Abstract
Sequence analysis of chloroplast and mitochondrial large subunit rRNA genes from over 75 green algae disclosed 28 new group I intron-encoded proteins carrying a single LAGLIDADG motif. These putative homing endonucleases form four subfamilies of homologous enzymes, with the members of each subfamily being encoded by introns sharing the same insertion site. We showed that four divergent endonucleases from the I-CreI subfamily cleave the same DNA substrates. Mapping of the 66 amino acids that are conserved among the members of this subfamily on the 3-dimensional structure of I-CreI bound to its recognition sequence revealed that these residues participate in protein folding, homodimerization, DNA recognition and catalysis. Surprisingly, only seven of the 21 I-CreI amino acids interacting with DNA are conserved, suggesting that I-CreI and its homologs use different subsets of residues to recognize the same DNA sequence. Our sequence comparison of all 45 single-LAGLIDADG proteins identified so far suggests that these proteins share related structures and that there is a weak pressure in each subfamily to maintain identical protein–DNA contacts. The high sequence variability we observed in the DNA-binding site of homologous LAGLIDADG endonucleases provides insight into how these proteins evolve new DNA specificity.
INTRODUCTION
Homing endonucleases are rare-cutting enzymes that promote site-specific transposition of their encoding genetic elements by generating double-stranded DNA breaks in alleles lacking these elements (1). Site specificity of this genetic phenomenon, termed homing, depends upon the capacity of the endonucleases to recognize and cleave long DNA sequences (14–40 bp). First identified in a mobile group I intron of yeast mitochondria (2), genes for homing endonucleases have been found in a wide range of organisms, where they are located not only in group I introns but also in group II introns, archaeal introns, intein coding sequences and free standing open reading frames (ORFs) (1,3–7).
Of the four subfamilies of homing endonucleases that can be distinguished on the basis of conserved amino acid motifs (LAGLIDADG, H-N-H, His-Cys box and GIY-YIG), the LAGLIDADG family is the largest, with more than 150 members reported to date (8,9). LAGLIDADG enzymes contain one or two copies of the consensus motif. Genes encoding single-LAGLIDADG enzymes have been found exclusively in group I introns inserted at four distinct sites in the large subunit (LSU) rRNA gene (see Table 1). Their presence has been documented in both the mitochondrial and chloroplast DNAs of green algae (8,10–13), in the mitochondrial DNA of the amoeboid protozoan Acanthamoeba castellanii (14) and in the chromosomal DNA of the bacterium Simkania negevensis (15) (see Table 1). In contrast, double-LAGLIDADG enzymes are more widespread; they are encoded by free standing ORFs as well as by ORFs in intein genes and in group I and archaeal introns disseminated in a wide range of host organisms and cellular compartments (9).
Table 1. List of all single-LAGLIDADG proteins identified to date.
| Designationa | Source | Length (amino acids) | Accession no.d | |
| |
Organismb |
Genomec |
|
|
| Proteins encoded by site 2593 LSU rDNA introns (IA3, L6)e | ||||
| I-CreI | Chlamydomonas reinhardtii (C) | cp | 163 | X01977 |
| Sob2593c | Scenedesmus obliquus (C) | cp | 167 | L43360* |
| Clu2593c | Carteria luzensis (C) | cp | 171 | L42986* |
| Col2593c | Carteria olivieri (C) | cp | 182 | L43500* |
| Ciy2593c | Chlamydomonas iyengarii (C) | cp | 212 | L43354* |
| Hla2593c | Haematococcus lacustris (C) | cp | 166 | L49151* |
| Cag2593c | Chlamydomonas agloeformis (C) | cp | 246 | L43351* |
| I-CvuI | Chlorella vulgaris (T) | cp | 161 | L43357*f |
| I-PakI | Pseudendoclonium akinetum (U) | cp | 168 | L44125* |
| Tmu2593c | Trichosarcina mucosa (U) | cp | 168 | AY008341* |
| Msp2593c | Monomastix sp. (P)g | cp | 167 | L44124* |
| I-MsoI | Monomastix sp. (P)h | cp | 170 | L49154* |
| Sdu2593c | Scherffelia dubia (P) | cp | 167 | L44126* |
| Mvi2593m | Mesostigma viride (P) | mt | 162 | AF323369* |
| Nol2593m | Nephroselmis olivacea (P) | mt | 164 | AF110138 |
| Aca2593m | Acanthamoeba castellanii | mt | 164 | U03732 |
| Proteins encoded by site 1923 LSU rDNA introns (IB4, L6)e | ||||
| I-CeuI | Chlamydomonas eugametos (C) | cp | 218 | Z17234 |
| I-CecI | Chlorococcum echinozigotum (C) | cp | 213 | L44123 |
| I-CmoI | Chlamydomonas monadina (C) | cp | 216 | L49149 |
| I-CelI | Chlorogonium elongatum (C) | cp | 229 | L42860 |
| I-CpaIII | Chlamydomonas pallidostigmatica (C) | cp | 214 | L43503 |
| I-CmuI | Chlamydomonas mutabilis (C) | cp | 219 | L42859 |
| I-CluI | Carteria luzensis (C) | cp | 225 | L42986 |
| I-SobI | Scenedesmus obliquus (C) | cp | 221 | L43360 |
| I-AstI | Ankistrodesmus stipitatus (C) | cp | 244 | L42984 |
| Proteins encoded by site 1931 LSU rDNA introns (IB4, L8)e | ||||
| I-CpaI | Chlamydomonas pallidostigmatica (C) | cp | 152 | L36830 |
| Cbr1931c | Chlorosarcina brevispinosa (T) | cp | 153 | L49150* |
| Cfr1931c | Chlamydomonas frankii (C) | cp | 154 | L43352* |
| Cme1931c | Chlamydomonas mexicana (C) | cp | 140 | L49148* |
| Cge1931c | Chlamydomonas geitleri (C) | cp | 177 | L43353* |
| Pcr1931c | Pterosperma cristatum (P) | cp | 141 | L43359* |
| Msp1931c | Monomastix sp. (P)g | cp | 140 | L44124* |
| Mso1931c | Monomastix sp. (P)h | cp | 138 | L49154* |
| Ptu1931c | Pedinomonas tuberculata (P) | cp | 145 | L43541* |
| Cvu1931m | Chlorella vulgaris (T) | mt | 144 | AY008337* |
| Msp1931m | Monomastix sp. (P)g | mt | 150 | AY008340* |
| Mso1931m | Monomastix sp. (P)h | mt | 202 | AY008339* |
| Nol1931m | Nephroselmis olivacea (P) | mt | 157 | AF110138 |
| Aca1931m | Acanthamoeba castellanii | mt | 142 | U03732 |
| Sne1931b | Simkania negevensis | ch | 143 | U68460 |
| Proteins encoded by site 1951 IA3 introns (IA3, L8)e | ||||
| Cbr1951c | Chlorosarcina brevispinosa (T) | cp | 163 | L49150* |
| Msp1951c | Monomastix sp. (P)g | cp | 165 | L44124* |
| Mso1951c | Monomastix sp. (P)h | cp | 161 | L49154* |
| Cvu1951m | Chlorella vulgaris (T) | mt | 166 | AY008338* |
| Aca1951m | Acanthamoeba castellanii | mt | 168 | U03732 |
aProteins are grouped according to the insertion sites of their encoding group I introns in the LSU rRNA gene. Those designated by I- followed by three letters and a roman number have been shown to be site-specific DNA endonucleases [I-CreI (21,37,38), I-CeuI and its homologs (8,39), I-CpaI (12), I-CvuI, I-PakI and I-MsoI (this study)].
