Abstract
Equine arteritis virus (EAV) is an enveloped, positive-strand RNA virus belonging to the family Arteriviridae of the order Nidovirales. EAV virions contain six different envelope proteins. The glycoprotein GP5 (previously named GL) and the unglycosylated membrane protein M are the major envelope proteins, while the glycoproteins GP2b (previously named GS), GP3, and GP4 are minor structural proteins. The unglycosylated small hydrophobic envelope protein E is present in virus particles in intermediate molar amounts compared to the other transmembrane proteins. The GP5 and M proteins are both essential for particle assembly. They occur as covalently linked heterodimers that constitute the basic protein matrix of the envelope. The GP2b, GP3, and GP4 proteins occur as a heterotrimeric complex in which disulfide bonds play an important role. The function of this complex has not been established yet, but the available data suggest it to be involved in the viral entry process. Here we investigated the role of the four cysteine residues of the mature GP2b protein in the assembly of the GP2b/GP3/GP4 complex. Open reading frames encoding cysteine-to-serine mutants of the GP2b protein were expressed independently or from a full-length infectious EAV cDNA clone. The results of these experiments support a model in which the cysteine residue at position 102 of GP2b forms an intermolecular cystine bridge with one of the cysteines of the GP4 protein, while the cysteine residues at positions 48 and 137 of GP2b are linked by an intrachain disulfide bond. In this model, another cysteine residue in the GP4 protein is responsible for the covalent association of GP3 with the disulfide-linked GP2b/GP4 heterodimer. In addition, our data highlight the importance of the correct association of the minor EAV envelope glycoproteins for their efficient incorporation into viral particles and for virus infectivity.
Equine arteritis virus (EAV) is an enveloped, positive-strand RNA virus which belongs to the genus Arterivirus. Other members of this single genus in the family Arteriviridae are Lactate dehydrogenase-elevating virus (LDV), Porcine reproductive and respiratory syndrome virus (PRRSV), and Simian hemorrhagic fever virus. On the basis of conserved amino acid sequence motifs in their polymerase (poly)proteins and similarities in their genome organization and gene expression strategy, the Arteriviridae have been grouped together with the Coronaviridae and the recently established family Roniviridae in the order Nidovirales (2, 3, 4).
The genome of EAV contains nine functional open reading frames (ORFs). ORF1a and -1b occupy the 5′ three-fourths of the genome and code for proteins involved in viral replication and transcription (6, 25, 29, 30, 32). The remaining one-fourth of the viral genome contains seven relatively small ORFs (ORF2a, -2b, and -3 through -7), which are expressed from a 3′-coterminal nested set of six leader-containing subgenomic mRNAs (RNA2 through -7) (8, 26) and code for the known viral structural proteins (9, 24, 34). In each of these subgenomic mRNAs, only the functional ORF positioned immediately downstream of the leader sequence is translated, with the exception of RNA2, which directs the synthesis of both the ORF2a and -2b products (24).
EAV virions have a diameter of 40 to 60 nm and are thought to possess an icosahedral core that is surrounded by a lipid-containing envelope with small surface protrusions (16, 19). The viral core consists of the EAV genome, an unsegmented RNA molecule of 12.7 kb with a 5′ cap and a 3′ poly(A) tail which is encapsidated by the 14-kDa phosphorylated nucleocapsid (N) protein (5, 17, 35). The N protein is encoded by ORF7 (9).
In the envelope of EAV, six different proteins have been identified so far. The two major envelope proteins are the GP5 glycoprotein (previously named GL), which varies in size from 30 to 42 kDa due to the addition of variable numbers of lactosamine repeats to its single N-linked glycan, and the 16-kDa nonglycosylated membrane protein (M). These proteins occur in virions in equimolar amounts and are encoded by ORF5 and −6, respectively (9). The third most abundant protein in the viral membrane is the envelope protein (E) of 10 kDa. This protein lacks N-linked oligosaccharide side chains and is encoded by ORF2a (24). The remaining envelope proteins are the 25-kDa GP2b glycoprotein (previously named GS), the heterogeneously N-glycosylated GP3 glycoprotein of 37 or 42 kDa, and the 28-kDa GP4 glycoprotein. These three polypeptides constitute the minor envelope proteins of EAV and are present in virus particles in equimolar amounts. They are encoded by ORF2b, -3, and -4, respectively (11, 34).
The M and GP5 proteins appear in EAV particles as disulfide-linked heterodimers (10). The GP2b, GP3, and GP4 proteins are present in virions as heterotrimeric complexes (see below for more details). The higher order structure of the E protein in virus particles is unknown, but there are indications that it is noncovalently associated with the GP2b/GP3/GP4 trimers (R. Wieringa et al., unpublished data). While M and GP5 are essential for EAV assembly (Wieringa et al., unpublished data), they do not determine the cell tropism of the virus. Exchange of the ectodomain of the EAV GP5 protein with that of LDV or PRRSV in the context of a full-length EAV cDNA clone did not alter the host cell range of the virus (12). Likewise, PRRSV mutants in which the ectodomain of the M protein was replaced by that of other arteriviruses retained their original cell tropism (31). Since noninfectious viral particles are produced in the absence of the GP2b, GP3, or GP4 protein (22, 24; Wieringa et al., unpublished data), it is likely that the complex of minor envelope glycoproteins mediates (initial) virus attachment to the host cell surface.
The GP2b and GP4 proteins are type I membrane glycoproteins, containing one and three functional N-glycosylation sites, respectively (11, 34). Both proteins possess three luminal cysteine residues after signal sequence removal. A fourth cysteine is located in the putative transmembrane segment of the GP2b protein and in the endodomain of the GP4 protein. The GP3 protein is a heavily glycosylated integral membrane protein with an uncleaved amino-terminal signal sequence and nine cysteine residues. The protein is inserted into the lipid bilayer by either or both of its hydrophobic terminal domains and has no parts that are detectably exposed cytoplasmically (15, 34).
In EAV-infected cells, the GP2b protein occurs in four monomeric conformations, which differ in their disulfide-bonded structures (11). In addition to these GP2b monomers, the GP2b protein assembles into a heterotrimeric complex with the GP3 and GP4 proteins (33). Due to the low stability of the GP2b/GP3/GP4 trimers, only the covalently linked GP2b/GP4 complexes of 45 kDa are detected after analysis of lysates from EAV-infected cells by immunoprecipitation and gel electrophoresis under nonreducing conditions (33). The minor envelope glycoproteins are incorporated into virions only as GP2b/GP3/GP4 complexes, i.e., incorporation of one of them requires the presence of the others (Wieringa et al., unpublished data). Interestingly, following the release of virus particles from infected cells, GP3 becomes disulfide linked to the GP2b/GP4 heterodimers, resulting in the formation of a 66-kDa complex consisting of covalently bound GP2b, GP3, and GP4 molecules. As a consequence of this postassembly modification, two different covalently linked GP2b complexes are observed in EAV particles, i.e., GP2b/GP4 heterodimers and GP2b/GP3/GP4 heterotrimers (33) (Fig. 1).
