Abstract
The primary receptor, the coxsackievirus and adenovirus receptor (CAR), and the secondary receptor, αv integrins, are the tropism determinants of adenovirus (Ad) type 5. Inhibition of the interaction of both the fiber with CAR and the penton base with the αv integrin appears to be crucial to the development of targeted Ad vectors, which specifically transduce a given cell population. In this study, we developed Ad vectors with ablation of both CAR and αv integrin binding by mutating the fiber knob and the RGD motif of the penton base. We also replaced the fiber shaft domain with that derived from Ad type 35. High transduction efficiency in the mouse liver was suppressed approximately 130- to 270-fold by intravenous administration of the double-mutant Ad vectors, which mutated two domains each of the fiber knob and shaft and the RGD motif of the penton base compared with those of conventional Ad vectors (type 5). Most significantly, the triple-mutant Ad vector containing the fiber knob with ablation of CAR binding ability, the fiber shaft of Ad type 35, and the penton base with a deletion of the RGD motif mediated a >30,000-fold lower level of mouse liver transduction than the conventional Ad vectors. This triple-mutant Ad vector also mediated reduced transduction in other organs (the spleen, kidney, heart, and lung). Viral DNA analysis showed that systemically delivered triple-mutant Ad vector was primarily taken up by liver nonparenchymal cells and that most viral DNAs were easily degraded, resulting in little gene expression in the liver. These results suggest that the fiber knob, fiber shaft, and RGD motif of the penton base each plays an important role in Ad vector-mediated transduction to the mouse liver and that the triple-mutant Ad vector exhibits little tropism to any organs and appears to be a fundamental vector for targeted Ad vectors.
Recombinant adenovirus (Ad) vectors are widely used for both in vitro and in vivo gene transfer. However, one of the hurdles confronting Ad-mediated gene transfer is their nonspecific distribution in tissue after in vivo gene transfer. This distribution is largely due to the relatively broad expression of the primary receptor, the coxsackievirus and adenovirus receptor (CAR), and the secondary receptor, αv integrin. This lack of specificity limits the utility of Ad vectors in gene therapy. Vector dissemination may lead to an increased risk of unwanted side effects of the gene therapy procedure, even when Ad vectors are locally administered to the tissue of interest. Targeted Ad vectors would represent a significant advance in the development of safer and more efficient gene delivery and gene therapy (20, 45).
The initial step of Ad infection involves at least two sequential steps. The first is attachment of the virus to the cell surface through binding of the knob domain of the fiber to CAR (5, 42). Following attachment, interaction between the RGD motif of the penton bases and secondary host cell receptors, αv integrins, facilitates internalization via receptor-mediated endocytosis (46, 47). Several strategies have been developed to construct targeted Ad vectors with ablation of CAR binding ability: Ad vectors containing an AB, DE, or FG loop mutation of the fiber knob (1, 6, 8, 16, 24, 32, 37, 40), Ad vectors containing the Ad type 40 short fiber, which has been hypothesized not to bind to any receptors (34), and Ad vectors containing an external trimerization motif instead of the fiber knob (13, 19, 26). Ad vectors with ablation of αv integrin binding have been developed by deleting the RGD motif of the penton base (32, 43).
Several groups have reported that Ad vectors with ablation of CAR or αv integrin binding do not change the biodistribution (especially natural tropism to the liver) and toxicity of Ad vectors (1, 24, 32, 40), although these Ad vectors indeed do not bind with CAR or αv integrin, respectively. Einfeld et al. reported that Ad vectors with both CAR-binding and αv integrin-binding ablation exhibit a >700-fold decrease in liver transduction (8). Ablation of both CAR and αv integrin binding is crucial to the development of targeted Ad vectors. More recently, the fiber shaft has been reported to be involved in the in vivo gene transfer properties of Ad vectors. Nakamura et al. reported that the natural tropism of Ad vectors in vivo is influenced not only by the fiber-CAR interaction but by the length of the fiber shaft (34). They replaced the tail, shaft, and knob domains of Ad type 5 fiber with those of Ad type 40 short fiber (their vectors contained the RGD motif of the penton base) and showed that the transduction efficiency in the liver for the chimeric Ad vectors was approximately 64-fold lower than for conventional Ad vectors. Furthermore, Smith et al. have shown that the KKTK (Lys-Lys-Thr-Lys) motif of the fiber shaft of Ad type 5 is involved in accumulation in the mouse liver of systemically administrated Ad vectors (41). Vigne et al. reported that shortening the Ad type 5 fiber shaft weakens the interactions of both fiber-CAR and penton base-αv integrin, possibly due to steric hindrance (43). Taking these results into account, the combination of the fiber-shaft change to a short fiber shaft without the KKTK motif and the ablation of both CAR and αv integrin binding might further reduce tropism to the liver.