bThe class to which each green alga belongs is indicated in parentheses. C, Chlorophyceae; T, Trebouxiophyceae; U, Ulvophyceae; P, Prasinophyceae.
ccp, chloroplast; mt, mitochondrial; ch, chromosomal.
dThe sequences determined in the present study are indicated by asterisks.
eFor each group of introns the intron insertion site is given according to the E.coli 23S rRNA. The intron subfamily (IA3 or IB4) as well as the position of the loop (L6 or L8) containing the LAGLIDADG protein gene in the intron secondary structure are indicated in parentheses.
fThe sequence reported by Wakasugi et al. (40) contains an arginine instead of a glutamine at position 43.
gThis Monomastix strain (M722) is from the private collection of M.Melkonian (University of Cologne).
hThis Monomastix strain is from the private collection of C.O’Kelly (Bigelow Laboratory for Ocean Sciences).
The crystal structures of a single-LAGLIDADG homing endonuclease, I-CreI, and of three double-LAGLIDADG enzymes, PI-SceI, I-DmoI and PI-PfuI, have been reported (16–20). These structures have revealed that single-motif enzymes function as homodimers, whereas double-motif enzymes are monomers with two separate domains, each resembling a subunit of a single-LAGLIDADG protein. The subunits of I-CreI and domains of PI-SceI, I-DmoI and PI-PfuI have identical αββαββα topologies, except that each I-CreI subunit contains two additional C-terminal α-helices. The first seven residues of the LAGLIDADG motif lie at the C-terminus of helix α1 and form the subunit (or domain) interface, whereas the remaining α-helices form a stabilizing hydrophobic core. The protein–DNA interface has been delineated for I-CreI bound to its 24 bp recognition sequence in the presence of Ca2+ (18), a cation that does not allow the cleavage reaction to proceed (21). As reported for other homing endonucleases, a divalent cation is required for I-CreI cleavage, with Mg2+ being the preferred metal ion (21). The structure of the I-CreI protein–DNA complex has revealed that the four β-strands of each subunit form a single β-sheet that binds to the major groove of the pseudo-palindromic DNA recognition sequence: in total, 21 residues of the DNA-binding site make contacts to nine bases and seven phosphate groups (see Fig. 5). In each active homodimer there are two Ca2+ ions bound near the scissile phosphates, suggesting the presence of two symmetry-related active sites. Each metal ion is predicted to be coordinated by residues G19, D20 and Q47, and either R51 or K98 has been proposed to participate in the cleavage reaction by stabilizing the pentacovalent transition state of the scissile phosphate or by activating the attacking water molecule (18). Only one of these I-CreI amino acids (D20) is conserved among the active sites of PI-SceI, I-DmoI and PI-PfuI. As residues Q47, R51 and K98 are partially conserved in PI-SceI and I-DmoI, it is still unknown whether the LAGLIDADG endonucleases share similar catalytic mechanisms (19).
Figure 5.
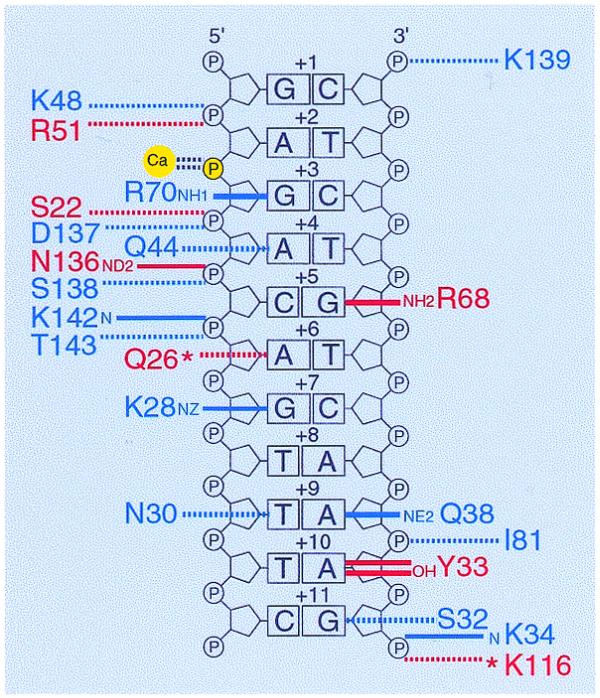
Conservation pattern of the I-CreI amino acid residues that interact with the DNA recognition sequence. To identify these residues, the coordinates of the I-CreI/DNA co-crystal (entry 1BP7) were analyzed using NUCPLOT 1.0 (25). For clarity, interactions are shown for one of the I-CreI subunits and half of the recognition sequence; contacts between the other subunit and the symmetry-related DNA sequence are almost identical. Base pairs are numbered as in Figure 2. The DNA backbones are drawn next to the bases, the sugars as pentagons and phosphates as circles. Interactions are plotted on either side of the strands. Interacting protein residues are represented by their atom name, residue name and number, which are colored red (similar residues) or blue (non-conserved residues) depending on the degree of conservation in other proteins encoded by site 2593 LSU rDNA introns (see Fig. 1). Hydrogen bonds and van der Waals interactions are denoted by solid and dotted lines, respectively. Water molecules are represented by asterisks. The calcium ion and scissile phosphate are highlighted in yellow; the residues binding the calcium ion (G19, D20 and Q47) are not shown. Note that the interactions shown in this figure differ slightly from those previously predicted by Jurica et al. (18) using QUANTA 96. The latter program identified exactly the same amino acid/base recognition profile as that predicted by NUCPLOT, except that the numbers of contacts made by Y30, Q38, R68 and R70 were found to be different. Concerning the interactions of protein residues with phosphate groups, QUANTA predicted no such interactions for I81, N136, D137, K139, K142 and T143 (18).