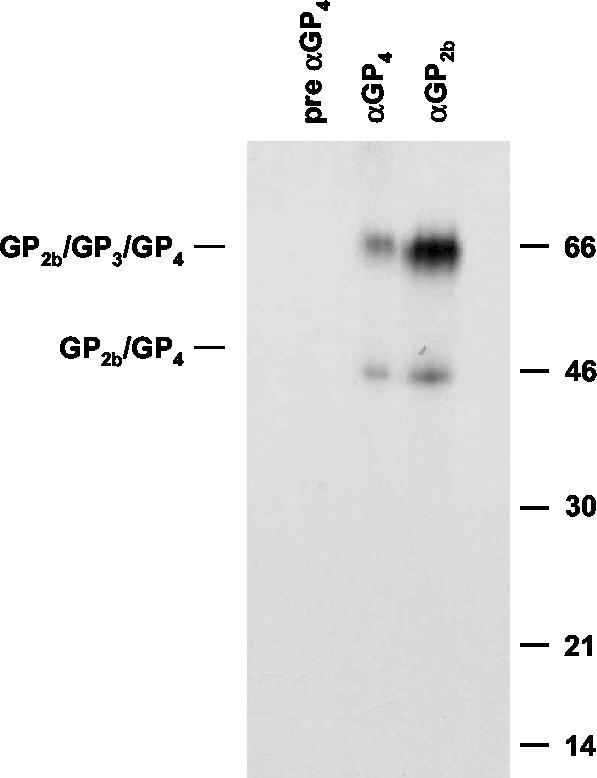
FIG. 1.
Covalently linked GP2b-containing complexes in EAV virions. [35S]cysteine-labeled EAV particles were pelleted through a sucrose cushion and further purified in an isopycnic sucrose gradient. Gradient fraction 5, the peak infectivity fraction, was subsequently incubated with αGP2b, αGP4, or preimmune αGP4 serum. The resulting immunoprecipitates were analyzed in an SDS-15% PAA gel under nonreducing conditions. The positions of the disulfide-linked GP2b/GP4 and GP2b/GP3/GP4 complexes are indicated on the left. The values on the right are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel.
Cystine bridges thus seem to play essential roles in the formation, stabilization, and probably also the functioning of the GP2b/GP3/GP4 trimers. We therefore performed a mutational analysis of the luminal cysteines of the GP2b protein by using the vaccinia virus-T7 RNA polymerase transfection system in the context of a full-length infectious EAV cDNA clone. These experiments established an important role for the cysteine residues of the GP2b protein in virus infectivity and allowed us to draw a model of the covalent interactions between the minor envelope glycoproteins in the 45- and 66-kDa complexes.
MATERIALS AND METHODS
Cells and antisera.
Baby hamster kidney (BHK-21 C13) cells (American Type Culture Collection) were grown and maintained in Glasgow minimal essential medium (GMEM; Invitrogen-Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 IU of penicillin per ml, and 100 μg of streptomycin per ml (GMEM-10% FBS). The production and characterization of rabbit antisera specific for the extreme carboxy termini of the EAV GP2b and M proteins (raised against synthetic peptides corresponding to residues 212 through 227 and 146 through 162 of the respective proteins) have been reported earlier (9).
Construction of mutant ORF2b expression plasmids.
Recombinant DNA techniques were performed essentially as described by Sambrook et al. (23). The plasmids pAVI02 C48S, pAVI02 C102S, pAVI02 C137S, and pAVI02 C195S, which code for cysteine-to-serine mutants of the EAV GP2b protein, were constructed by oligonucleotide-mediated site-directed mutagenesis using the method described by Kunkel et al. (18). First, the 0.8-kb BamHI-EcoRV fragment of plasmid pAVI02 (9) containing EAV ORF2b was cloned into BamHI- and EcoRV-digested M13BM20 (Roche), yielding M13BM20-ORF2b. Next, uracil-containing single-stranded DNA was prepared from M13BM20-ORF2b in the dut ung Escherichia coli strain CJ236 (Roche). Five picomoles of the mutagenic oligonucleotide 288, 289, 290, or 291 (Table 1) was then annealed to 1 μg of the single-stranded M13BM20-ORF2b DNA in a total volume of 10 μl of 20 mM Tris-HCl (pH 7.5)-10 mM MgCl2-50 mM NaCl-1 mM dithiothreitol (DTT). Before hybridization to the single-stranded DNA template, the oligonucleotides were phosphorylated with bacteriophage T4 polynucleotide kinase (Amersham Pharmacia Biotech). The annealing products were converted into double-stranded covalently closed circles by the addition of 20 μl of 20 mM Tris-HCl (pH 7.5)-10 mM MgCl2-10 mM DTT containing 1 mM ATP, 1 mM (each) deoxynucleoside triphosphates, 5 U of bacteriophage T7 DNA polymerase (Amersham Pharmacia Biotech), and 7 U of bacteriophage T4 DNA ligase (Amersham Pharmacia Biotech) and incubation for 2 h at 37°C. The second-strand synthesis was terminated by the addition of 1 μl of 0.5 M EDTA (pH 8.0), and the reaction mixtures were used to transform E. coli strain PC2495 (Phabagen). Individual plaques were picked and mixed with 2 ml of Luria-Bertani medium and 100 μl of an overnight culture of PC2495. After incubation for 5 h at 37°C, with constant agitation, single-stranded DNA was purified and subjected to nucleotide sequence analysis with a bacteriophage T7 DNA polymerase sequencing kit (Amersham Pharmacia Biotech) and [α-35S]dATP (>1,000 Ci/mmol) (Amersham Pharmacia Biotech). Next, replicative-form DNA was prepared from M13BM20-ORF2b clones carrying the intended point mutations and incubated with BamHI and EcoRV. The resulting 0.8-kb BamHI-EcoRV fragments containing the mutant ORF2b sequences were purified from gels and inserted into BamHI- and EcoRV-digested pBluescript KS(+) (Stratagene), yielding pAVI02 C48S, pAVI02 C102S, pAVI02 C137S, and pAVI02 C195S. In these plasmids, a bacteriophage T7 RNA polymerase promoter sequence is positioned upstream of the (mutant) ORF2b for expression studies by the vaccinia virus-T7 RNA polymerase transfection system.
TABLE 1.
Oligonucleotides used for this study
| Name (polarity) | Sequence (5′→3′)a | Positionsb | Plasmid generated |
|---|---|---|---|
| 288 (−) | GGTTGTCGCTCTGCAAATC | 9956-9974 | pAVI02 C48S |
| 289 (−) | GGTCAAAGCTATCAGGAG | 10199-10136 | pAVI02 C102S |
| 290 (−) | GCAATTTACTGTTGATGGC | 10223-10241 | pAVI02 C137S |
| 291 (−) | GTGGTATGCTTGCAAATAG | 10397-10415 | pAVI02 C195S |
Mutant nucleotides are indicated in boldface.
Based on EAV Utr (6).
Construction of mutant full-length EAV cDNA clones.
To generate full-length EAV cDNA clones in which the codon for Cys-102 or Cys-137 of the GP2b protein was changed to a serine codon, the 3.7-kb BamHI-XhoI fragment of pEAV WT was inserted into BamHI- and XhoI-digested pBluescript KS(+) (Stratagene), creating pEAVBX-sh. The construct pEAV WT is a derivative of the full-length infectious EAV cDNA clone pEAV030 (28) in which the thymidylate corresponding to genome position 5117 of the EAV untranslated repeat was replaced by a deoxycytidylate and the sequence ACAATCTGTTTTCGTG at positions 8941 through 8956 was replaced by the sequence CCAAAGCGTCTTCGTC to remove a BamHI recognition site and a cryptic bacteriophage T7 RNA polymerase terminator sequence, respectively. Next, the 0.4-kb MscI-NsiI fragment of pAVI02 C102S and of pAVI02 C137S was cloned into MscI- and NsiI-digested pEAVBX-sh. The 2.3-kb BamHI-EcoRI fragments of the resulting plasmids were used to replace the corresponding fragment of pEAV WT, generating pEAV GP2b C102S and pEAV GP2b C137S, respectively.