In the present study, we developed a triple-mutant Ad vector containing a mutant fiber knob derived from Ad type 5, with CAR binding ablated, the fiber shaft derived from Ad type 35 (the fiber shaft of Ad type 35, without the KKTK motif, is shorter than that of Ad type 5 [Ad type 35 fiber shaft, 6 β-repeats; Ad type 5 fiber shaft, 22 β-repeats]), the fiber tail derived from Ad type 5, and the mutant penton base of Ad type 5 without the RGD motif. This vector was coupled with a simple method for generating fiber-modified Ad vectors in which oligonucleotides corresponding to the peptide of interest can be introduced into the coding region of both the HI loop and the C terminus of the fiber knob by a simple plasmid construction based on in vitro ligation (18, 33). A unique restriction site was introduced into the HI loop and C-terminal coding region of the fiber knob. We also generated a newer packaging cell line based on 293 cells to amplify the triple-mutant Ad vectors. Wild-type Ad type 5 fiber was stably expressed in the packaging cells. The in vitro and in vivo gene transfer properties of mutant Ad vectors were evaluated.
MATERIALS AND METHODS
Cells.
SK HEP-1 (endothelial cell line derived from the human liver) (10), LN319 (human anaplastic astrocytoma), LN444 (human glioblastoma multiforme), SF295 (human glioblastoma multiforme) (kindly provided by M. Tada, Hokkaido University, Hokkaido, Japan) (4), and 293 cells were cultured with Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
Fiber-293 cells were stable transformants generated by transfection of pCMVmfiber-Hyg (described below) into 293 cells and selection with hygromycin (GIBCO-BRL, Rockville, Md.).
Plasmids.
The plasmid pCMVmfiber-Hyg, which contains a wild-type fiber gene and a hygromycin resistance gene, was constructed as follows. pEco-ITR5 (33), which contains the EcoRI/ClaI fragment of an Ad type 5 genome (from bp 27331 to the right end of the Ad type 5 genome [Δ28133-30818]), was digested with MunI and ClaI after an NsiI site located downstream of the 3′ inverted terminal repeat (ITR) of the Ad genome was changed into a ClaI site by using NsiI phosphorylated linkers (New England Biolabs, Beverly, Mass.) and was ligated with oligonucleotides 5′-AATTGCCGAGAACTTCAAGTCCTTCTTCATCCAGTAGGCGGCCGCAT-3′ and 5′-CGATGCGGCCGCCTACTGGATGAAGAAGGACTTGAAGTTCTCGGC-3′. The resulting plasmid, pFiber1-1, was digested with StuI and HindIII and ligated with oligonucleotides 5′-aCTgTACcTGTTcACcGCcTCcAACAAcagCAAgA-3′ and 5′-AGCTTcTTGctgTTGTTgGAgGCgGTgAACAgGTAcAGt-3′ (silent mutation sequences are shown in lowercase). The resulting plasmid, pFiber1-2, was digested with XbaI and NdeI and ligated with oligonucleotides 5′-CTAGGAATTCGCCCACCATGAAGCGCGCcAGACCcTCcGAgGAcACCTTCAACCCCGTGTAcCCA-3′ and 5′-TATGGgTACACGGGGTTGAAGGTgTCcTCgGAgGGTCTgGCGCGCTTCATGGTGGGCGAATTC-3′. The resulting plasmid, pFiber1-3, was digested with PpuMI and NheI and ligated with oligonucleotides 5′-GACCCCTgACcGTGTCcGAgGGcAAa-3′ and 5′-CTAGtTTgCCcTCgGACACgGTcAGGGG-3′. The resulting plasmid, pFiber1-4, was digested with BstXI and BglII and ligated with oligonucleotides 5′-TTGGAAtTTcAGgAAcGGc-3′ and 5′-GATCgCCgTTcCTgAAaTTCCAATATT-3′. The resulting plasmid, pFiber1-5, was digested with EcoRI and NotI and ligated with the EcoRI/NotI fragment of pCMVSL3 (50). The resulting plasmid, pCMVmfiber, contained the sequence of the cytomegalovirus (CMV) promoter-enhancer, intron A, Ad type 5 fiber (with a total of 28 bp changed as follows: 6 mutations between bp 1 and 45 of the fiber gene, 6 mutations between bp 447 and 472 of the fiber gene, 11 mutations between bp 918 and 954 of the fiber gene, and 5 mutations between bp 1437 and 1449 of the fiber gene), simian virus 40 (SV40) poly(A), and the SV40 enhancer. The plasmid vector containing the sequence of the CMV promoter-enhancer, intron A, SV40 poly(A), and the SV40 enhancer is much more efficient than the conventional vector containing the sequence of the CMV promoter-enhancer and SV40 or bovine growth hormone poly(A) (50). Finally, the hygromycin resistance gene derived from pCMVLacI(Stratagene, La Jolla, Calif.) was inserted downstream from SV40 poly(A) and the SV40 enhancer of pCMVmfiber, resulting in pCMVmfiber-Hyg.