Comparative sequence analyses of single-LAGLIDADG endonucleases offer a unique opportunity to understand how homing endonucleases recognize and cleave their recognition sequence and also how they evolved different DNA specificities. Evolutionarily related endonucleases from each of the four subfamilies of single-LAGLIDADG proteins can be readily identified, as LSU rRNA introns are widespread among the chloroplasts and mitochondria of green algae (8,11,12). Considering that proteins belonging to the same subfamily are likely to recognize the same DNA substrate (8), comparative sequence analysis of these proteins should reveal the amino acids that are most important for dimerization and for formation of the hydrophobic core, the protein–DNA interface and the active sites. In the case of the I-CreI subfamily the availability of the I-CreI/DNA co-crystal structure makes this comparative approach particularly powerful and allows the putative roles of the conserved residues to be predicted with high confidence.
In the present study we have identified 28 new single-LAGLIDADG proteins, 13 of which are encoded by LSU rRNA introns found at the same insertion site as the I-CreI-encoding intron (site corresponding to position 2593 in the 23S rRNA gene of Escherichia coli). Analysis of the conserved amino acids in the 16 proteins encoded by site 2593 LSU rRNA introns allowed us to infer that all of these potential endonucleases have comparable 3-dimensional structures. These proteins can recognize and cleave the same DNA substrate but, surprisingly, they appear to use different protein–DNA interactions to achieve substrate recognition.
MATERIALS AND METHODS
DNA sequencing and sequence analyses
PCR products encompassing the mitochondrial and chloroplast LSU rRNA genes from various green algae were generated with primers complementary to highly conserved regions of the LSU rRNA gene (8). These PCR products were sequenced directly and the resulting sequences were analyzed as described previously (8). Alignments of intron-encoded proteins were generated using CLUSTAL W 1.81 (22), analyzed with AMAS 1.67b (23) and displayed using ALSCRIPT 2.07a (24). Protein–DNA interactions were predicted using NUCPLOT 1.0 (25) and the coordinates of the I-CreI/DNA co-crystal (18).
Cloning of endonuclease genes
The genes encoding I-CreI, I-MsoI, I-PakI and I-CvuI were amplified by PCR from chloroplast DNA-enriched preparations of Chlamydomonas reinhardtii, Monomastix sp., Pseudendoclonium akinetum and Chlorella vulgaris, respectively. For these amplifications primers were designed to introduce a NcoI site at the initiation codon of each endonuclease gene and a BamHI (I-MsoI, I-PakI and I-CvuI) or XhoI (I-CreI) site downstream of the stop codon. The creation of the NcoI site resulted in the following changes of codon at the second position: N→D (I-CreI), T→A (I-MsoI), S→G (I-PakI) and Q→E (I-CvuI). Appropriately digested PCR products were cloned into pET30a (Novagen, Madison, WI) downstream of the region specifying an N-terminal six-histidine tag and the resulting constructs were introduced into the E.coli BL21/DE3 Codon Plus strain (Stratagene, La Jolla, CA). Plasmid inserts of the four clones used in gene expression experiments (pET-ICreI, pET-IMsoI, pET-IPakI and pET-ICvuI) were sequenced to ensure that no mutations occurred in the endonuclease genes and that each endonuclease gene was in-frame with the histidine tag.
Protein purification
A culture of each clone carrying a homing endonuclease gene was grown at 37°C in 400 ml of L broth. At mid-exponential phase (0.6 OD600 nm) gene expression was induced by adding IPTG to a final concentration of 1 mM. Cells were further incubated for 3 h and collected by centrifugation. All subsequent steps were performed at 4°C unless indicated otherwise. Cells were resuspended in 5 ml of ice-cold buffer A [50 mM sodium phosphate pH 8.0, 0.5 M NaCl, 1 mM phenylmethylsulfonide fluoride (PMSF)] containing 10 mM imidazole and disrupted in a French Pressure Cell (SLM Aminco, Urbana, IL). The cell lysate was centrifuged for 15 min at 10 000 g and the supernatant mixed with 1 ml of Ni–NTA resin (Qiagen, Mississauga, Ontario, Canada) for 1 h. The resin was collected by centrifugation, washed three times with 4 ml of buffer A containing 20 mM imidazole and equilibrated with 2 ml of buffer B (20 mM Tris–HCl pH 7.4, 50 mM NaCl, 0.1 mM CaCl2). Enterokinase (5 U; Novagen) was added to the resin and the mixture was incubated at room temperature for 16 h. After centrifugation the supernatant containing the cleaved proteins (i.e. the proteins free of the histidine tag) was recovered and enterokinase was removed using an EKapture-agarose kit (Novagen). Protein concentration was estimated using the MicroBCA kit (Pierce, Rockford, IL) and protein purity was assessed by 15% SDS–PAGE. Aliquots of purified proteins were used immediately or stored at –20°C in 50% glycerol and 4 mM EGTA.
Endonuclease assays
DNA substrates containing the natural recognition sequences of I-CreI (substrate S1) and of I-MsoI, I-Pak1 and I-CvuI (substrate S2) were generated by PCR from chloroplast DNA preparations of Chlamydomonas zebra and Neochloris pseudoalveolaris, respectively, using the following primers that had been previously labeled at their 5′-end with T4 polynucleotide kinase and [γ-33P]ATP: for S1, 5′-TGTCGGCTTATCGCATCCTG-3′ and 5′-TCCATGCATAGCTACCCAGC-3′; for S2, 5′-GGAAGGTTTGGCACCTCGATG-3′ and 5′-GTACTCATCTTGGGGTGGGCTT-3′. Each purified endonuclease (150 pM) was incubated at 37°C for 1 h in 200 µl of reaction buffer (10 mM TAPS–KOH pH 8.5, 1 mM dithiothreitol, 10 µg/ml bovine serum albumin and either 10 mM MgCl2 or 5 mM MnCl2) containing substrate S1 or S2 (30 pM). The reaction was stopped by addition of SDS to 0.5% and proteinase K to 0.2 mg/ml, followed by incubation at 50°C for 1 h. DNA was ethanol precipitated in the presence of 20 µg glycogen and 0.75 M ammonium acetate and dissolved in 4 µl of loading buffer (2% Ficoll 400, 10 mM EDTA, 0.02% bromophenol blue). DNA samples were electrophoresed in an 8% polyacrylamide–1× TBE (90 mM Tris–borate, pH 8.0, 2 mM EDTA) gel. The gel was fixed in a solution containing 10% ethanol and 10% acetic acid, dried and exposed to a Fuji imaging plate (Fuji Photo System, Japan). To precisely map the positions of the breaks introduced by the endonucleases into the top and bottom strands of each substrate the reaction products were electrophoresed in a 5% polyacrylamide–7 M urea gel alongside sequencing ladders that were derived from both strands of the substrate (12). After electrophoresis the gel was fixed as described above, dried and exposed to X-ray film.