To make the full-length EAV cDNA clone pEAV GP2b C48S, which codes for a GP2b mutant carrying a cysteine-to-serine substitution at amino acid position 48, the 1.3-kb BamHI-SphI fragment of pEAV WT was inserted into BamHI- and SphI-digested pUCBM20 (Roche), resulting in pEAVBS-sh. The 0.3-kb EagI-SacI fragment of pAVI02 C48S was then cloned into EagI- and SacI-digested pEAVBS-sh. The 1.3-kb BamHI-SphI fragment of the resulting construct was inserted into the BamHI-SphI-digested pEAVBX-sh. The 2.3-kb BamHI-EcoRI fragment of the latter construct was swapped into pEAV WT as described above to produce pEAV GP2b C48S. For all these cloning steps, E. coli strain PC2495 was used.
Independent expression studies using the vaccinia virus-T7 RNA polymerase transfection system.
Subconfluent monolayers of BHK-21 C13 cells were washed with GMEM and inoculated with vTF7.3, a recombinant vaccinia virus expressing the bacteriophage T7 RNA polymerase gene, in GMEM for 50 min at 37°C at a multiplicity of infection of ≥10 PFU per cell. The cells were then washed with GMEM, subjected to treatment with Lipofectamine with plasmid DNA as previously described (33), and subsequently incubated at 37°C. At 3 h postinfection (p.i.), 2 ml of prewarmed GMEM-10% FBS was added to the cells.
In vitro synthesis and transfection of genome-length EAV RNA.
After digestion with XhoI, the linearized plasmid DNAs were purified by phenol-chloroform extraction, ethanol precipitated, and dissolved in water. The in vitro transcription reactions were carried out by using the T7 mMESSAGE mMACHINE kit (Ambion) according to the manufacturer's instructions. After a 2-h incubation at 37°C, the mixtures were kept on ice until the start of the transfection procedure. Confluent monolayers of 2 × 107 BHK-21 cells were digested with trypsin, resuspended in GMEM, and pelleted at 400 × g for 5 min. The cells were washed twice in phosphate-buffered saline (PBS) containing 50 mM CaCl2 and 50 mM MgCl2 (PBS Ca/Mg) and resuspended in 800 μl of PBS Ca/Mg. A 10-μl aliquot of the transcription mixture was added to the cell suspension, which was then transferred into electroporation cuvettes (BTX) with a 4-mm gap size. The cells were pulsed twice at 850 V and 50 μF, with the pulse controller set at “infinite.” The cells were resuspended in GMEM-10% FBS, seeded into 10-cm2 wells, and incubated at 39°C.
Metabolic labeling of intracellular proteins.
At the indicated time points after transfection, the cells were washed with PBS and incubated in prewarmed starvation medium (Dulbecco's modified Eagle's medium without l-cysteine and l-methionine [Invitrogen-Life Technologies] supplemented with 5% dialyzed FBS and 10 mM HEPES-KOH [pH 7.4]). Following an incubation period of 30 min, 80 μCi of l-[35S]methionine (ICN) was added per 10-cm2 culture dish and the cells were labeled at 37°C (independent expression studies) or 39°C (EAV RNA transfection experiments) for the indicated times. Alternatively, the cells were incubated in starvation medium containing 0.2 mM l-methionine and labeled with 80 μCi of l-[35S]cysteine (ICN) instead of l-[35S]methionine. After the labeling period, the cells were put on ice and washed with ice-cold PBS containing 50 mM CaCl2, 50 mM MgCl2, and, to block reactive thiol groups, 20 mM N-ethylmaleimide (NEM; Sigma-Aldrich) or 50 mM iodoacetamide (IAM; Sigma-Aldrich) as indicated in the figure legends. Next, the cells were lysed in ice-cold lysis buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with 1 μg each of aprotinin, leupeptin, and pepstatin A per ml and containing either 20 mM NEM or 50 mM IAM. The cell lysate was cleared by centrifugation for 15 min at 14,000 rpm in a microcentrifuge. The pellet was discarded, and the supernatant was supplemented with EDTA to a final concentration of 5 mM.
Preparation of radiolabeled viral particles.
BHK-21 C13 cells were transfected with synthetic EAV RNAs and labeled at 7 h posttransfection (p.t.) with l-[35S]methionine or l-[35S]cysteine as described above. After a 12-h labeling period at 39°C, the culture supernatant was harvested and the cell debris was removed by low-speed centrifugation (10 min at 4,000 rpm and room temperature in a microcentrifuge). The viral particles were then pelleted through a cushion of 20% (wt/wt) sucrose in 20 mM Tris-HCl (pH 7.6)-20 mM MgCl2 by centrifugation for 2 h in an SW 50.1 rotor (Beckman) at 28,000 rpm and 4°C. The resulting pellet was dissolved in 1 ml of ice-cold lysis buffer containing 20 mM NEM and further processed as a true cell lysate.
Immunoprecipitation and gel electrophoresis.
Proteins were immunoprecipitated from cell lysates and dissolved virions and analyzed in SDS-15% polyacrylamide (PAA) gels as described previously (33). The samples were heated for 5 min at 95°C before being applied to the gels, except for immunoprecipitates prepared with the M-specific antiserum (αM), which were analyzed without heating to avoid aggregation of the EAV M and GP5 proteins. After electrophoresis, the gels were processed for fluorography as previously reported (34) and exposed at −80°C to Kodak X-ray films or, for quantitative analyses, to storage phosphorimaging plates (Molecular Dynamics).
Sucrose density gradients.
[35S]cysteine-labeled EAV particles were concentrated through a 20% (wt/wt) sucrose cushion, as described above. The resulting pellet was gently resuspended in TM buffer (20 mM Tris-HCl [pH 7.0], 20 mM MgCl2). Next, a 400-μl aliquot of the virus suspension was carefully mixed with 1.2 ml of 67% (wt/wt) sucrose in TM buffer. A 10 to 50% (wt/wt) sucrose gradient was then prepared by overlaying the virus-sucrose mixture with 5.4 ml of 40% (wt/wt), 2 ml of 35% (wt/wt), 1 ml of 30% (wt/wt), and finally 1.5 ml of 10% (wt/wt) sucrose in TM buffer. The gradient was subjected to centrifugation for 36 h at 36,000 rpm and 4°C with an SW41 rotor (Beckman) and was collected in 12 serial fractions of approximately 1 ml from the top of the centrifugation tube. The different fractions were transferred to SW50.1 tubes and diluted with TM buffer to a final volume of 4 ml. Virus particles present in these fractions were subsequently pelleted by centrifugation for 1 h at 24,000 rpm and 4°C with an SW50.1 rotor (Beckman). The resulting pellets were dissolved in 1 ml of ice-cold lysis buffer containing 20 mM NEM and further processed as described above. The EAV particles ended up in fractions 4 through 6, as determined by 50% tissue culture infective dose (TCID50) assay and by immunoprecipitation analysis using a GP5-specific rabbit antiserum (10).