The vector plasmid, pAdHM54, which we used for the generation of Ad vectors with ablation of CAR binding of the Ad type 5 fiber knob, the Ad type 35 fiber shaft, and the Ad type 5 penton base with a deletion of the RGD motif, was constructed as follows. pF35-2.3 was constructed by self-ligation of AgeI/XbaI-digested pF35-2.2 (28), which contains the sequence surrounding the Ad type 35 fiber gene, after an AgeI site of pF35-2.2 was changed into an XbaI site by use of XbaI phosphorylated linkers (New England Biolabs). An AseI site was created between the fiber shaft and the knob coding sequence by use of the QuikChange site-directed mutagenesis kit (Stratagene) with oligonucleotides 5′-AAAGGATAGTATTAAtACCTTATGGACTGGA-3′ and 5′-TCCAGTCCATAAGGTaTTAATACTATCCTTT-3′ (silent mutation sequences are lowercase and the AseI site sequence is underlined), resulting in pF35-2.3(AseI). pGEM-Teasy-knobCAR(−)-F was constructed by inserting PCR fragments generated by using primers (sense, 5′-AATAATACTTTGTGGACCACACCAGCT-3′; antisense: 5′-TTCGAAGTTGTGTCTCCTGT-3′) and pAdHM42 (as a template), which is a derivative of pAdHM41 (18), into pGEM-T Easy (Promega, Madison, Wis.). pHM-S35-K5-CAR(−) was constructed by four-piece ligation of the following fragments: (i) AflII/AseI fragment of pF35-2.3(AseI), (ii) AscI/Csp45I fragment of pGEM-Teasy-knobCAR(−)-F, (iii) Csp45I/MunI fragment of pHM14-Eco3 (18), and (iv) MunI/AflII fragment of pHMCMV6 (31). Next, pHM14-Eco2-S35 was generated by the ligation of SrfI/MunI fragments of pS35-K5-2.2CAR(−) and pHM14-Eco2 (18). Finally, the EcoRI/ClaI fragment of pHM14-Eco2-S35 was ligated with the EcoRI/ClaI fragment of pAdHM43, which has chimeric fiber sequences derived from pAdHM26 and deletion of the coding region of the RGD motif of the penton base derived from pAdHM32 (29), resulting in pAdHM54. pAdHM54 carries a complete Ad genome with deletions of the E1 and E3 regions, with I-CeuI, SwaI, and PI-SceI sites in the E1 deletion region, the deletion of the RGD peptide-coding sequence of the penton base (MNDHAIRGDTFATRAE was changed to MNDTSRAE), the chimeric fiber-coding sequence of the Ad type 5 fiber knob with CAR binding ablated (deletion of the FG-loop-coding region of the fiber protein [T489, A490, Y491, and T492 of the fiber knob protein]), and the Ad type 35 fiber shaft sequences. pAdHM54 also contains unique Csp45I and ClaI sites in the HI loop and the C-terminal end of the fiber knob-coding sequence, respectively. The vector plasmid pAdHM52, which contains the same chimeric fiber (knob, shaft, and tail)-coding sequence as pAdHM54 and the wild-type penton base-coding sequence, and pAdHM43, which contains the same chimeric fiber knob-coding sequence as pAdHM54, the Ad type 5 fiber shaft-coding sequence, and deletion of the RGD peptide-coding sequence of the penton base, were constructed similarly. All mutations of the fiber and penton base-coding sequences were checked by sequencing.
Virus.
The Ad vectors were constructed by means of an improved in vitro ligation method that was described previously (30, 31). Luciferase-expressing Ad vector plasmids (pAdHM54-CMVL2, pAdHM43-CMVL2, and pAdHM52-CMVL2) were constructed by ligating I-CeuI/PI-SceI-digested pAdHM54, pAdHM43, or pAdHM52, respectively, with I-CeuI/PI-SceI-digested pCMVL1 (33), in which the luciferase gene is cloned into pHMCMV6 (31). pAdHM54-CMVL2, pAdHM43-CMVL2, and pAdHM52-CMVL2 were digested with PacI and purified by phenol-chloroform extraction and ethanol precipitation. Linearized DNAs were transfected into 293 cells (in the case of pAdHM52-CMVL2) or Fiber-293 cells (in the case of pAdHM54-CMVL2 and pAdHM43-CMVL2) with SuperFect (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Viruses (Ad/ΔFΔP-S35-L2, Ad/ΔFΔP-L2, and Ad/ΔF-S35-L2, respectively) were prepared by standard methods, with the exceptions that Ad/ΔFΔP-S35-L2 and Ad/ΔFΔP-L2 were amplified in Fiber-293 cells and that only the last step of viral amplification was performed by the infection of normal 293 cells. A conventional luciferase-expressing Ad vector, Ad-L2, had been constructed previously (33). Determination of virus particle titers was accomplished spectrophotometrically by the methods of Maizel et al. (27). Virus particle titers of the vector stocks, prepared from five 150-mm-diameter dishes (approximately 8 × 107 cells), were as follows: Ad-L2, 3.9 × 1012 vector particles (VP)/ml; Ad/ΔFΔP-S35-L2, 3.4 × 1012 VP/ml; Ad/ΔFΔP-L2, 1.4 × 1012 VP/ml; and Ad/ΔF-S35-L2, 2.5 × 1012 VP/ml.
Western blotting.
Protein samples were prepared by the incubation of cell pellets of Fiber-293 or 293 cells in the presence of 20 mM HEPES (pH 7.5), 2 mM EGTA, 10% glycerol, 1% Triton X-100, 5 mM dithiothreitol, and 2 mM phenylmethylsulfonyl fluoride on ice for 30 min. After boiling for 5 min, 10 μg of total protein in 1× sample buffer with 4% β-mercaptoethanol was separated in a sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE) gel, followed by electrotransfer to a nitrocellulose membrane. In the case of virus samples, 200 ng of virus in 1× sample buffer containing 4% β-mercaptoethanol was loaded on the SDS-PAGE gel after boiling for 5 min, followed by electrotransfer to a nitrocellulose membrane. After blocking in Block Ace (Dainippon Pharmaceuticals, Osaka, Japan), the filters were incubated with a rabbit fiber knob polyclonal antibody (1:3,000) (kindly provided by R. D. Gerard [Southwestern Medical Center, The University of Texas, Dallas])(12), followed by incubation in the presence of peroxidase-labeled anti-rabbit antibody (1:10,000). Filters were developed by chemiluminescence (ECL Western blotting detection system; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). The signals were read by using an LAS-3000 machine (FUJIFILM, Tokyo, Japan) and were quantified by using Image Gauge software (FUJIFILM).