RESULTS
Identification of green algal single-LAGLIDADG proteins
To identify homologs of I-CreI and of other previously identified single-LAGLIDADG proteins we analyzed the mitochondrial and chloroplast LSU rRNA gene sequences of more than 75 green algae, investigated the presence of group I introns in these genes and determined if they encode single-LAGLIDADG proteins. The selected green algae belong to the four main classes (Prasinophyceae, Ulvophyceae, Trebouxiophyceae and Chlorophyceae) of the Chlorophyta. Table 1 presents the 28 single-LAGLIDADG proteins identified in the present study and the 17 cognate proteins previously documented. Thirteen of the newly identified single-LAGLIDADG proteins are encoded by introns that are inserted at the same site as the I-CreI-encoding intron (site 2593), whereas the others are encoded by introns occupying the same sites as the I-CpaI-encoding intron (site 1931) and the second intron in the Acanthamoeba mitochondrial LSU rRNA gene (site 1951). As previously reported for site 1923 LSU rRNA introns (8), all of the introns sharing a common insertion site appear to be evolutionarily related; they share a remarkably similar core structure that is typical of subgroup IA3 or IB4 introns (see Table 1) and display an ORF with a similar sequence (see Figs 1 and 6) in the same loop (L6 or L8) of the intron RNA secondary structure.
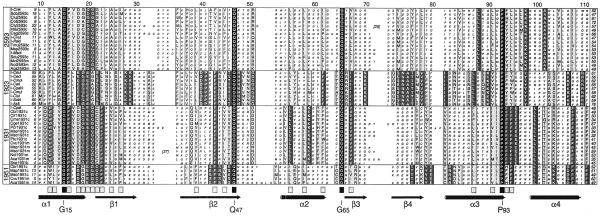
Figure 1.
Sequence alignment of the predicted proteins encoded by group I introns at site 2593 of the LSU rRNA gene. Identical amino acids in all of the sequences are shown on a black background, whereas conserved sets of amino acids sharing at least eight of the 10 features in the property matrix of AMAS (23) and possibly containing one atypical residue are shown on a gray background. Secondary structure elements of I-CreI (18) are depicted as cylinders (α-helices) and arrows (β-strands) and are shown below the alignment. Proteins are designated as in Table 1 and numbers refer to the amino acid coordinates of I-CreI.
Figure 6.
Sequence alignment of all known single-LAGLIDADG proteins. The proteins are arranged into four groups according to the positions of their encoding group I introns. The numbers on the extreme left of the alignment indicate these positions. Identical amino acids in all of the four groups are displayed on a black background, whereas identical residues in at least one of the groups are shown on a dark gray background. In each group sets of residues sharing eight of the 10 features in the property matrix of AMAS are shown on a light gray background. Gray and black squares below the aligned sequences denote the positions of the strictly conserved and similar residues, respectively. Note that positions were considered to be conserved in amino acid similarity when one of the subfamilies contained a single protein with an atypical residue. Cylinders (α-helices) and arrows (β-strands) denote the secondary structure elements found in I-CreI (18). Numbers above the alignment refer to the amino acid coordinates of I-CreI. For each protein the numbers of amino acids preceding and following those in the aligned region are indicated on the left and right sides of the alignment, respectively. In two proteins the sequences of distinct regions containing more than 25 residues are not shown; the numbers of residues in these sequences are indicated in brackets. Proteins are designated as in Table 1.
Site 2593 LSU rRNA introns encode homologous endonucleases
As shown in Figure 1, the sequences of I-CreI and of its 15 homologs can be aligned over their entire length, except for the positions corresponding to the first 11 and last 18 residues. Six distinct regions of the alignment feature gaps and, interestingly, all correspond to loops joining secondary structure elements in the crystal structure of I-CreI. Three loops, those connecting α1 to β1, β4 to α3 and α3 to α4, correspond to regions that are identical in size in all homologous proteins. Nine positions in the alignment show strictly conserved amino acids and 57 show residues conserved in similarity. Conserved residues are found in all of the α-helices and β-strands of I-CreI as well as in all of the loops of this enzyme, with the exception of the short loop connecting β3 to β4.
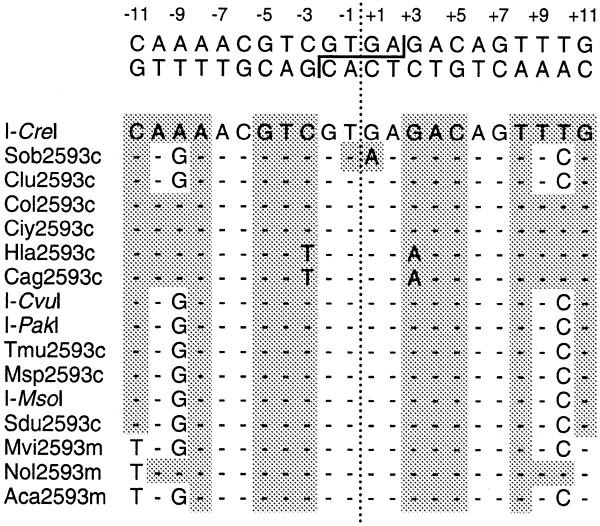
To determine whether I-CreI and the 15 potential endonucleases that are homologous to this protein are likely to recognize the same DNA substrate we assessed the level of sequence identity displayed by their natural substrates by comparing the LSU rRNA gene sequences that flank their encoding introns (Fig. 2). Comparison of the 22 bp regions corresponding to the I-CreI recognition sequence in the 16 green algal genes indicates that the sequences of Carteria olivieri (Chlorophyceae), Chlamydomonas iyengarii (Chlorophyceae) and Nephroselmis olivacea (Prasinophyceae) are identical to that of C.reinhardtii. The predicted DNA recognition sequences of nine of the 12 other I-CreI homologs differ from the natural substrate of C.reinhardtii at positions –9 and +9, those of two I-CreI homologs show substitutions at positions –3 and +3, while the natural substrate of the remaining I-CreI homolog is predicted to differ at positions –9, +1 and +10. All of the six identified substitutions (all transitions) do not prevent DNA cleavage by I-CreI (26,27), suggesting that the 15 potential endonucleases homologous to this enzyme are likely to cleave the I-CreI recognition sequence.
Figure 2.
Alignment of the predicted DNA recognition sequences of proteins encoded by group I introns at site 2593 of the LSU rRNA gene. On top of the figure is shown the I-CreI recognition sequence; the staggered line denotes the site of cleavage by I-CreI. The lower portion of the figure shows an alignment of the I-CreI recognition sequence and the corresponding LSU rRNA gene sequences found at the borders of site 2593 group I introns encoding homologous proteins. Dashes represent bases that are identical to those found at the same positions in the I-CreI recognition sequence. The bases forming palindromic sequences relative to the axis of symmetry denoted by the dotted line are shown on a gray background.