RESULTS
Disulfide-bonded structure of the GP2b monomers.
It was previously shown by SDS-PAA gel electrophoresis (SDS-PAGE) under nonreducing conditions that the GP2b protein adopts four different monomeric conformations when expressed by a recombinant vaccinia virus vector or in EAV-infected cells (11). The three fastest migrating of these monomeric forms most likely contain different intrachain disulfide bonds, since under reducing conditions the GP2b protein migrates as a single band and has the same electrophoretic mobility as the slowest migrating GP2b species resolved under nonreducing conditions (11).
To investigate the folding of the GP2b protein in more detail, we infected BHK-21 cells with vTF7.3 and transfected them with plasmids directing the synthesis of the wild-type GP2b protein or of the GP2b cysteine mutants GP2b C48S, GP2b C102S, GP2b C137S, and GP2b C195S. In each of these GP2b mutants, one of the cysteine residues downstream of the signal sequence was replaced by a serine residue. At 4 h p.i., the proteins in the cells were metabolically labeled with [35S]methionine for 15 min. Next, the cells were dissolved in lysis buffer containing a 50 mM concentration of the alkylating agent IAM, immunoprecipitations with a GP2b-specific antiserum (αGP2b) were performed, and the immunoprecipitates were analyzed in an SDS-15% PAA gel under nonreducing conditions. Under these conditions, both the wild-type GP2b protein and the GP2b C195S mutant showed the familiar pattern of four distinct monomeric conformations (Fig. 2). The other three GP2b mutants each displayed a unique pattern of two bands. For the GP2b C102S mutant, the fastest migrating of its two folding variants had a very similar electrophoretic mobility as the fastest migrating form of the wild-type GP2b protein. The most compact of the GP2b C48S and GP2b C137S species comigrated with the second fastest and second slowest migrating forms of the wild-type GP2b protein, respectively. The slowest migrating form of all mutant GP2b proteins comigrated with that of wild-type GP2b and hence must lack intramolecular disulfide bridges. The subtle differences in electrophoretic mobility between corresponding forms of the wild-type and mutant GP2b proteins are most likely direct consequences of the cysteine-to-serine mutations. These results confirm that the multiple band appearance of the GP2b protein under nonreducing conditions is indeed due to the formation of different intrachain disulfide bonds between the luminal cysteine residues and allow us to assign a specific disulfide bridge to three of the four monomeric forms, as depicted in Fig. 7A.
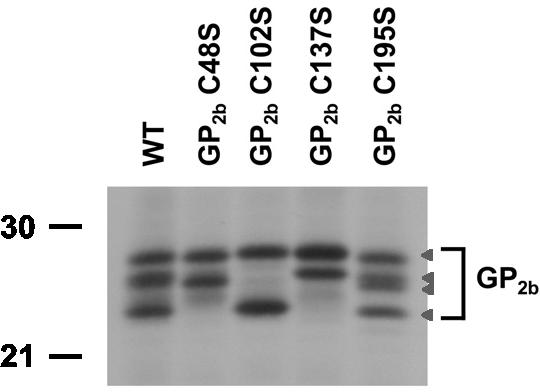
FIG. 2.
Conformational variants of the GP2b monomers. vTF7.3-infected BHK-21 C13 cells were transfected with plasmids encoding the wild-type GP2b protein or the GP2b cysteine mutants. At 4 h p.i., the cells were labeled for 15 min with [35S]methionine. Next, cell lysates were prepared in the presence of IAM and immunoprecipitations were performed with αGP2b. The immunoprecipitates were analyzed under nonreducing conditions in an SDS-15% PAA gel. The arrowheads indicate the positions of the different GP2b conformations. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel.
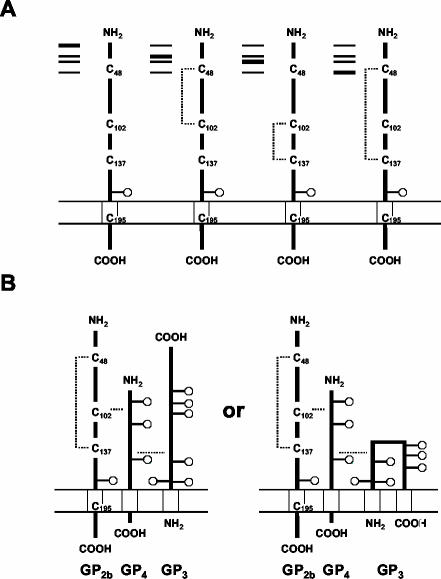
FIG. 7.
Disulfide-bonded structure of the GP2b monomers and the disulfide-bonded GP2b/GP3/GP4 heterotrimers. (A) Disulfide-bonded structures of each of the GP2b monomeric forms resolved by SDS-PAGE under nonreducing conditions are shown. (B) Two alternative models for the disulfide-bonded structure of the covalently linked GP2b/GP3/GP4 heterotrimers are drawn. The only difference between these models is the membrane topology of the GP3 protein, which is currently unknown. Since the disulfide-bonded structures of the GP3 and GP4 proteins have not yet been determined, the positions of the interchain cystine bridges relative to the ectodomains of these proteins are arbitrary. Disulfide bonds are indicated by dotted lines. The boxes symbolize the predicted transmembrane domains and the circles represent N-linked oligosaccharide side chains.
Fate of the GP2b cysteine mutants in cells transfected with synthetic full-length EAV RNAs.
In EAV-infected cells, the GP2b protein occurs in a multimeric complex with the GP3, GP4, and possibly E proteins (33). Due to the low stability of this complex, only disulfide-linked GP2b/GP4 heterodimers are detected after SDS-PAGE under nonreducing conditions (33). To study the role of the luminal cysteine residues of the GP2b protein in complex formation with GP3 and GP4 as well as their significance for virus infectivity, the GP2b mutations C48S, C102S, and C137S were introduced into a full-length EAV cDNA clone. The GP2b C195S mutant was not included in this study, as Cys-195 resides in the transmembrane domain of GP2b and is therefore most likely not involved in intermolecular disulfide bond formation. Moreover, its cloning was hampered by the overlap of the 3′ end of ORF2b with the 5′ end of ORF3 in the viral genome.
First, we investigated the intracellular fate of the GP2b cysteine mutants. BHK-21 cells were transfected with in vitro transcripts of the full-length EAV cDNA clones pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, pEAV GP2b C137S, and pEAV KO2b. The latter construct, in which the GP2b gene is inactivated by a point mutation in its initiation codon (24), served as a negative control. At 7 h p.t., the cells were labeled for 12 h with [35S]methionine, after which the cells were in a progressive state of cytopathic effect. Following lysis of the cells, the GP2b protein was immunoprecipitated with αGP2b and analyzed by SDS-PAGE under reducing and nonreducing conditions (Fig. 3).
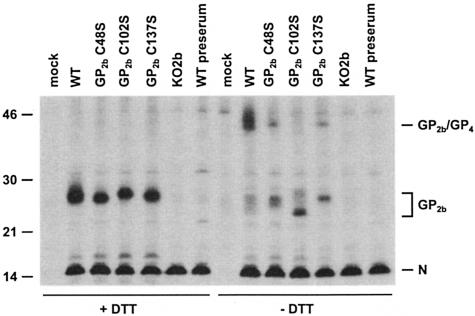
FIG. 3.