Adenovirus-mediated gene transduction into cultured cells.
Cells (104 cells) were seeded into a 96-well dish. On the following day, they were transduced with Ad-L2, Ad/ΔFΔP-S35-L2, Ad/ΔFΔP-L2, or Ad/ΔF-S35-L2 (300 or 3,000 VP/cell) for 1.5 h. After a 48-h culture period, luciferase production in the cells was measured by using a luciferase assay system (PicaGene LT2.0; Toyo Inki, Tokyo, Japan).
Adenovirus-mediated gene transduction in vivo.
Ad-L2, Ad/ΔFΔP-S35-L2, Ad/ΔFΔP-L2, or Ad/ΔF-S35-L2 (3.0 × 1010 VP) was intravenously injected into C57BL/6 mice (5-week-old males; Nippon SLC, Shizuoka, Japan). Forty-eight hours later, the hearts, lungs, livers, kidneys, and spleens were isolated and homogenized as previously described (50). Luciferase production was determined by using a luciferase assay system (PicaGene 5500; Toyo Inki). Protein content was measured with a Bio-Rad assay kit (Bio-Rad, Hercules, Calif.), with bovine serum albumin as a standard.
Slot-blot assay to determine blood clearance of Ad vectors.
Blood samples were collected by retro-orbital bleeding at the indicated times (2, 5, 10, 15, 30, and 60 min) following intravenous administration of Ad-L2 or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP/mouse). Total DNA, including the Ad vector DNA, was isolated from whole blood by use of the QIAamp DNA blood Mini kit (Qiagen). The Ad vector DNA standards were similarly isolated from mouse whole blood mixed with equivalent amounts of Ad vectors. The estimated total blood volume of each mouse was 1.26 ml (7.3% of body weight) (2). Six hundred nanograms of the total DNA was mixed with 0.8 N NaOH, and the mixture was mixed vigorously on a vortex machine. After a 10-min incubation at room temperature, 2 N ammonium acetate was added to neutralize the mixture. The mixtures were slot blotted onto a positively charged nylon membrane (Hybond N+; Amersham Pharmacia Biotech) with Bio-Dot SF (Bio-Rad). The membrane was submerged in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for washing, and the DNA was cross-linked to the membrane. A luciferase-specific probe, which is an XbaI/NotI fragment of pCMVL1 (33), was labeled by AlkPhos Direct (Amersham Pharmacia Biotech). Prehybridization and hybridization were performed according to the manufacturer's instructions. The signals were read with a FluorImager 595 (Molecular Dynamics) and quantified by using ImageQuant software (Molecular Dynamics).
Slot-blot assay to determine liver accumulation of Ad vectors.
Livers were recovered from mice 1 or 48 h after intravenous injection of Ad-L2 or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP/mouse). Total DNA, including the Ad vector DNA, was isolated from the livers with a Tissue DNeasy kit (Qiagen). Two micrograms of the total DNA was subjected to slot-blot analysis as described above. The Ad vector DNA standards were similarly prepared from an aliquot of naive mouse liver mixed with equivalent amounts of Ad vectors.
Amounts of Ad vector DNA in liver PC and NPC.
Mice were intravenously administered Ad-L2 or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP/mouse). Mice were anesthetized by peritoneal administration of pentobarbital sodium 1 or 48 h after Ad vector injection. The liver cells were separated into parenchymal cells (PC) (hepatocytes) and nonparenchymal cells (NPC) (Kupffer cells and endothelial cells) as described previously (35). In brief, the liver was perfused with HEPES buffer (pH 7.5) containing collagenase. The dispersed cells were separated into PC and NPC fractions by differential centrifugation. Semiquantitative PCR was performed to examine the amounts of Ad vector DNA in the PC and NPC. Total DNA, including the Ad vector DNA, was isolated from the PC and NPC with the Tissue DNeasy kit (Qiagen). The DNA was subjected to semiquantitative PCR. PCR analysis of luciferase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed in a 50-μl reaction mixture containing 10 ng of total DNA, 1.25 U of AmpliTaq DNA polymerase, 1.5 mM MgCl2, and 0.2 mM deoxynucleoside triphosphates, using GeneAmp PCR core reagents (Perkin-Elmer, Norwalk, Conn.). The sequences of the primers for luciferase (3) and GAPDH (36) were as follows: for luciferase, forward (5′-GCGCCATTCTATCCGCTGGA-3′) and reverse (5′-CTATCGAAGGACTCTGGCAC-3′); for GAPDH, forward (5′-ACCACAGTCCATGCCATCAC-3′) and reverse (5′-TCCACCACCCTGTTGCTGTA-3′). The following parameters were used: (i) for luciferase, 60 s at 94°C, 30 s at 60°C, and 120 s at 72°C for 25, 30, or 35 cycles; (ii) for GAPDH, 45 s at 94°C, 60 s at 55°C, and 90 s at 72°C for 25 cycles. The PCR products were electrophoresed in 2.0% agarose gels.
