C3—C4 | 121.1 (2) | C14—C15—C16 | 120.91 (19) |
| C2—C3—H3 | 119.4 | C14—C15—H15 | 119.5 |
| C4—C3—H3 | 119.4 | C16—C15—H15 | 119.5 |
| C5—C4—C3 | 118.9 (2) | C17—C16—C15 | 121.71 (19) |
| C5—C4—H4 | 120.5 | C17—C16—H16 | 119.1 |
| C3—C4—H4 | 120.5 | C15—C16—H16 | 119.1 |
| C4—C5—C6 | 121.40 (19) | C16—C17—C18 | 117.16 (18) |
| C4—C5—H5 | 119.3 | C16—C17—H17 | 121.4 |
| C6—C5—H5 | 119.3 | C18—C17—H17 | 121.4 |
| C5—C6—C1 | 118.71 (18) | N3—C18—C17 | 129.29 (17) |
| C5—C6—C7 | 122.54 (17) | N3—C18—C13 | 108.07 (16) |
| C1—C6—C7 | 118.73 (18) | C17—C18—C13 | 122.64 (18) |
| O2—C7—N1 | 121.25 (17) | N3—C19—H19A | 109.5 |
| O2—C7—C6 | 122.64 (17) | N3—C19—H19B | 109.5 |
| N1—C7—C6 | 116.05 (17) | H19A—C19—H19B | 109.5 |
| O3—C8—N2 | 119.69 (18) | N3—C19—H19C | 109.5 |
| O3—C8—C9 | 123.09 (18) | H19A—C19—H19C | 109.5 |
| N2—C8—C9 | 117.21 (17) | H19B—C19—H19C | 109.5 |
| C8—C9—H9A | 109.5 | O4—C20—H20A | 109.5 |
| C8—C9—H9B | 109.5 | O4—C20—H20B | 109.5 |
| H9A—C9—H9B | 109.5 | H20A—C20—H20B | 109.5 |
| C8—C9—H9C | 109.5 | O4—C20—H20C | 109.5 |
| H9A—C9—H9C | 109.5 | H20A—C20—H20C | 109.5 |
| H9B—C9—H9C | 109.5 | H20B—C20—H20C | 109.5 |
| O4—C10—N2 | 111.40 (15) | ||
| C7—N1—N2—C8 | −84.8 (2) | N1—N2—C10—C11 | 48.99 (19) |
| C7—N1—N2—C10 | 89.2 (2) | C18—N3—C11—C12 | 0.6 (2) |
| O1—C1—C2—C3 | −177.23 (18) | C19—N3—C11—C12 | 176.65 (18) |
| C6—C1—C2—C3 | 2.8 (3) | C18—N3—C11—C10 | −176.91 (16) |
| C1—C2—C3—C4 | 0.2 (3) | C19—N3—C11—C10 | −0.9 (3) |
| C2—C3—C4—C5 | −1.6 (3) | O4—C10—C11—C12 | 11.2 (3) |
| C3—C4—C5—C6 | −0.1 (3) | N2—C10—C11—C12 | −109.8 (2) |
| C4—C5—C6—C1 | 3.0 (3) | O4—C10—C11—N3 | −171.84 (16) |
| C4—C5—C6—C7 | −178.81 (18) | N2—C10—C11—N3 | 67.2 (2) |
| O1—C1—C6—C5 | 175.68 (17) | N3—C11—C12—C13 | −0.6 (2) |
| C2—C1—C6—C5 | −4.3 (3) | C10—C11—C12—C13 | 176.68 (18) |
| O1—C1—C6—C7 | −2.6 (3) | C11—C12—C13—C14 | −178.7 (2) |
| C2—C1—C6—C7 | 177.42 (17) | C11—C12—C13—C18 | 0.3 (2) |
| N2—N1—C7—O2 | 19.8 (3) | C18—C13—C14—C15 | −0.2 (3) |
| N2—N1—C7—C6 | −162.92 (15) | C12—C13—C14—C15 | 178.6 (2) |
| C5—C6—C7—O2 | −163.41 (18) | C13—C14—C15—C16 | 1.0 (3) |
| C1—C6—C7—O2 | 14.8 (3) | C14—C15—C16—C17 | −0.2 (3) |
| C5—C6—C7—N1 | 19.4 (3) | C15—C16—C17—C18 | −1.3 (3) |
| C1—C6—C7—N1 | −162.41 (17) | C11—N3—C18—C17 | −179.76 (19) |
| N1—N2—C8—O3 | 171.07 (16) | C19—N3—C18—C17 | 4.0 (3) |
| C10—N2—C8—O3 | −2.5 (3) | C11—N3—C18—C13 | −0.4 (2) |
| N1—N2—C8—C9 | −10.1 (2) | C19—N3—C18—C13 | −176.59 (17) |
| C10—N2—C8—C9 | 176.32 (16) | C16—C17—C18—N3 | −178.60 (19) |
| C20—O4—C10—N2 | −71.2 (2) | C16—C17—C18—C13 | 2.1 (3) |
| C20—O4—C10—C11 | 168.97 (15) | C14—C13—C18—N3 | 179.21 (16) |
| C8—N2—C10—O4 | 104.46 (19) | C12—C13—C18—N3 | 0.1 (2) |
| N1—N2—C10—O4 | −69.42 (19) | C14—C13—C18—C17 | −1.4 (3) |
| C8—N2—C10—C11 | −137.13 (17) | C12—C13—C18—C17 | 179.47 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1o···O2 | 0.87 (1) | 1.81 (2) | 2.599 (2) | 150 (3) |
| N1—H1n···O3i | 0.85 (1) | 2.01 (1) | 2.812 (2) | 157 (2) |
Symmetry codes: (i) −x+1/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2298).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Jin, L.-H., Chen, J., Song, B.-A., Chen, Z., Yang, S., Li, Q.-Z., Hu, D.-Y. & Xu, R.-Q. (2006). Bioorg. Med. Chem. Lett.16, 5036–5040. [DOI] [PubMed]
- Joshi, S. D., Vagdevi, H. M., Vaidya, V. P. & Gadaginamath, G. S. (2008). Eur. J. Med. Chem. In the press. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Szczepankiewicz, B. G., Liu, G., Jae, H.-S., Tasker, A. S., Gunawardana, I. W., von Geldern, T. W., Gwaltney, S. L., Wu-Wong, R., Gehrke, L., Chiou, W. J., Credo, R. B., Alder, J. D., Nukkala, M. A., Zielinski, N. A., Jarvis, K., Mollison, K. W., Frost, D. J., Bauch, J. L., Hui, Y. H., Claiborne, A. K., Li, Q. & Rosenberg, S. H. (2001). J. Med. Chem.44, 4416–4430. [DOI] [PubMed]
- Westrip, S. P. (2008). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026846/tk2298sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026846/tk2298Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report