Fate of the GP2b cysteine mutants in cells transfected with synthetic full-length EAV RNAs. BHK-21 C13 cells were mock transfected or transfected with in vitro transcripts of the full-length EAV cDNA clone pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, pEAV GP2b C137S, or pEAV KO2b. At 7 h p.t., the cells were labeled with [35S]methionine for 12 h. Next, cell lysates were prepared in the presence of NEM and immunoprecipitations were performed with αGP2b or the corresponding preimmune serum. The immunoprecipitates were analyzed in SDS-15% PAA gels under reducing (+DTT) and nonreducing (−DTT) conditions. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. The positions of the N protein, the GP2b monomers, and the disulfide-linked GP2b/GP4 heterodimers are indicated on the right.
Not only the GP2b but also the N protein was precipitated from each of the cell lysates (Fig. 3; +DTT). The N protein is not coprecipitated through an interaction with the GP2b protein but binds directly to the Staphylococcus aureus cells used to collect the specific immune complexes (27) (lanes KO2b and WT preserum). Quantitative analysis of the part of the gel corresponding to the left half of Fig. 3 yielded comparable GP2b/N ratios for pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, and pEAV GP2b C137S, indicating that the cysteine-to-serine mutations did not markedly alter the stability of the GP2b protein. When analyzed under nonreducing conditions (Fig. 3; −DTT), the four monomeric forms of the wild-type GP2b protein and the two typical monomeric conformations of the GP2b mutants C48S, C102S, and C137S were again observed, though they were less distinct than in the previous experiment. This is largely due to the use of NEM instead of IAM as the sulfhydryl-modifying agent (7). In addition, the 45-kDa GP2b-containing complex was clearly observed in the sample derived from the pEAV WT RNA-transfected cells. Recently, we established that this 45-kDa complex is a covalently linked GP2b/GP4 heterodimer (33) rather than a disulfide-bonded GP2b/GP2b homodimer (11). Due to protein maturation, these GP2b/GP4 heterodimers appear as a doublet (11). Also, from lysates of cells transfected with pEAV GP2b C48S or pEAV GP2b C137S RNA, protein species were precipitated with an electrophoretic mobility similar to that of the GP2b/GP4 heterodimers, but the fraction of GP2b present in these complexes was relatively small. As discussed below, the single-band appearance of these complexes probably results from a block in the formation of intrachain disulfide bonds in GP2b C48S and GP2b C137S and a concomitant change in the processing of one or more of the N-linked oligosaccharide side chains attached to the GP2b/GP4 heterodimers. No covalently linked GP2b/GP4 proteins were observed in cells transfected with pEAV GP2b C102S RNA. These observations suggest that Cys-102 of GP2b is involved in the formation of the cystine bridge with the GP4 protein.
Particle formation by EAV GP2b cysteine mutants.
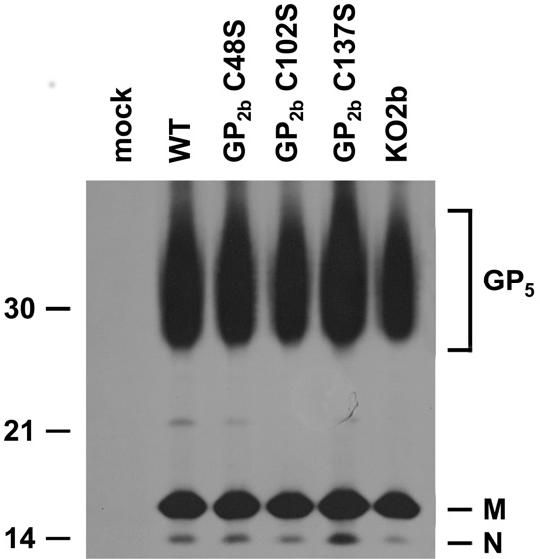
Recently, we demonstrated that GP5, M, and N are the only EAV proteins required for the production of viral particles. In the absence of any one of these proteins, no viral particles are assembled and no structural proteins are released into the culture supernatants of infected cells (Wieringa et al., unpublished data). To investigate the effect of the cysteine-to-serine mutations in GP2b on virion formation, we transfected BHK-21 cells with synthetic RNAs of the different full-length EAV cDNA clones as described above and labeled them with [35S]methionine for 12 h. The particulate material in the culture supernatants was then concentrated by sedimentation through a 20% (wt/wt) sucrose cushion, dissolved in lysis buffer, and incubated with αM antibodies. In the absence of a reducing agent, this antiserum precipitates M monomers, M homodimers, and GP5/M heterodimers (10). The resulting immune complexes were analyzed by SDS-PAGE under reducing conditions. As shown in Fig. 4, the GP5, M, and N proteins were present in all samples, with the exception of the control sample derived from mock-transfected BHK-21 cells. Apparently, each of the synthetic full-length EAV RNAs directed the formation of viral particles, indicating that the mutations in the GP2b protein did not interfere with particle assembly.
FIG. 4.
Particle formation by the EAV GP2b cysteine mutants. BHK-21 C13 cells were mock transfected or transfected with in vitro transcripts of the full-length EAV cDNA clone pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, pEAV GP2b C137S, or pEAV KO2b. At 7 h p.t., the cells were labeled with [35S]methionine for 12 h. After removal of cell debris by low-speed centrifugation, the viral particles present in the cell culture medium were pelleted through a cushion of 20% (wt/wt) sucrose. The pellets were then dissolved in lysis buffer and subjected to immunoprecipitation with αM antibody. The immunoprecipitates were analyzed under reducing conditions in an SDS-15% PAA gel. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. The positions of the EAV GP5, M, and N proteins are indicated on the right.
Composition of EAV GP2b cysteine mutant particles.
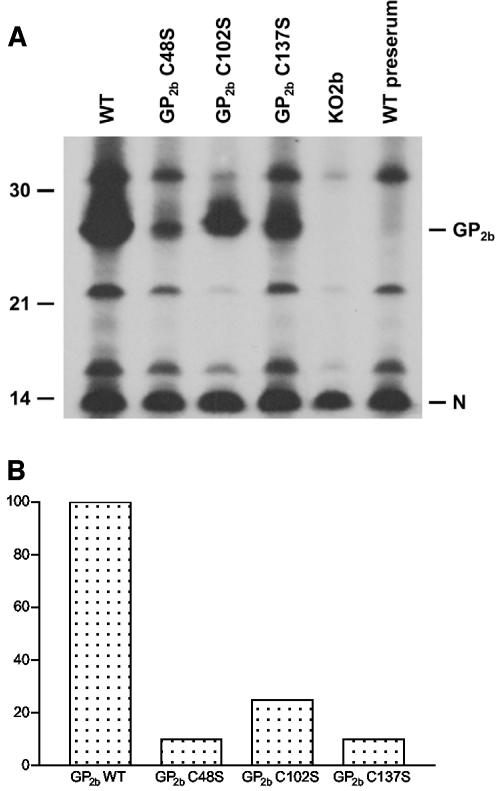
Next, we determined whether the viral particles purified from the culture supernatants of cells transfected with pEAV GP2b C48S, pEAV GP2b C102S, or pEAV GP2b C137S RNA contained (mutant) GP2b proteins. To this end, aliquots of the extracellular protein preparations from the previous experiment were subjected to immunoprecipitation with αGP2b in the presence of 5 mM DTT. Analysis by SDS-PAGE under reducing conditions revealed the presence of clearly detectable, albeit different, amounts of the GP2b protein in the viral particles released from cells that had received pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, or pEAV GP2b C137S RNA. As expected, no GP2b molecules were detected in the sample derived from pEAV KO2b RNA-transfected cells (Fig. 5A). To more closely study the efficiency with which different GP2b proteins are incorporated into EAV particles, we determined the GP2b/N ratio of each sample by phosphorimager analysis of the gel depicted in Fig. 5A. As shown in Fig. 5B, the viral particles obtained with pEAV GP2b C48S and pEAV GP2b C137S contained about 10 times less GP2b protein than the virions released from pEAV WT RNA-transfected cells. The GP2b content of the viral particles secreted by pEAV GP2b C102S RNA-transfected cells was approximately fourfold lower than that of wild-type EAV particles. Reproduction of this experiment yielded essentially the same results.