RESULTS
Generation of several mutant Ad vectors.
To examine the contribution of the Ad fiber knob, fiber shaft, and RGD motif of the penton base to gene transfer in vitro and in vivo, we constructed several mutant Ad vectors expressing luciferase. Ad/ΔFΔP-L2 contains the Ad type 5 fiber knob with a four-amino-acid deletion of the FG loop and has a deletion of the RGD motif of the penton base. Ad/ΔF-S35-L2 contains the Ad type 5 fiber knob with deletion of the FG loop and the Ad type 35 fiber shaft. Ad/ΔFΔP-S35-L2 contains the Ad type 5 fiber knob with deletion of the FG loop and the Ad type 35 fiber shaft and has a deletion of the RGD motif of the penton base. Ad-L2 is a conventional Ad vector (Table 1). All mutations of the triple-mutant Ad vector, Ad/ΔFΔP-S35-L2, are summarized in Table 2 (also see Materials and Methods).
TABLE 1.
Ad vectors used for the present study
| Vector | Status of FG loop of fiber knob | Status of RGD motif of penton base | Type of fiber shaft |
|---|---|---|---|
| Ad-L2 | Intact | Intact | 5 |
| Ad/ΔFΔP-L2 | Mutation | Mutation | 5 |
| Ad/ΔF-S35-L2 | Mutation | Intact | 35 |
| Ad/ΔFΔP-S35-L2 | Mutation | Mutation | 35 |
TABLE 2.
Amino acid changes in triple-mutant Ad vector
| Vector | Sequence of penton base | Ad type of tail | Ad type of shaft | Amino acid sequence (nucleotide sequence) of knob domain
|
||
|---|---|---|---|---|---|---|
| FG loop | HI loop | C terminus | ||||
| Conventional Ad | MND-HAIRGDTFAT-RAE | 5 | 5 (22 β-repeats) | TEGTAYTNAV | DTTPSA (GAC ACA ACT CCA AGT GCA) | QE-Stop (CAA GAA TAA) |
| Triple-mutant Ad | MND-TS-RAE (RGD motif deleted) | 5 | 35 (6 β-repeats) | TEGNAV (aa 489 to 492 deleted) | DTTSNPSA (GAC ACA ACT TCG AAC CCA AGT GCA)a | QEID-Stop (CAA GAA ATC GAT TAA)b |
Underlining indicates an added Csp45I site. Two amino acids (S and N) were added to the sequence.
Underlining indicates an added ClaI site. Two amino acids (I and D) were added to the sequence.
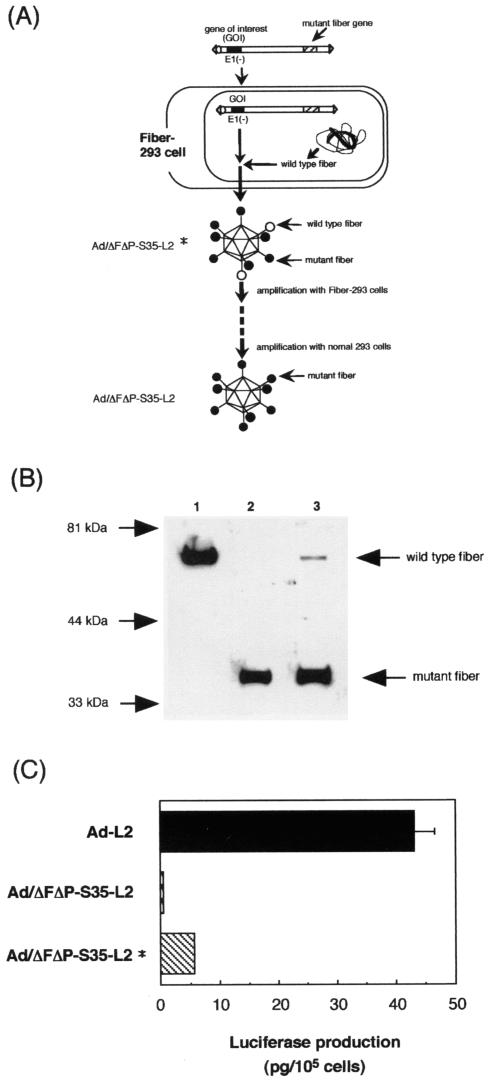
Because Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2 could not be generated in normal 293 cells due to the lack of interaction of the virus and cellular receptors (CAR or αv integrin), 293 cells expressing Ad type 5 fiber protein (Fiber-293 cells) were used as the packaging cell line. Fiber-293 cells were constructed by transfection of fiber-expressing plasmids (pCMVmfiber-Hyg) into 293 cells. Western blot analysis showed that Fiber-293 cells expressed fiber proteins (Fig. 1). The fiber gene (1,746 bp) in pCMVmfiber-Hyg contains a total of 28 silent mutations at least every 500 bp to reduce the possibility of reversion of the mutant fiber gene in the virus to the wild-type fiber gene due to recombination of the mutant fiber gene in the virus and the fiber gene integrated into Fiber-293 cells. When Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2 are amplified in Fiber-293 cells, some of the fiber proteins of Ad/ΔFΔP-L2* and Ad/ΔFΔP-S35-L2* produced from Fiber-293 cells should be wild-type Ad type 5 fiber proteins which were derived from Fiber-293 cells (the viruses produced by Fiber-293 cells were named Ad/ΔFΔP-L2* and Ad/ΔFΔP-S35-L2*). Ad/ΔFΔP-L2* and Ad/ΔFΔP-S35-L2* should infect Fiber-293 cells through the interaction of the wild-type fiber protein and CAR. Ad/ΔFΔP-L2* and Ad/ΔFΔP-S35-L2* were successfully amplified in the Fiber-293 cells. At the final stage of viral amplification, Ad/ΔFΔP-L2* and Ad/ΔFΔP-S35-L2* were allowed to infect normal 293 cells. The recovered viruses, Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2, should contain only mutant fiber proteins (Fig. 2A).