To ascertain that site 2593 LSU rRNA intron-encoded proteins are endonucleases that share the same DNA specificity we overproduced the proteins of four distantly related green algae in E.coli and assayed them in DNA cleavage reactions. The intron-encoded genes of Chlorella vulgaris (Trebouxiophyceae), Pseudendoclonium akinetum (Ulvophyceae) and Monomastix sp. (Prasinophyceae) as well the I-CreI gene sequence (Chlorophyceae) were expressed as fusion proteins carrying a six-histidine tag at the N-terminus. These proteins accumulated to levels representing 5–10% of total cell proteins after induction of transcription by IPTG. The four recombinant proteins were purified to ∼75% homogeneity by affinity chromatography and the N-terminal histidine tag was removed by proteolytic digestion. For the DNA cleavage reactions we prepared a 232 bp DNA fragment containing the recognition sequence of I-CreI (substrate S1) and also a 302 bp fragment containing the natural recognition sequence of I-MsoI, I-PakI and I-CvuI (substrate S2). These recognition sequences differ at positions –9 and +10 (Fig. 2). All four proteins cleaved both substrates in the presence of Mg2+ or Mn2+, yielding fragments of the same size (Fig. 3A). Precise mapping of the cleavage sites in the top and bottom strands of each substrate revealed that I-MsoI, I-PakI and I-CvuI introduce breaks at exactly the same positions as does I-CreI (Fig. 3B). The four endonucleases showed different cleavage efficiencies depending on the metal ion used as cofactor and the substrate employed (Fig. 3A). I-CreI and I-MsoI cleaved the two substrates with similar efficiency and were more active in the presence of Mg2+ than in the presence of Mn2+ when the I-CreI substrate was used. In contrast, I-PakI and I-CvuI preferred their natural substrate; I-PakI showed more activity with Mg2+ as the cofactor, whereas I-CvuI was more active in the presence of Mn2+. Overall, our results suggest that all proteins encoded by site 2593 LSU rRNA introns recognize and cleave the same or very similar DNA sequences. The observed differences in cleavage efficiencies probably reflect differences in the biochemical properties of the endonucleases examined.
Figure 3.
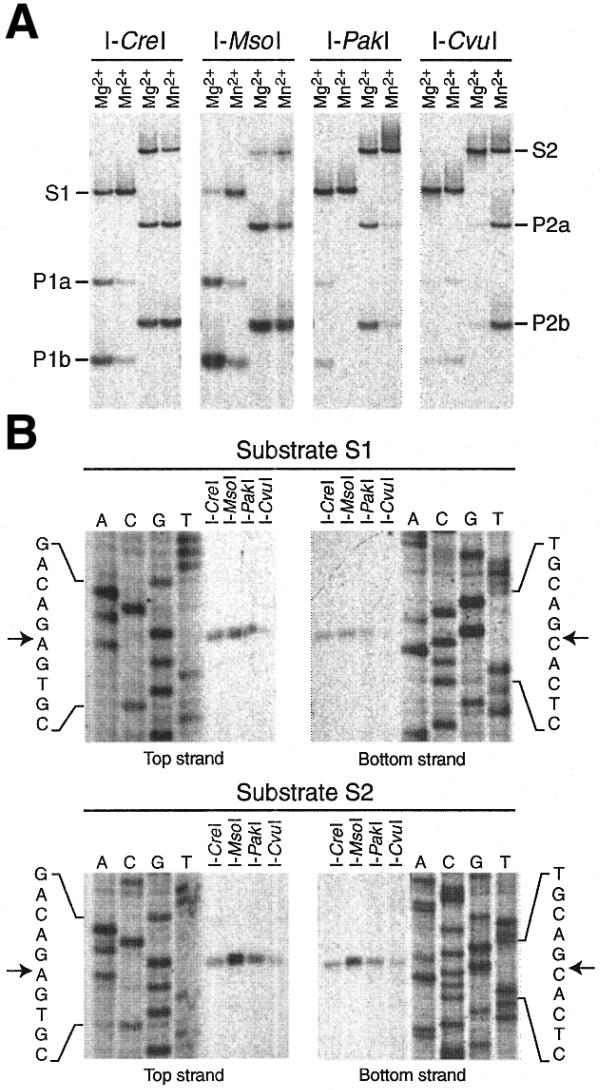
Endonuclease activities of four proteins encoded by group I introns at site 2593 of the LSU rRNA gene. Each protein was incubated at 37°C for 1 h in a 200 µl reaction mixture containing substrate S1 (232 bp) or S2 (302 bp) in the presence of either Mg2+ or Mn2+ as cofactor. (A) Electrophoresis pattern of the reaction products in an 8% polyacrylamide gel. P1a and P1b, products of the reaction with substrate S1; P2a and P2b, products of the reaction with substrate S2. (B) Mapping of the top and bottom strand cleavage sites in substrates S1 and S2. The cofactor in the cleavage reactions was Mg2+. The products of each cleavage reaction were electrophoresed in a 5% polyacrylamide–7 M urea gel alongside sequencing ladders (A, C, G and T lanes) that were produced from the substrate used in the reaction. Arrows indicate the previously reported positions of the I-CreI cleavage sites (26,38). Note that there is a slight difference between the mobility of each I-CreI cleavage product and that of the sequencing product displaying the same number of bases. This difference is most likely due to the presence of 7-deaza-dGTP instead of dGTP in the sequencing products.
Predicted roles of the conserved amino acids in I-CreI and its homologs
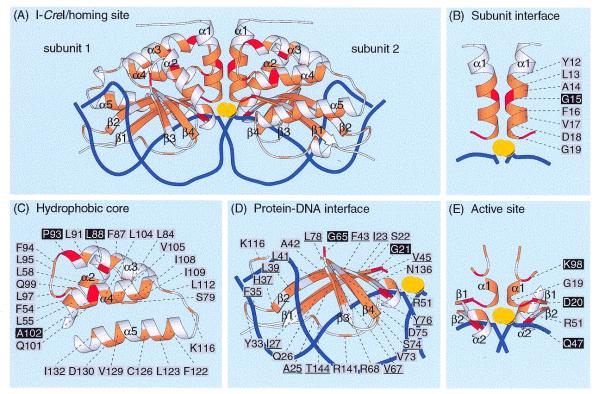
To determine how the conserved amino acids between I-CreI and its homologs contribute to the make-up of functional endonucleases we localized these amino acids on the I-CreI/DNA co-crystal structure (Fig. 4). In Figure 4B it can be seen that all residues of helix α1 participating in homodimerization are conserved in identity or similarity. This conservation pattern not only highlights the essential role of these amino acids in the assembly of the subunits but also indicates that I-CreI and its homologs employ the same strategy for subunit assembly.