FIG. 5.
Incorporation of the EAV GP2b cysteine mutants into viral particles. (A) [35S]methionine-labeled viral particles were produced, concentrated, and dissolved in lysis buffer as described in the legend for Fig. 4. Subsequently, immunoprecipitations were performed with αGP2b or the corresponding preimmune serum in the presence of 5 mM DTT. The immunoprecipitates were analyzed under reducing conditions in an SDS-15% PAA gel. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. The positions of the EAV GP2b and N proteins are indicated on the right. (B) The amounts of radiolabel incorporated into the EAV GP2b and N proteins were determined by phosphorimager analysis. Subsequently, the GP2b/N ratio was calculated and plotted, setting the ratio obtained for the wild-type virus at 100.
Complexes of the minor envelope glycoproteins in EAV GP2b cysteine mutant particles.
In EAV particles, the GP2b protein occurs in a complex with GP3 and GP4. To study the role of the luminal cysteine residues of the GP2b protein in the formation of this complex, we again prepared radiolabeled viral particles, using the wild-type and mutant full-length EAV cDNA clones. To improve the detection of the GP3 and GP4 proteins, we performed the labeling procedure with [35S]cysteine instead of [35S]methionine. The solubilized virus particles were incubated with αGP2b and the resulting immunoprecipitates were analyzed by SDS-PAGE under reducing and nonreducing conditions.
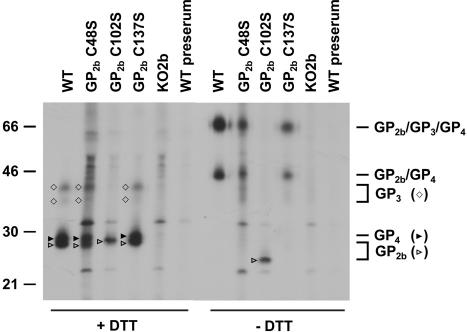
As shown in Fig. 6 (−DTT), the protein profiles obtained after analysis under nonreducing conditions were similar for EAV WT, EAV GP2b C48S, and EAV GP2b C137S. In all cases, covalently linked GP2b/GP4 dimers and GP2b/GP3/GP4 trimers were observed, though the complexes containing the mutant GP2b proteins migrated slightly slower than those comprising wild-type GP2b. In contrast, neither of these complexes was observed in viral particles produced by pEAV GP2b C102S RNA-transfected cells. Instead, a single protein species of 23 kDa was detected, with apparently the same electrophoretic mobility as the GP2b monomeric species inferred to contain an intramolecular disulfide bond between Cys-48 and Cys-137 (cf. Fig. 2, 3, and 8). The latter observation indicates that the GP2b species lacking Cys-102 is incorporated into viral particles but does not form a cystine bridge with GP4 and/or GP3.
FIG. 6.
Complexes of the minor envelope glycoproteins in GP2b cysteine mutant particles. Radiolabeled viral particles were produced, concentrated, and dissolved in lysis buffer as described in the legend for Fig. 4, except that the proteins were labeled with [35S]cysteine rather than [35S]methionine. Subsequently, immunoprecipitations were performed with αGP2b or the corresponding preimmune serum. The immunoprecipitates were analyzed in SDS-15% PAA gels under reducing (+DTT) and nonreducing (−DTT) conditions. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. The positions of the GP2b, GP3, and GP4 monomers and the disulfide-linked GP2b/GP4 heterodimers and GP2b/GP3/GP4 heterodimers are indicated on the right.
FIG. 8.
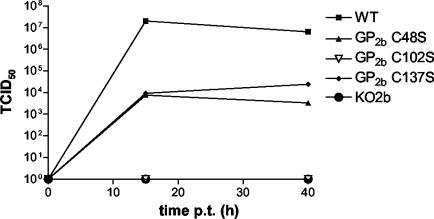
Infectivity of the EAV GP2b cysteine mutant particles. BHK-21 C13 cells were transfected with synthetic RNAs transcribed from pEAV WT, pEAV KO2b, pEAV GP2b C48S, pEAV GP2b C102S, or pEAV GP2b C137S. Next, the infectious titers in the culture supernatants of these cells at 15 and 40 h p.t. were determined by end-point dilution. Times p.t. (in hours) are indicated on the abscissa, while TCID50 values (in infectious units per milliliter) are plotted on the ordinate.
Also, under reducing conditions similar protein patterns were seen for EAV WT, EAV GP2b C48S, and EAV GP2b C137S (Fig. 6, +DTT). The two characteristic species of 38 and 42 kDa correspond to the GP3 protein, while the 28-kDa product represents the GP4 protein (34). Under reducing conditions, relatively little GP3 seems to be recovered compared to GP2b or GP4. It should be noted, however, that the GP3 protein originates from the disulfide-bonded GP2b/GP3/GP4 heterotrimers only, while the GP2b and GP4 proteins are derived from both the GP2b/GP4 and GP2b/GP3/GP4 complexes. Moreover, when analyzed under reducing conditions, the GP3 protein migrates as two species due to variable glycosylation (34). Since GP3 and GP4 molecules were not coprecipitated with the GP2b protein using the material derived from pEAV GP2b C102S-transfected cells, GP2b Cys-102 must be involved in the formation of the disulfide bond with the GP4 protein. The other two luminal cysteine residues of GP2b seem not to be involved in intermolecular disulfide bond formation between the minor envelope glycoproteins, consistent with our earlier interpretation that these residues form an intramolecular cystine bridge. This is further supported by the slightly reduced electrophoretic mobilities of the GP2b/GP4 and GP2b/GP3/GP4 complexes in EAV GP2b C48S and EAV GP2b C137S particles compared to those in the wild-type virus. In addition, the single-band appearance of the covalently linked GP2b/GP4 dimers in cells transfected with pEAV GP2b C48S or pEAV GP2b C137S RNA (Fig. 3) could be explained by the absence of an intrachain disulfide in GP2b C48S and C137S.
Both GP2b C48S and GP2b C137S are able to form cystine-linked GP2b/GP3/GP4 heterotrimers. This implies that the covalent interaction of the disulfide-bonded GP2b/GP4 dimers with the GP3 protein in the GP2b/GP3/GP4 complex has to occur through the GP4 protein. A schematic model for the disulfide-bonded structure in EAV particles of the covalently linked GP2b/GP3/GP4 trimers is presented in Fig. 7. This model is based on the assumption that the covalent association of GP3 with the disulfide-bonded GP2b/GP4 dimers does not affect the intramolecular cystine bridge in GP2b.