FIG. 1.
Western blot analysis of 293 cells transduced with Ad-L2, normal 293 cells, and Fiber-293 cells. The protein sample was prepared from normal 293 (lane 2) and Fiber-293 (lane 3) cells. The protein sample was also prepared from 293 cells with CPE, which were transduced with Ad-L2 and cultured for 1 day, as a positive control (lane 1). Samples were separated on an SDS-12% PAGE gel and analyzed by Western blotting using a rabbit fiber knob polyclonal antibody.
FIG. 2.
Diagram of the generation of mutant Ad vectors. (A) Diagram of the generation of Ad/ΔFΔP-S35-L2 using Fiber-293 cells. PacI-linearized pAdHM54-CMVL2 (a vector plasmid for generation of Ad/ΔFΔP-S35-L2) was transfected into Fiber-293 cells and cultured for 12 days. Virus was recovered from cells with CPE and amplified in Fiber-293 cells. Some of the fiber proteins of Ad/ΔFΔP-S35-L2* prepared from Fiber-293 cells are wild-type Ad type 5 fiber, which is derived from Fiber-293 cells. Ad/ΔFΔP-S35-L2* should infect Fiber-293 cells or normal 293 cells through the wild-type Ad type 5 fiber. At the final stage of viral amplification, Ad/ΔFΔP-S35-L2* was allowed to infect normal 293 cells. Recovered virus (Ad/ΔFΔP-S35-L2) should contain only mutant fiber proteins. Ad/ΔFΔP-S35-L2* is the virus recovered from Fiber-293 cells. Ad/ΔFΔP-S35-L2 is the virus recovered from normal 293 cells. (B) Western blot analysis of Ad-L2, Ad/ΔFΔP-S35-L2, and Ad/ΔFΔP-S35-L2*. Two hundred nanograms of virus was separated on an SDS-12% PAGE gel and analyzed by Western blotting using a rabbit fiber knob polyclonal antibody as described in Materials and Methods. Lane 1, Ad-L2; lane 2, Ad/ΔFΔP-S35-L2; lane 3, Ad/ΔFΔP-S35-L2*. (C) Comparison of luciferase production in SK HEP-1 cells transduced with Ad-L2, Ad/ΔFΔP-S35-L2*, or Ad/ΔFΔP-S35-L2. SK HEP-1 cells were transduced with 300 VP of Ad-L2, Ad/ΔFΔP-S35-L2*, or Ad/ΔFΔP-S35-L2 per cell for 1.5 h. After incubation for 48 h, luciferase production was measured by a luminescent assay. The data are expressed as means ± standard deviations (SD) (n = 4).
In order to determine the ratio of the mutant fibers, which are derived from viral DNA, to the wild-type Ad type 5 fibers, which are derived from Fiber-293 cells, on the virus particles of Ad/ΔFΔP-S35-L2*, Western blot analysis of the viral protein was performed (Fig. 2B). The mutant and wild-type fibers are easily distinguished because mutant fibers are smaller than wild-type fibers due to the small size of the Ad type 35 fiber shaft. Quantified analysis showed that the ratio of mutant fibers to wild-type fibers of Ad/ΔFΔP-S35-L2* was 1 to 50, suggesting that about one fiber protein per one or two virions is the wild type. Ad/ΔFΔP-S35-L2 did not contain the wild-type fiber.
Next, in order to examine whether Ad/ΔFΔP-S35-L2* indeed has a higher transduction efficiency than does Ad/ΔFΔP-S35-L2, luciferase production in SK HEP-1 cells (CAR positive) transduced with Ad/ΔFΔP-S35-L2*, Ad/ΔFΔP-S35-L2, or Ad-L2 was compared (Fig. 2C). The data indicate that Ad/ΔFΔP-S35-L2* mediated approximately 10 times more luciferase production than did Ad/ΔFΔP-S35-L2, although its production was less than that mediated by Ad-L2. These results suggest that only some of the fiber protein in Ad/ΔFΔP-S35-L2* is wild type and that the higher luciferase production in the cells transduced with Ad/ΔFΔP-S35-L2* is likely mediated via wild-type fiber-CAR interactions.
Ad/ΔF-S35-L2 was generated in normal 293 cells, probably because Ad/ΔF-S35-L2 infected the cells via interaction of the RGD motif of the penton base and αv integrin. All of the mutant Ad vectors used for this study were readily propagated, with similar particle titers to those of the control virus, Ad-L2 (see Materials and Methods), although in the mutant vectors a delay was observed before full cytopathic effect (CPE) was reached (CPE of Ad-L2 was observed at 2 days postinfection, while that of the mutant vectors was usually observed at 3 days postinfection.).
