Abstract
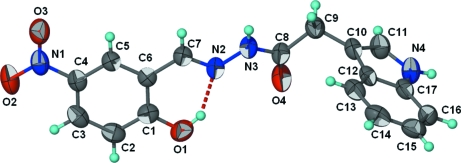
The molecule of the title compound, C17H14N4O4, uses its amide –NH– group to form a hydrogen bond to the amido –C(=O)– group of an adjacent molecule to furnish a linear chain structure. The hydroxy group forms an intramolecular hydrogen bond; the indolyl –NH– unit does not engage in any strong hydrogen-bonding interactions.
Related literature
For similar compounds, see: Martin Reyes et al. (1986 ▶); Martin Zarza et al. (1989 ▶).
Experimental
Crystal data
C17H14N4O4
M r = 338.32
Orthorhombic,

a = 9.5387 (2) Å
b = 11.2724 (3) Å
c = 29.7796 (7) Å
V = 3202.0 (1) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 100 (2) K
0.30 × 0.25 × 0.20 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
47721 measured reflections
3679 independent reflections
2059 reflections with I > 2σ(I)
R int = 0.052
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.160
S = 1.02
3679 reflections
228 parameters
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.21 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026044/bq2091sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026044/bq2091Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1o⋯N2 | 0.84 | 1.85 | 2.583 (2) | 146 |
| N3—H3n⋯O4i | 0.88 | 2.07 | 2.827 (2) | 144 |
Symmetry code: (i)  .
.
Acknowledgments
We thank the Science Fund (12–02-03–2031, 12–02-03–2051) and the University of Malaya (PJP) for supporting this study. We are grateful to the University of Malaya for the purchase of the diffractometer.
supplementary crystallographic information
Comment
There are many examples of Schiff bases derived from the condensation of salicylaldehyde and substituted salicyldehydes with hydrazides such as the ones reported by Martin Reyes et al. (1986) and Martin Zarza et al. (1989). The title compound (Fig. 1) is another example. The molecule uses its amido –NH– group to form a hydrogen bond to the amido –C(=O)– group of an adjacent molecule to furnish a linear chain structure.
Experimental
The Schiff base was prepared by refluxing a solution of indole-3-acetic acid hydrazide (0.34 g, 1.80 mmol) and 5-nitrosalicylaldehyde (0.30 g, 1.80 mmol) in acidified ethanol (25 ml) for 2 h. On cooling to room temperature, yellow crystals separated out.
Refinement
All H-atoms were placed in calculated positions (C—H 0.95, N—H 0.88, O–H 0.84 Å) and were included in the refinement in the riding model approximation, with U(H) set to 1.2 to 1.5Ueq(C,N,O).
Figures
Fig. 1.
Thermal ellipsoid plot of (I) (Barbour, 2001) at the 50% probability level. Dashed line indicates H-bonding.