Infectivity of the EAV GP2b cysteine mutant particles.
To investigate whether the viral particles generated with the mutant full-length EAV cDNA clones were infectious, BHK-21 cells were transfected with synthetic RNAs transcribed from pEAV WT, pEAV KO2b, pEAV GP2b C48S, pEAV GP2b C102S, or pEAV GP2b C137S. An aliquot of the cells was seeded on glass coverslips and processed for immunofluorescence at 15 h p.t. Labeling with αM antibodies indicated that a transfection efficiency of approximately 90% had been achieved in each case (data not shown). The rest of the cells were transferred to dishes. The infectious titers in the culture supernatants of these dishes at 15 and 40 h p.t. were determined by end-point dilution (Fig. 8). At 15 h p.t., the titer in the culture supernatant of pEAV WT RNA-transfected cells was approximately 2 × 107 TCID50/ml. This titer decreased to 6.3 × 106 TCID50/ml at 40 h p.t. EAV GP2b C48S and EAV GP2b C137S showed comparable growth kinetics but reached much lower maximum titers (between 3 × 103 and 2 ×104 TCID50/ml) than the wild-type virus. No infectivity was found in the culture supernatants of the cells transfected with pEAV GP2b C102S or pEAV KO2b RNA.
DISCUSSION
The EAV GP2b protein is a typical class I membrane glycoprotein, containing three cysteine residues in its ectodomain and one in its transmembrane anchor (7, 11). In EAV-infected cells, the bulk of the GP2b molecules are present as four different monomeric species while a small fraction occurs in a heteromeric complex with GP3 and GP4 (33). Only this heteromeric complex appears in virus particles, albeit in relatively small amounts, whereas the GP2b monomers stay behind in the endoplasmic reticulum of the infected cells. It was already clear from previous experiments performed in our laboratory (11) that disulfide bonds are involved in the formation of the different GP2b monomers and the GP2b-containing heterooligomers. In this study, the importance of the intra- and intermolecular disulfide bonds of the GP2b protein for the assembly of complete and infectious virus particles was demonstrated by use of cysteine-to-serine mutants of the GP2b protein.
Expression of the ORFs coding for these GP2b cysteine mutants from plasmids or full-length EAV cDNA clones confirmed that three of the four monomeric GP2b species contained alternative intrachain disulfide bonds between the luminal cysteine residues. As might perhaps be expected from a linkage bringing together distant parts of a polypeptide, the most compacted and thus fastest migrating GP2b monomers possess a disulfide bond between Cys-48 and Cys-137. This simplification does not hold for the other monomeric forms of GP2b, as the species with the linkage between Cys-48 and Cys-102 (54 residues apart) runs slower than the protein in which Cys-102 and Cys-137 (35 residues apart) are connected. The fully reduced molecule has the lowest electrophoretic mobility.
The data obtained with the full-length EAV cDNA clones encoding GP2b cysteine mutants resulted in a model for the intra- and intermolecular disulfide bonds of the GP2b protein (Fig. 7B). In this model, GP2b Cys-102 is responsible for the cystine bridge with the GP4 protein while an intrachain disulfide bond links Cys-48 and Cys-137 of GP2b. The structure proposed in Fig. 7B is primarily based on the observation that both EAV GP2b C48S and EAV GP2b C137S particles contained disulfide-bonded GP2b/GP4 dimers as well as disulfide-bonded GP2b/GP3/GP4 trimers but that neither of these complexes was present in GP2b C102S mutant particles. Furthermore, the electrophoretic mobility under nonreducing conditions of the GP2b C102S molecules that were incorporated into viral particles corresponded with that of the GP2b monomers containing an intramolecular cystine bridge between Cys-48 and Cys-137. Finally, the single-band appearance of the covalently linked GP2b/GP4 dimers extracted from cells transfected with pEAV GP2b C48S or pEAV GP2b C137S RNA and the slightly lower electrophoretic mobility of the heteromeric complexes in virus particles comprising GP2b C48S and GP2b C137S than of those in virus particles containing the wild-type GP2b protein are, presumably, both the result of the absence of intrachain disulfide bonds in these GP2b mutants.
In our model, all luminal cysteine residues of GP2b are used for the formation of intra- and interchain disulfide bonds. This implies that the covalent association of GP3 with the cystine-linked GP2b/GP4 heterodimer occurs via the GP4 protein. In this respect, it is interesting that we recently obtained preliminary evidence for the presence in EAV GP2b C102S particles of a protein species with an apparent molecular mass compatible with that of a covalently bound GP3/GP4 heterodimer (data not shown). It is at present difficult to assign the cysteine residues that are involved in the intermolecular disulfide bond formation between GP4 and GP3. All three luminal cysteines of GP4 and most of the nine cysteines of GP3 are conserved among the sequenced EAV isolates (1). Likewise, we do not know which cysteine residue in GP4 interacts with Cys-102 of GP2b. The GP2b protein of EAV has many features in common with its homologs in other arteriviruses. Each of these proteins seems to be a class I membrane protein (13), and the relative positions of their luminal cysteine residues, N-glycosylation sites, and transmembrane anchors are highly conserved (14). However, the GP2 and VP-3 M proteins of PRRSV and LDV, respectively, do not contain an EAV GP2b Cys-102 counterpart. Little is known about complex formation of the EAV GP2b, GP3, and GP4 homologs in other arteriviruses. It has been reported that the GP4 protein of the IAF-Klop strain of PRRSV is coprecipitated with the GP3 protein from lysates of virus-infected cells (20) and that the GP2 protein of the Lelystad strain of PRRSV occurs in virions only in its fully reduced conformation (21). In contrast, approximately half of the GP2 molecules present in lysates of PRRSV-infected CL2621 cells contain an intramolecular disulfide bond. The intrachain disulfide bond in the GP2 protein of PRRSV particles may hence have been lost during the analytical procedures. Nevertheless, these observations do not exclude the existence of noncovalently linked complexes of the minor envelope glycoprotein in these viruses. Especially given the crucial role of EAV GP2b Cys-102 in the formation of the disulfide-bonded GP2b/GP4 dimers and GP2b/GP3/GP4 trimers and the absence of this residue from the GP2 proteins of LDV and PRRSV, covalently linked heterotrimeric complexes of the minor envelope glycoproteins are not expected for the latter viruses.
The different cysteine-to-serine mutations introduced into the GP2b protein did not hamper the assembly of viral particles, i.e., similar amounts of GP5, M, and N were detected in the culture supernatants of cells transfected with synthetic RNAs transcribed from pEAV WT, pEAV GP2b C48S, pEAV GP2b C102S, or pEAV GP2b C137S. Nonetheless, the amino acid substitutions resulted in a reduced incorporation of the GP2b protein into viral particles, i.e., EAV GP2b C48S and EAV GP2b C137S particles each contained approximately 10 times less GP2b protein than the wild-type virus. Thus, the cysteine-to-serine mutations at positions 48 and 137 of the GP2b protein not only have a similar effect on the generation of the intrachain disulfide bond and the covalently linked GP2b/GP4 heterodimer but also affect the incorporation of these complexes into virions. It is quite plausible that the formation of disulfide-bonded GP2b/GP4 heterodimers is a limiting step in the assembly of complete virus particles. This seems, however, in conflict with the observation that GP2b C102S does not covalently interact with the GP4 protein but is nevertheless present in larger amounts in virions than the other two GP2b mutants. Obviously, the C102S mutation has a less dramatic effect on the conformation of GP2b, and the GP2b C102S molecules incorporated into virus particles are likely to be associated with the GP4 protein in a noncovalent way. The incorporation of GP3 and GP4 in EAV GP2b C102S particles would be consistent with a previous observation that the inclusion of the GP2b protein into virus particles requires the GP3, GP4, and E proteins (Wieringa et al., unpublished data). Unfortunately, because of the high background signals obtained with the GP3- and GP4-specific antisera, we were unable to clearly detect the probably rather small amounts of GP3 and GP4 monomers in EAV GP2b C102S particles.