Gene transfer in vitro.
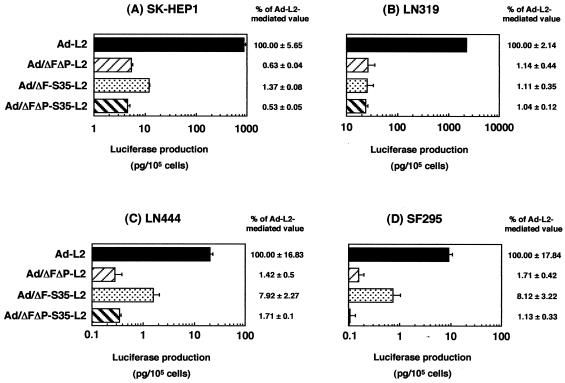
We compared the gene transfer activity in various types of human cells of Ad/ΔFΔP-L2, Ad/ΔF-S35-L2, and Ad/ΔFΔP-S35-L2 with the activity of conventional Ad vector Ad-L2 (Fig. 3). SK HEP-1 and LN319 cells are CAR positive, while SF295 and LN444 cells are CAR negative. All cell types expressed αv integrin (4, 17, 33). In the CAR-positive cells, Ad-L2 mediated higher levels of luciferase production than Ad/ΔFΔP-S35-L2, Ad/ΔFΔP-L2, and Ad/ΔF-S35-L2, which mediated only approximately 1% of the luciferase production of Ad-L2. In CAR-negative cells, Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2 mediated 1 to 2% of Ad-L2 luciferase production and Ad/ΔF-S35-L2 mediated approximately 8% of Ad-L2 luciferase production (note that the absolute level of luciferase production of Ad/ΔF-S35-L2 in CAR-negative cells was lower than that in CAR-positive cells) (Fig. 3). The relatively higher luciferase production of Ad/ΔF-S35-L2 in CAR-negative cells suggested that the interaction of the RGD motif of the penton base and αv integrin plays a role in gene transfer. Since Ad/ΔF-L2, which contains a fiber knob with CAR binding activity ablated and an Ad type 5 fiber shaft, showed 30 to 46% of the luciferase production of Ad-L2 in CAR-negative cells in our previous report (32), substitution of the Ad type 35 fiber shaft for that of Ad type 5 also effects reduced gene transfer. No significant difference was observed between Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2, suggesting that the substitution of the fiber shaft domain has little effect on gene transfer in vitro when the fiber shaft exchange is performed in Ad vectors with both CAR and αv integrin binding ablated.
FIG. 3.
Comparison of luciferase production in several human cell types transduced with Ad-L2, Ad/ΔFΔP-L2, Ad/ΔF-S35-L2, or Ad/ΔFΔP-S35-L2. SK HEP-1, LN319, LN444, or SF295 cells were transduced with 3,000 VP of Ad-L2, Ad/ΔFΔP-L2, Ad/ΔF-S35-L2, or Ad/ΔFΔP-S35-L2 per cell for 1.5 h. After incubation for 48 h, luciferase production was measured by a luminescent assay. The data are expressed as means ± SD (n = 4). The relative expression levels were calculated by designating the value of Ad-L2 as 100. Mean background values of luciferase production in each cell type are as follows: SK HEP-1, 0.004; LN319, 0.023; LN444, 0.001; SF295, 0.001 (pg/105 cells).
Gene transfer in vivo.
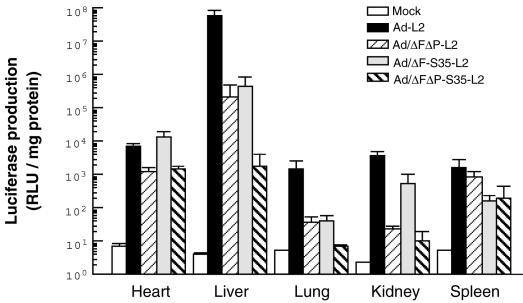
Next, in order to examine whether natural Ad tropism to the liver can be suppressed by the mutant Ad vectors, we intravenously administered each Ad vector (3.0 × 1010 VP) to mice and measured luciferase production in the organ (Fig. 4). Although we previously reported that Ad vectors with CAR and αv integrin binding ablated do not reduce liver transduction (32), Ad vectors with ablation of both CAR binding and αv integrin binding (Ad/ΔFΔP-L2) mediated approximately 270-fold lower liver transduction than Ad-L2. This finding is consistent with the report of Einfeld et al. (8). Ad/ΔF-S35-L2 also exhibited approximately 130-fold lower luciferase production in the liver than Ad-L2. More interestingly, Ad/ΔFΔP-S35-L2 mediated >30,000-fold lower liver transduction than did Ad-L2, suggesting that the fiber shaft domain plays some role in Ad tropism to the liver. A similar pattern was observed for the lung, although the absolute level of luciferase production was much lower. For the kidney, the penton base modification appears to be the most important. Ad/ΔFΔP-S35-L2 also mediated 1-log reduced luciferase production in the spleen and heart compared with Ad-L2. These results indicate that the fiber knob, fiber shaft, and RGD motif of the penton base each play an important role in Ad vector-mediated transduction to the mouse liver and that the triple-mutant Ad vector exhibits little tropism to any other organs.