Crystal data
| C17H14N4O4 | F000 = 1408 |
| Mr = 338.32 | Dx = 1.404 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3679 reflections |
| a = 9.5387 (2) Å | θ = 2.5–22.2º |
| b = 11.2724 (3) Å | µ = 0.10 mm−1 |
| c = 29.7796 (7) Å | T = 100 (2) K |
| V = 3202.0 (1) Å3 | Irregular block, yellow |
| Z = 8 | 0.30 × 0.25 × 0.20 mm |
Data collection
| Bruker SMART APEX diffractometer | 2059 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.053 |
| Monochromator: graphite | θmax = 27.5º |
| T = 100(2) K | θmin = 1.4º |
| ω scans | h = −12→12 |
| Absorption correction: None | k = −14→13 |
| 47721 measured reflections | l = −38→38 |
| 3679 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.044 | w = 1/[σ2(Fo2) + (0.0885P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.160 | (Δ/σ)max = 0.001 |
| S = 1.02 | Δρmax = 0.18 e Å−3 |
| 3679 reflections | Δρmin = −0.21 e Å−3 |
| 228 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.007 (1) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.76143 (16) | 0.34962 (15) | 0.66810 (5) | 0.0747 (5) | |
| H1O | 0.7344 | 0.4058 | 0.6845 | 0.112* | |
| O2 | 0.50825 (18) | 0.27110 (16) | 0.47819 (5) | 0.0896 (6) | |
| O3 | 0.36688 (19) | 0.40496 (16) | 0.50246 (5) | 0.0853 (5) | |
| O4 | 0.78329 (15) | 0.61201 (15) | 0.75230 (5) | 0.0790 (5) | |
| N1 | 0.4670 (2) | 0.33901 (17) | 0.50757 (6) | 0.0647 (5) | |
| N2 | 0.60812 (15) | 0.52866 (14) | 0.69094 (5) | 0.0522 (4) | |
| N3 | 0.56843 (16) | 0.61209 (15) | 0.72184 (5) | 0.0543 (5) | |
| H3N | 0.4821 | 0.6397 | 0.7223 | 0.065* | |
| N4 | 0.79603 (19) | 0.75966 (17) | 0.89103 (6) | 0.0712 (5) | |
| H4N | 0.8598 | 0.7879 | 0.9095 | 0.085* | |
| C1 | 0.6858 (2) | 0.34823 (18) | 0.63004 (7) | 0.0563 (5) | |
| C2 | 0.7229 (2) | 0.26733 (18) | 0.59698 (8) | 0.0651 (6) | |
| H2 | 0.7980 | 0.2137 | 0.6022 | 0.078* | |
| C3 | 0.6523 (2) | 0.26386 (18) | 0.55689 (7) | 0.0625 (6) | |
| H3 | 0.6787 | 0.2090 | 0.5342 | 0.075* | |
| C4 | 0.5426 (2) | 0.34134 (17) | 0.55005 (6) | 0.0538 (5) | |
| C5 | 0.5011 (2) | 0.42048 (16) | 0.58253 (6) | 0.0511 (5) | |
| H5 | 0.4237 | 0.4715 | 0.5772 | 0.061* | |
| C6 | 0.57211 (19) | 0.42584 (16) | 0.62300 (6) | 0.0473 (5) | |
| C7 | 0.53086 (19) | 0.51242 (17) | 0.65658 (6) | 0.0505 (5) | |
| H7 | 0.4468 | 0.5567 | 0.6530 | 0.061* | |
| C8 | 0.6644 (2) | 0.65087 (18) | 0.75150 (6) | 0.0555 (5) | |
| C9 | 0.6133 (2) | 0.7485 (2) | 0.78219 (6) | 0.0643 (6) | |
| H9A | 0.6427 | 0.8262 | 0.7699 | 0.077* | |
| H9B | 0.5095 | 0.7472 | 0.7832 | 0.077* | |