Figure 4.
Positions of conserved amino acid residues within the I-CreI/DNA co-crystal structure. (A) Global pattern of amino acid conservation. A ribbon diagram of the I-CreI/DNA structure was obtained using MOLSCRIPT 2.1.2 (41) and the coordinates in the Brookhaven Protein Data Bank entry 1BP7. Strictly conserved, similar and non-conserved residues, as defined in Figure 1, are colored in red, orange and white, respectively. Calcium ions are colored yellow and DNA strands are represented as dark blue lines. (B–E) Conserved amino acids in the subunit interface, the hydrophobic core, the protein–DNA interface and the active site, respectively. Strictly conserved residues are identified on a black background, whereas similar residues are identified on a gray background. The residues with underlined coordinates have their side chains oriented towards the hydrophobic core.
As expected, the central core of the I-CreI subunit formed by helix α2 and the region spanning α3, α4 and α5 contains a large number of conserved hydrophobic residues. Of the 27 conserved residues in this region, 22 are hydrophobic (Figs 1 and 4C) and almost all of their side chains are oriented towards the inner part of the protein, supporting the idea that they are essential for formation of the hydrophobic core of each subunit. As the strictly conserved P93 residue in the loop connecting α3 to α4 produces a strong curvature at the C-terminus of helix α3, this residue appears to be required to initiate a loop that properly positions the strictly conserved K98 residue in the active site.
The region of each I-CreI subunit comprising the two pairs of β-strands, their two joining loops and the unfolded segment at the C-terminus displays 25 residues that are conserved in similarity and only two (G21 and G65) that are strictly conserved (Fig. 4D). As the two strictly conserved glycine residues immediately precede each pair of β-strands, they are likely to confer flexibility to these structural elements during binding to DNA. Fourteen of the 25 residues conserved in similarity have their side chains oriented towards the hydrophobic core (see Fig. 4D), suggesting that they interact with this core to allow residues contacting the DNA recognition sequence to adopt appropriate positions. The remaining 11 residues conserved in similarity have their side chains pointing towards the DNA substrate.
As shown in Figure 5, 14 of the 21 residues binding the DNA recognition sequence in I-CreI are not conserved in homologs of this enzyme. The remaining seven residues are only conserved in similarity: four (S22, Q26, R51 and N136) interact with phosphate groups and three (R68, Q26 and Y33) contact DNA bases. As the adenine (at position +10) interacting with residue Y33 is substituted by a guanine in the natural recognition sequences of 10 I-CreI homologs (Fig. 2), this conserved tyrosine may be involved in purine-specific interactions. Overall, these observations suggest that I-CreI and its homologs have evolved different protein–DNA interfaces while retaining the ability to recognize closely related DNA sequences.
All of the five residues that have been proposed to participate in the active sites of I-CreI are conserved in the 15 other members of the protein subfamily. The D20, Q47 and K98 residues are strictly conserved, whereas G19 and R51 are replaced by similar residues in some proteins (Fig. 4D). This observation suggests that the active sites of I-CreI and its 15 homologs are very similar, if not identical.
Common features among all known single-LAGLIDADG homing endonucleases
To determine whether some of the conserved features reported here for I-CreI homologs are also shared by single-LAGLIDADG homing endonucleases having different DNA specificities we compared the predicted sequences of all 45 putative single-LAGLIDADG endonucleases identified to date. As observed for I-CreI and its homologs (this study) and for site 1923 LSU rRNA intron-encoded endonucleases (8), the single-LAGLIDADG proteins specified by the LSU rRNA introns inserted at sites 1931 and 1951 display a high level of sequence identity and similarity over most of their length (Fig. 6). In each of the four subfamilies of homologous proteins the majority of conserved amino acids are hydrophobic and map to regions corresponding to structural elements identified in I-CreI.
The sequences of the 45 single-LAGLIDADG proteins markedly differ at the N- and C-termini, but can be aligned over a segment of ∼100 amino acids which begins with the LAGLIDADG motif (Fig. 6). This segment spans all of the secondary structure elements found in I-CreI, except α5, and exhibits five regions containing gaps, all of which correspond to loops. Four of the regions with gaps have been described above for the group of endonucleases containing I-CreI and its homologs; the remaining one coincides with the loop connecting β4 to α3. The absence of gaps in the loops connecting α1 to β1 and α3 to α4 suggests that extra amino acids cannot be tolerated in these regions participating in formation of the active sites. In the alignment of the 45 protein sequences four positions show strictly conserved residues and 22 show residues conserved in similarity (Fig. 6). Twenty-one of these residues, including the strictly conserved G15, Q47 and P93, fall within six of the nine structural elements displayed by I-CreI (α1, β1, β2, α2, β3 and α4). The five remaining conserved residues, including the strictly conserved G65, correspond to the loops connecting α1 to β1, α2 to β3 and α3 to α4. None of the residues that are known to contact DNA in I-CreI are conserved in all 45 single-LAGLIDADG proteins. The highest density of conserved residues (12/26) is found in the region spanning α1 and β1.
DISCUSSION
Conserved residues of LAGLIDADG endonucleases: roles in protein folding and catalysis
Our comparative sequence analysis of the 45 single-LAGLIDADG homing endonucleases identified to date has revealed a large number of conserved amino acid residues that appear to be involved in protein folding. These residues are located within an internal segment of 100 amino acids, which begins with the LAGLIDADG motif and overlaps with all but one (α5) of the I-CreI secondary structure elements. In the region corresponding to helix α1 of I-CreI the aromatic residue preceding the LAGLIDADG motif (Y12 in I-CreI) as well as the first conserved glycine (G15) and acidic (D18) residues of this motif are probably essential for homodimerization and active site structuring. In the regions equivalent to α2, α3 and α4 the residues conserved with respect to their hydrophobic character are likely to form a stabilizing core and to interact with other conserved hydrophobic residues in the regions corresponding to the β-strands, thus allowing proper positioning of the protein–DNA interfaces. A conserved proline (P93) terminates the region corresponding to α3 and initiates a loop containing a putative active site residue (K98). The glycine residues (G21 and G65) preceding the segments equivalent to the two pairs of β-strands may provide a signal for folding of these regions and favor binding to DNA by conferring flexibility to the β-strands. Overall, the structural similarities shared by single-LAGLIDADG endonucleases strongly suggest that all of these enzymes adopt a similar αββαββα fold over an internal region of 100 amino acids. Both the N- and C-termini of these proteins are highly divergent in sequence and likely to differ markedly in structure. In agreement with structural and mutational studies of I-CreI and I-CeuI (8,18,28), this observation suggests that the terminal sequences of single-LAGLIDADG endonucleases do not play a critical role in the function of these enzymes.