In view of these considerations, it was remarkable that EAV GP2b C102S particles were noninfectious. Apparently the disulfide bond between GP2b and GP4 is an absolute requirement for the functioning of the GP2b/GP3/GP4/(E) complex. As we argued before, this complex is most likely involved in viral targeting and cell entry. Conversely, EAV GP2b C48S and EAV GP2b C137S particles exhibited a low but significant level of infectivity even though they contain less of the GP2b protein than the EAV GP2b C102S particles. This infectivity, which was remarkably similar for both mutants, might be due to reversion of the mutation. However, we find this possibility very unlikely, since all GP2b cysteine mutants were generated by U-to-A transversions and no infectivity was found in the culture supernatant of cells transfected with pEAV GP2b C102S or pEAV KO2b RNA. The 3-log difference in infectivity between EAV GP2b C48S or EAV GP2b C137S particles and the wild-type virus was not proportional to their approximately 10-fold difference in GP2b content. There are many possible reasons for this apparent discrepancy; one interpretation is that the intramolecular disulfide bond in GP2b is also important for the functioning of the GP2b/GP3/GP4/(E) complex. However, as the GP2b protein is not essential for viral RNA replication and transcription (22, 24), it is unlikely that the point mutations introduced by us directly affected EAV replication instead of being related to the biosynthesis, folding, and incorporation into virus particles of the minor envelope glycoproteins of EAV. The unraveling of this function is one of the major challenges of future arterivirus research.
Acknowledgments
We are grateful to Catherine Saunier for technical assistance. In addition, we thank Bernard Moss for providing the recombinant vaccinia virus vTF7.3 and Eric Snijder for making available the full-length EAV cDNA clones pEAV WT and pEAV KO2b.
REFERENCES
- 1.Archambault, D., G. Laganière, and G. St.-Laurent. 1998. Genetic variation and phylogenetic analysis of open reading frames 3 and 4 of various equine arteritis virus isolates. Adv. Exp. Med. Biol. 440:813-819. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 3.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 4.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 5.den Boon, J. A., M. F. Kleijnen, W. J. M. Spaan, and E. J. Snijder. 1996. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J. Virol. 70:4291-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. F. de Vries, M. C. Horzinek, and W. J. M. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries, A. A. F. 1994. The molecular biology of equine arteritis virus. Ph.D. thesis. Utrecht University, Utrecht, The Netherlands.
- 8.de Vries, A. A. F., E. D. Chirnside, P. J. Bredenbeek, L. A. Gravestein, M. C. Horzinek, and W. J. M. Spaan. 1990. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 18:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries, A. A. F., E. D. Chirnside, M. C. Horzinek, and P. J. M. Rottier. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries, A. A. F., S. M. Post, M. J. B. Raamsman, M. C. Horzinek, and P. J. M. Rottier. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 69:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, A. A. F., M. J. B. Raamsman, H. A. van Dijk, M. C. Horzinek, and P. J. M. Rottier. 1995. The small envelope glycoprotein (GS) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J. Virol. 69:3441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbe, J. C., Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288:283-294. [DOI] [PubMed] [Google Scholar]
- 13.Faaberg, K. S., and P. G. W. Plagemann. 1995. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology 212:512-525. [DOI] [PubMed] [Google Scholar]
- 14.Godeny, E. K., A. A. F. de Vries, X. C. Wang, S. L. Smith, and R. J. de Groot. 1998. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 72:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedges, J. F., U. B. R. Balasuriya, and N. J. MacLachlan. 1999. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology 264:92-98. [DOI] [PubMed] [Google Scholar]
- 16.Horzinek, M. C., J. Maess, and R. Laufs. 1971. Studies on the substructure of togaviruses. II. Analysis of equine arteritis, rubella, bovine viral diarrhea, and hog cholera viruses. Arch. Gesamte Virusforsch. 33:306-318. [PubMed] [Google Scholar]
- 17.Hyllseth, B. 1973. Structural proteins of equine arteritis virus. Arch. Gesamte Virusforsch. 40:177-188. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 19.Magnusson, P., B. Hyllseth, and H. Marusyk. 1970. Morphological studies on equine arteritis virus. Arch. Gesamte Virusforsch. 30:105-112. [DOI] [PubMed] [Google Scholar]
- 20.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meulenberg, J. J. M., and A. Petersen-den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated into virus particles. Virology 225:44-51. [DOI] [PubMed] [Google Scholar]
- 22.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. B. Raamsman, and A. A. F. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snijder, E. J., A. L. M. Wassenaar, and W. J. M. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Berlo, M. F., P. J. M. Rottier, W. J. M. Spaan, and M. C. Horzinek. 1986. Equine arteritis virus-induced polypeptide synthesis. J. Gen. Virol. 67:1543-1549. [DOI] [PubMed] [Google Scholar]
- 27.van Berlo, M. F., J. J. W. Zeegers, M. C. Horzinek, and B. A. M. van der Zeijst. 1983. Antigenic comparison of equine arteritis virus (EAV) and lactic dehydrogenase virus (LDV); binding of staphylococcal protein A to the nucleocapsid protein of EAV. Zentbl. Veterinarmed. B 30:297-304. [DOI] [PubMed] [Google Scholar]
- 28.van Dinten, L. C., J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dinten, L. C., S. Rensen, W. J. M. Spaan, A. E. Gorbalenya, and E. J. Snijder. 1999. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dinten, L. C., A. L. M. Wassenaar, A. E. Gorbalenya, W. J. M. Spaan, and E. J. Snijder. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 70:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verheije, M. H., T. J. M. Welting, H. T. Jansen, P. J. M. Rottier, and J. J. M. Meulenberg. 2002. Chimeric arteriviruses generated by swapping of the M protein ectodomain rule out a role of this domain in viral targeting. Virology 303:364-373. [DOI] [PubMed] [Google Scholar]
- 32.Wassenaar, A. L. M., W. J. M. Spaan, A. E. Gorbalenya, and E. J. Snijder. 1997. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 71:9313-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieringa, R., A. A. F. de Vries, and P. J. M. Rottier. 2003. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 77:6216-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieringa, R., A. A. F. de Vries, M. J. B. Raamsman, and P. J. M. Rottier. 2002. Characterization of two new structural glycoproteins, GP3 and GP4, of equine arteritis virus. J. Virol. 76:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeegers, J. J. W., B. A. M. van der Zeijst, and M. C. Horzinek. 1976. The structural proteins of equine arteritis virus. Virology 73:200-205. [DOI] [PubMed] [Google Scholar]