| C10 | 0.6695 (2) | 0.73543 (17) | 0.82885 (6) | 0.0546 (5) | |
| C11 | 0.7735 (2) | 0.7979 (2) | 0.84821 (7) | 0.0684 (6) | |
| H11 | 0.8242 | 0.8600 | 0.8340 | 0.082* | |
| C12 | 0.62245 (19) | 0.65262 (16) | 0.86168 (7) | 0.0514 (5) | |
| C13 | 0.5173 (2) | 0.56695 (18) | 0.86298 (8) | 0.0626 (6) | |
| H13 | 0.4599 | 0.5530 | 0.8374 | 0.075* | |
| C14 | 0.4978 (3) | 0.50326 (19) | 0.90151 (9) | 0.0745 (7) | |
| H14 | 0.4262 | 0.4447 | 0.9024 | 0.089* | |
| C15 | 0.5803 (3) | 0.5221 (2) | 0.93952 (8) | 0.0753 (7) | |
| H15 | 0.5640 | 0.4760 | 0.9657 | 0.090* | |
| C16 | 0.6847 (2) | 0.6062 (2) | 0.93974 (7) | 0.0667 (6) | |
| H16 | 0.7411 | 0.6194 | 0.9655 | 0.080* | |
| C17 | 0.7037 (2) | 0.67047 (18) | 0.90073 (7) | 0.0561 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0634 (10) | 0.0901 (11) | 0.0707 (10) | 0.0201 (8) | −0.0089 (8) | 0.0030 (8) |
| O2 | 0.0991 (14) | 0.1023 (13) | 0.0672 (11) | −0.0122 (10) | 0.0139 (9) | −0.0333 (10) |
| O3 | 0.0835 (12) | 0.0954 (12) | 0.0771 (11) | 0.0045 (10) | −0.0245 (9) | −0.0215 (9) |
| O4 | 0.0393 (9) | 0.1232 (13) | 0.0745 (11) | 0.0156 (8) | −0.0106 (7) | −0.0360 (9) |
| N1 | 0.0663 (12) | 0.0712 (12) | 0.0566 (11) | −0.0201 (10) | 0.0062 (9) | −0.0134 (10) |
| N2 | 0.0406 (9) | 0.0683 (10) | 0.0477 (9) | −0.0033 (8) | 0.0012 (7) | −0.0058 (8) |
| N3 | 0.0360 (8) | 0.0762 (11) | 0.0507 (10) | 0.0042 (8) | 0.0002 (7) | −0.0134 (8) |
| N4 | 0.0664 (12) | 0.0860 (13) | 0.0612 (11) | −0.0182 (10) | −0.0085 (9) | −0.0171 (10) |
| C1 | 0.0460 (11) | 0.0607 (12) | 0.0623 (13) | −0.0011 (9) | 0.0046 (10) | 0.0044 (10) |
| C2 | 0.0548 (13) | 0.0590 (13) | 0.0814 (16) | 0.0065 (10) | 0.0103 (12) | 0.0004 (11) |
| C3 | 0.0592 (14) | 0.0557 (12) | 0.0724 (15) | −0.0079 (10) | 0.0201 (12) | −0.0112 (10) |
| C4 | 0.0512 (12) | 0.0533 (11) | 0.0568 (12) | −0.0139 (9) | 0.0058 (10) | −0.0048 (9) |
| C5 | 0.0460 (11) | 0.0536 (11) | 0.0536 (11) | −0.0039 (9) | 0.0030 (8) | −0.0029 (9) |
| C6 | 0.0409 (10) | 0.0498 (10) | 0.0513 (11) | −0.0052 (8) | 0.0050 (8) | 0.0011 (9) |
| C7 | 0.0416 (11) | 0.0583 (11) | 0.0515 (11) | −0.0014 (9) | 0.0018 (9) | −0.0006 (9) |
| C8 | 0.0428 (12) | 0.0750 (13) | 0.0488 (11) | 0.0012 (10) | −0.0009 (9) | −0.0068 (10) |
| C9 | 0.0573 (13) | 0.0742 (14) | 0.0613 (13) | 0.0078 (11) | −0.0020 (10) | −0.0119 (11) |
| C10 | 0.0496 (12) | 0.0617 (12) | 0.0524 (11) | −0.0006 (9) | 0.0004 (9) | −0.0154 (9) |
| C11 | 0.0671 (15) | 0.0730 (14) | 0.0650 (14) | −0.0150 (12) | 0.0039 (11) | −0.0092 (11) |
| C12 | 0.0443 (11) | 0.0532 (11) | 0.0568 (12) | 0.0047 (9) | 0.0009 (9) | −0.0189 (9) |