Considering that the residues equivalent to D20 and Q47 in I-CreI are conserved in all of the 45 proteins examined, there is little doubt that these residues are part of the active sites in all single-LAGLIDADG endonucleases. This interpretation is supported by the structural data available for LAGLIDADG endonucleases (16–20) and also by biochemical data indicating that the I-CreI D20 and Q47 residues as well as the corresponding I-CeuI residues (E66 and Q93) are critical for endonuclease activity (8,28; our unpublished results). Of the two amino acids (R51 and K98) that have been proposed to participate in the cleavage reaction as either a Lewis acid or a proton donor activator (18), K98 is the most likely to assume one of these functions because it is strictly conserved among the 15 homologs of I-CreI. Although K98 is not conserved in all non-homologs of I-CreI, it is replaced by a residue that can also act as a Lewis acid or a proton donor activator (e.g. a tyrosine in I-CeuI homologs). Moreover, the hypothesis that the residue at this site plays the same role in all single-LAGLIDADG enzymes is supported by our prediction that its localization is conserved in the 3-dimensional structures of all enzymes.
Comparative sequence analysis of double-LAGLIDADG endonucleases has revealed that these enzymes are much less conserved than their single-motif counterparts (9). Sequence conservation is restricted to the LAGLIDADG motif in alignments of all double-motif endonucleases and only a small subset of enzymes share all of the conserved residues reported here for single-LAGLIDADG enzymes (8). The double-motif enzymes PI-SceI, I-DmoI and PI-PfuI differ markedly in primary sequences yet they share obvious similarities at the structural level (16,19,20); whether they use the same catalytic mechanism remains to be determined (19). Given that genes for double-LAGLIDADG enzymes are likely to have originated from duplication of genes encoding single-LAGLIDADG enzymes (1), it is surprising that the latter proteins exhibit a higher level of sequence conservation. A possible reason for this paradox is that more divergent single-motif enzymes remain to be identified. An alternative explanation is that the evolution of single-LAGLIDADG endonucleases may be more constrained than that of double-motif enzymes because the former enzymes must function as homodimers and cleave DNA substrates with symmetrical or pseudo-symmetrical sequences. Their coding sequences may have remained confined to introns within the LSU rRNA gene because this gene contains a number of palindromic sequences that can serve as potential substrates.
DNA recognition by LAGLIDADG endonucleases: mechanism and evolution
The low level of sequence conservation we observed in the I-CreI DNA-binding site contrasts sharply with the high conservation of the homing site and other functional domains of this endonuclease. None of the 21 I-CreI residues interacting with the half-recognition sequence is strictly conserved and seven are conserved in similarity. Only three of the latter residues (Q26, Y33 and R68) contact bases and are thus involved in specific DNA recognition; the four remaining conserved residues contact phosphate groups and interact non-specifically with DNA. Given that more than one interaction is required for unambiguous recognition of a base (6), the single bonds established by the conserved residues Q26 and R68 are expected to make a limited contribution to specific recognition of the target DNA sequence. Considering that the overall cleavage frequency of I-CreI across random sequences is estimated at 10–8–10–9 (a value corresponding to specific recognition of 9–10 consecutive base pairs) (18) and that I-CreI homologs most probably share a similar level of specificity in order to avoid host lethality, it is evident that the conserved residues interacting with the target DNA sequence are insufficient to account for the high specificity of these enzymes. Therefore, interactions involving non-conserved amino acid residues most likely play a major role in DNA recognition.
Our results suggest that I-CreI and its homologs use different subsets of amino acid residues to recognize identical or very similar DNA sequences. The same strategy is most probably employed by members of the three other subfamilies of single-LAGLIDADG endonucleases, as most of the residues predicted to interact with DNA are not conserved. In a recent study it was shown that the restriction enzymes BglII and BamHI recognize almost the same DNA recognition sequences by establishing different protein–DNA contacts (29). Unlike the result reported here, this observation was expected, as BglII and BamHI exhibit strikingly different structures (29).
Our hypothesis that there exists a weak pressure to maintain identical protein–DNA contacts in each subfamily of single-LAGLIDADG endonucleases is consistent with one of the most fundamental properties of homing endonucleases, namely their relaxed specificity. These endonucleases can still cleave their target sequences following the introduction of single mutations across the homing sites [I-CreI (26,27), I-CeuI (30), I-SceI (2), I-SceII (31), I-TevI (32), I-DmoI (33) and I-PpoI (27,34)]. In the case of I-CreI it has been proposed that different subsets of the large number of protein–DNA contacts are sufficient to maintain a high degree of sequence-specific DNA recognition and cleavage (27). This hypothesis is in agreement with our evidence that the DNA-binding site of single-LAGLIDADG endonucleases is evolving at a fast rate.
The high sequence variability we observed in the DNA-binding site of homologous LAGLIDADG enzymes recognizing the same DNA sequence provides clues as to how these enzymes can rapidly gain new DNA specificity and as to how their encoding genes can move to new genomic sites. Because LAGLIDADG endonucleases are not constrained to maintain identical protein–DNA contacts, a diversity of enzyme variants specific for a given substrate can coexist in populations. Occasionally, a variant will arise which cleaves a sequence found at a new genetic locus in addition to the cognate site. If the gene encoding this enzyme is inserted into an intron whose insertion site is cleaved by the variant, these events may result not only in spreading of the endonuclease gene to a new site, but also in long-term survival of the intron found at this site through the newly acquired homing ability. Although there is evidence that endonuclease genes can transpose to introns at new genetic locations (7), the mechanism underlying such events remains unknown. Transposition could be initiated following cleavage of the target intron by the endonuclease encoded by the gene to be transposed, as it has been shown for some group I introns that the insertion sites of the intron and of the endonuclease gene share significant sequence similarities (35,36).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Michael Melkonian and Debashish Bhattacharya for providing DNA samples from prasinophytes and Charles O’Kelly for providing one of the Monomastix strains and for his help in the selection of the green algae analyzed in this study. M.T. and C.L. are Associates in the Program in Evolutionary Biology of the Canadian Institute for Advanced Research. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (GP0003293 to M.T. and GP0002830 to C.L.).