| C13 | 0.0534 (13) | 0.0564 (12) | 0.0781 (15) | −0.0004 (10) | −0.0012 (11) | −0.0154 (11) |
| C14 | 0.0665 (15) | 0.0541 (12) | 0.103 (2) | −0.0025 (11) | 0.0122 (14) | −0.0064 (13) |
| C15 | 0.0858 (18) | 0.0594 (13) | 0.0808 (17) | 0.0180 (13) | 0.0207 (14) | 0.0039 (12) |
| C16 | 0.0717 (15) | 0.0718 (14) | 0.0566 (13) | 0.0192 (13) | −0.0007 (11) | −0.0090 (11) |
| C17 | 0.0523 (12) | 0.0578 (12) | 0.0583 (12) | 0.0059 (10) | 0.0013 (10) | −0.0171 (10) |
Geometric parameters (Å, °)
| O1—C1 | 1.344 (2) | C5—H5 | 0.9500 |
| O1—H1O | 0.8400 | C6—C7 | 1.452 (3) |
| O2—N1 | 1.227 (2) | C7—H7 | 0.9500 |
| O3—N1 | 1.220 (2) | C8—C9 | 1.512 (3) |
| O4—C8 | 1.216 (2) | C9—C10 | 1.497 (3) |
| N1—C4 | 1.456 (3) | C9—H9A | 0.9900 |
| N2—C7 | 1.274 (2) | C9—H9B | 0.9900 |
| N2—N3 | 1.369 (2) | C10—C11 | 1.347 (3) |
| N3—C8 | 1.345 (2) | C10—C12 | 1.424 (3) |
| N3—H3N | 0.8800 | C11—H11 | 0.9500 |
| N4—C11 | 1.363 (3) | C12—C13 | 1.393 (3) |
| N4—C17 | 1.367 (3) | C12—C17 | 1.412 (3) |
| N4—H4N | 0.8800 | C13—C14 | 1.366 (3) |
| C1—C2 | 1.388 (3) | C13—H13 | 0.9500 |
| C1—C6 | 1.409 (3) | C14—C15 | 1.395 (3) |
| C2—C3 | 1.371 (3) | C14—H14 | 0.9500 |
| C2—H2 | 0.9500 | C15—C16 | 1.375 (3) |
| C3—C4 | 1.378 (3) | C15—H15 | 0.9500 |
| C3—H3 | 0.9500 | C16—C17 | 1.381 (3) |
| C4—C5 | 1.374 (3) | C16—H16 | 0.9500 |
| C5—C6 | 1.384 (3) | ||
| C1—O1—H1O | 109.5 | O4—C8—C9 | 123.48 (18) |
| O3—N1—O2 | 122.85 (19) | N3—C8—C9 | 114.46 (18) |
| O3—N1—C4 | 119.01 (18) | C10—C9—C8 | 111.95 (17) |
| O2—N1—C4 | 118.1 (2) | C10—C9—H9A | 109.2 |
| C7—N2—N3 | 118.59 (16) | C8—C9—H9A | 109.2 |
| C8—N3—N2 | 118.45 (16) | C10—C9—H9B | 109.2 |
| C8—N3—H3N | 120.8 | C8—C9—H9B | 109.2 |
| N2—N3—H3N | 120.8 | H9A—C9—H9B | 107.9 |
| C11—N4—C17 | 109.21 (17) | C11—C10—C12 | 106.33 (18) |
| C11—N4—H4N | 125.4 | C11—C10—C9 | 127.6 (2) |
| C17—N4—H4N | 125.4 | C12—C10—C9 | 126.11 (18) |
| O1—C1—C2 | 117.97 (19) | C10—C11—N4 | 110.5 (2) |
| O1—C1—C6 | 122.12 (18) | C10—C11—H11 | 124.7 |
| C2—C1—C6 | 119.91 (19) | N4—C11—H11 | 124.7 |
| C3—C2—C1 | 120.8 (2) | C13—C12—C17 | 118.12 (19) |
| C3—C2—H2 | 119.6 | C13—C12—C10 | 134.45 (19) |
| C1—C2—H2 | 119.6 | C17—C12—C10 | 107.40 (17) |
| C2—C3—C4 | 118.89 (19) | C14—C13—C12 | 119.1 (2) |
| C2—C3—H3 | 120.6 | C14—C13—H13 | 120.5 |
| C4—C3—H3 | 120.6 | C12—C13—H13 | 120.5 |
| C5—C4—C3 | 121.73 (19) | C13—C14—C15 | 121.7 (2) |
| C5—C4—N1 | 118.73 (19) | C13—C14—H14 | 119.2 |
| C3—C4—N1 | 119.54 (18) | C15—C14—H14 | 119.2 |
| C4—C5—C6 | 120.04 (18) | C16—C15—C14 | 121.2 (2) |
| C4—C5—H5 | 120.0 | C16—C15—H15 | 119.4 |
| C6—C5—H5 | 120.0 | C14—C15—H15 | 119.4 |
| C5—C6—C1 | 118.63 (17) | C15—C16—C17 | 116.9 (2) |
| C5—C6—C7 | 119.78 (17) | C15—C16—H16 | 121.5 |
| C1—C6—C7 | 121.58 (18) | C17—C16—H16 | 121.5 |
| N2—C7—C6 | 119.53 (17) | N4—C17—C16 | 130.4 (2) |
| N2—C7—H7 | 120.2 | N4—C17—C12 | 106.52 (18) |
| C6—C7—H7 | 120.2 | C16—C17—C12 | 123.1 (2) |
| O4—C8—N3 | 122.03 (18) | ||
| C7—N2—N3—C8 | −163.35 (18) | N3—C8—C9—C10 | 142.14 (19) |
| O1—C1—C2—C3 | −178.10 (19) | C8—C9—C10—C11 | 103.7 (2) |
| C6—C1—C2—C3 | 1.7 (3) | C8—C9—C10—C12 | −76.0 (3) |
| C1—C2—C3—C4 | −0.7 (3) | C12—C10—C11—N4 | 0.4 (2) |
| C2—C3—C4—C5 | −0.9 (3) | C9—C10—C11—N4 | −179.39 (19) |
| C2—C3—C4—N1 | 179.78 (17) | C17—N4—C11—C10 | −0.8 (2) |
| O3—N1—C4—C5 | −1.9 (3) | C11—C10—C12—C13 | 177.9 (2) |
| O2—N1—C4—C5 | 177.32 (17) | C9—C10—C12—C13 | −2.4 (3) |
| O3—N1—C4—C3 | 177.38 (18) | C11—C10—C12—C17 | 0.1 (2) |
| O2—N1—C4—C3 | −3.4 (3) | C9—C10—C12—C17 | 179.90 (18) |
| C3—C4—C5—C6 | 1.6 (3) | C17—C12—C13—C14 | −0.6 (3) |
| N1—C4—C5—C6 | −179.14 (16) | C10—C12—C13—C14 | −178.1 (2) |
| C4—C5—C6—C1 | −0.5 (3) | C12—C13—C14—C15 | 0.1 (3) |
| C4—C5—C6—C7 | 177.88 (16) | C13—C14—C15—C16 | 0.2 (3) |
| O1—C1—C6—C5 | 178.74 (17) | C14—C15—C16—C17 | −0.1 (3) |
| C2—C1—C6—C5 | −1.0 (3) | C11—N4—C17—C16 | −178.7 (2) |
| O1—C1—C6—C7 | 0.3 (3) | C11—N4—C17—C12 | 0.8 (2) |
| C2—C1—C6—C7 | −179.43 (17) | C15—C16—C17—N4 | 179.0 (2) |
| N3—N2—C7—C6 | 178.97 (15) | C15—C16—C17—C12 | −0.4 (3) |
| C5—C6—C7—N2 | −170.09 (17) | C13—C12—C17—N4 | −178.75 (16) |
| C1—C6—C7—N2 | 8.3 (3) | C10—C12—C17—N4 | −0.6 (2) |
| N2—N3—C8—O4 | −1.8 (3) | C13—C12—C17—C16 | 0.8 (3) |
| N2—N3—C8—C9 | 176.17 (17) | C10—C12—C17—C16 | 178.93 (18) |
| O4—C8—C9—C10 | −39.9 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1o···N2 | 0.84 | 1.85 | 2.583 (2) | 146 |
| N3—H3n···O4i | 0.88 | 2.07 | 2.827 (2) | 144 |
| N4—H4n···O2ii | 0.88 | 2.49 | 3.216 (2) | 140 |
Symmetry codes: (i) x−1/2, y, −z+3/2; (ii) −x+3/2, −y+1, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BQ2091).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Martin Reyes, M. G., Gili, P., Zarza, P. M., Medina Ortega, A. & Diaz Gonzalez, M. C. (1986). Inorg. Chim. Acta, 116, 153–156.
- Martin Zarza, P., Gili, P., Mederos, A. & Medina, A. (1989). Thermochim. Acta, 156, 231–238.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2008). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026044/bq2091sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026044/bq2091Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report