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 418 656 2131; Fax: +1 418 656 5036; Email: AF323369, AY008337–AY008341, L42986, L43351–L43354, L43357, L43359, L43360, L43500, L43541, L44124–L44126, L49148, L49150, L49151, L49154
References
- 1.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colleaux L., D’Auriol,L., Galibert,F. and Dujon,B. (1988) Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl Acad. Sci. USA, 85, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dujon B. (1989) Group I introns as mobile genetic elements: facts and mechanistic speculations. Gene, 82, 91–114. [DOI] [PubMed] [Google Scholar]
- 4.Lambowitz A.M. and Belfort,M. (1993) Introns as mobile genetic elements. Annu. Rev. Biochem., 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 5.Belfort M. and Perlman,P.S. (1995) Mechanisms of intron mobility. J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- 6.Jurica M.S. and Stoddard,B.L. (1999) Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci., 55, 1304–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimble F.S. (2000) Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett., 185, 99–107. [DOI] [PubMed] [Google Scholar]
- 8.Turmel M., Otis,C., Côté,V. and Lemieux,C. (1997) Evolutionarily conserved and functionally important residues in the I-CeuI homing endonuclease. Nucleic Acids Res., 25, 2610–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rochaix J.D., Rahire,M. and Michel,F. (1985) The chloroplast ribosomal intron of Chlamydomonas reinhardii codes for a polypeptide related to mitochondrial maturases. Nucleic Acids Res., 13, 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turmel M., Gutell,R.R., Mercier,J.P., Otis,C. and Lemieux,C. (1993) Analysis of the chloroplast large subunit ribosomal RNA gene from 17 Chlamydomonas taxa. Three internal transcribed spacers and 12 group I intron insertion sites. J. Mol. Biol., 232, 446–467. [DOI] [PubMed] [Google Scholar]
- 12.Turmel M., Côté,V., Otis,C., Mercier,J.P., Gray,M.W., Lonergan,K.M. and Lemieux,C. (1995) Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion). Mol. Biol. Evol., 12, 533–545. [DOI] [PubMed] [Google Scholar]
- 13.Turmel M., Lemieux,C., Burger,G., Lang,B.F., Otis,C., Plante,I. and Gray,M.W. (1999) The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell, 11, 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonergan K.M. and Gray,M.W. (1994) The ribosomal RNA gene region in Acanthamoeba castellanii mitochondrial DNA. A case of evolutionary transfer of introns between mitochondria and plastids? J. Mol. Biol., 239, 476–499. [DOI] [PubMed] [Google Scholar]
- 15.Everett K.D., Kahane,S., Bush,R.M. and Friedman,M.G. (1999) An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts and mitochondria. J. Bacteriol ., 181, 4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan X., Gimble,F.S. and Quiocho,F.A. (1997) Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell, 89, 55–64. [DOI] [PubMed] [Google Scholar]
- 17.Heath P.J., Stephens,K.M., Monnat,R.J.,Jr and Stoddard,B.L. (1997) The structure of I-Crel, a group I intron-encoded homing endonuclease. Nature Struct. Biol., 4, 468–476. [DOI] [PubMed] [Google Scholar]
- 18.Jurica M.S., Monnat,R.J. and Stoddard,B.L. (1998) DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell, 2, 469–476. [DOI] [PubMed] [Google Scholar]
- 19.Silva G.H., Dalgaard,J.Z., Belfort,M. and Van Roey,P. (1999) Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol., 286, 1123–1136. [DOI] [PubMed] [Google Scholar]
- 20.Ichiyanagi K., Ishino,Y., Ariyoshi,M., Komori,K. and Morikawa,K. (2000) Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol., 300, 889–901. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Kim,H.H., Yuan,X. and Herrin,D.L. (1997) Purification, biochemical characterization and protein–DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res., 25, 3767–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livingstone C.D. and Barton,G.J. (1993) Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci., 9, 745–756. [DOI] [PubMed] [Google Scholar]
- 24.Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- 25.Luscombe N.M., Laskowski,R.A. and Thornton,J.M. (1997) NUCPLOT: a program to generate schematic diagrams of protein–nucleic acid interactions. Nucleic Acids Res., 25, 4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dürrenberger F. and Rochaix,J.D. (1993) Characterization of the cleavage site and the recognition sequence of the I-CreI DNA endonuclease encoded by the chloroplast ribosomal intron of Chlamydomonas reinhardtii. Mol. Gen. Genet., 236, 409–414. [DOI] [PubMed] [Google Scholar]
- 27.Argast G.M., Stephens,K.M., Emond,M.J. and Monnat,R.J. (1998) I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol., 280, 345–353. [DOI] [PubMed] [Google Scholar]
- 28.Seligman L.M., Stephens,K.M., Savage,J.H. and Monnat,R.J.,Jr (1997) Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics, 147, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 Å resolution. Nature Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 30.Marshall P. and Lemieux,C. (1992) The I-CeuI endonuclease recognizes a sequence of 19 bp and preferentially cleaves the coding strand of the Chlamydomonas moewusii chloroplast large subunit rRNA gene. Nucleic Acids Res., 20, 6401–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernette C., Saldanha,R., Smith,D., Ming,D., Perlman,P.S. and Butow,R.A. (1992) Complex recognition site for the group I intron-encoded endonuclease I-SceII. Mol. Cell. Biol., 12, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryk M., Quirk,S.M., Mueller,J.E., Loizos,N., Lawrence,C. and Belfort,M. (1993) The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J., 12, 2141–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aagaard C., Awayez,M.J. and Garrett,R.A. (1997) Profile of the DNA recognition site of the archaeal homing endonuclease I-DmoI. Nucleic Acids Res., 25, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittmayer P.K., McKenzie,J.L. and Raines,R.T. (1998) Degenerate DNA recognition by I-PpoI endonuclease. Gene, 206, 11–21. [DOI] [PubMed] [Google Scholar]
- 35.Loizos N., Tillier,E.R. and Belfort,M. (1994) Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc. Natl Acad. Sci. USA, 91, 11983–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall P., Davis,T.B. and Lemieux,C. (1994) The I-CeuI endonuclease: purification and potential role in the evolution of Chlamydomonas group I introns. Eur. J. Biochem., 220, 855–859. [DOI] [PubMed] [Google Scholar]
- 37.Dürrenberger F. and Rochaix,J.D. (1991) Chloroplast ribosomal intron of Chlamydomonas reinhardtii: in vitro self-splicing, DNA endonuclease activity and in vivo mobility. EMBO J., 10, 3495–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson A.J., Yuan,X., Kudlicki,W. and Herrin,D.L. (1992) Cleavage and recognition pattern of a double-strand-specific endonuclease (I-CreI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene, 119, 247–251. [DOI] [PubMed] [Google Scholar]
- 39.Gauthier A., Turmel,M. and Lemieux,C. (1991) A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr. Genet., 19, 43–47. [DOI] [PubMed] [Google Scholar]
- 40.Wakasugi T., Nagai,T., Kapoor,M., Sugita,M., Ito,M., Ito,S., Tsudzuki,J., Nakashima,K., Tsudzuki,T., Suzuki,Y., Hamada,A., Ohta,T., Inamura,A., Yoshinaga,K. and Sugiura,M. (1997) Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl Acad. Sci. USA, 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]