FIG. 4.
Luciferase production in mice after the systemic administration of Ad-L2, Ad/ΔFΔP-L2, Ad/ΔF-S35-L2, or Ad/ΔFΔP-S35-L2. Ad-L2, Ad/ΔFΔP-L2, Ad/ΔF-S35-L2, or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP) was intravenously injected into mice. Forty-eight hours later, the hearts, lungs, livers, kidneys, and spleens were isolated, and luciferase production was measured by a luminescent assay. All data represent the means ± standard errors for five mice.
The fate of Ad vectors after intravenous administration.
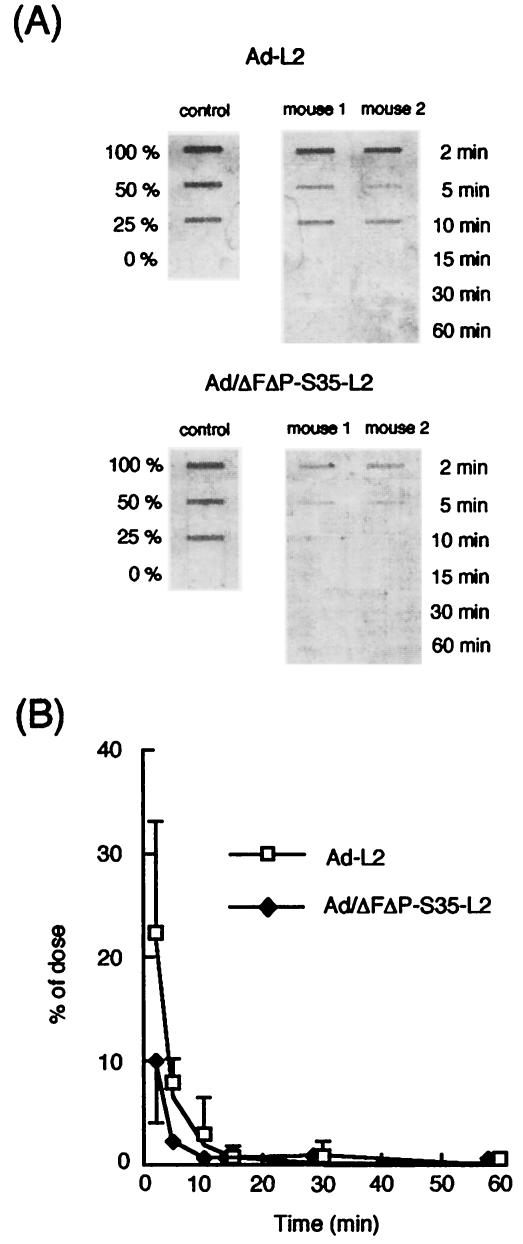
In order to examine the fate of systemically administered Ad/ΔFΔP-S35-L2, blood clearance rates of Ad-L2 and Ad/ΔFΔP-S35-L2 for mice were evaluated by slot-blot analysis, with luciferase cDNA as a probe. Blood clearance curves for Ad-L2 and Ad/ΔFΔP-S35-L2 were similar and showed a rapid decrease of the Ad vectors in the blood (Fig. 5). The half-life of Ad-L2 and Ad/ΔFΔP-S35-L2 in the blood was <2 min. Negligible levels of the Ad vectors remained in the blood 30 min after injection. The clearance rate of Ad-L2 was consistent with those observed in previous studies (2, 15) in which blood clearances of Ad type 5 vectors were determined by measuring the titers of Ad type 5 vectors circulating in the blood after injection. These results suggest that Ad/ΔFΔP-S35-L2 as well as Ad-L2 is rapidly cleared from the bloodstream and should be delivered to certain organs.
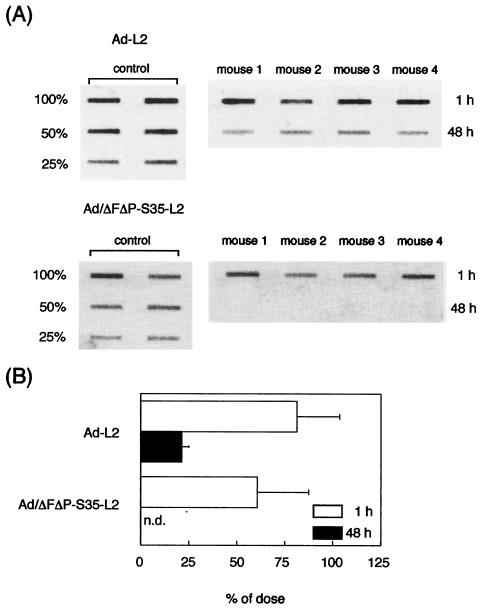
FIG. 5.
Blood clearance of Ad-L2 and Ad/ΔFΔP-S35-L2 after intravenous administration into mice. (A) Fluorimager image for determination of concentrations in blood of Ad-L2 and Ad/ΔFΔP-S35-L2. (B) Concentrations in blood of Ad-L2 and Ad/ΔFΔP-S35-L2 after intravenous administration into mice. Ad-L2 and Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP) were intravenously injected, and blood was drawn from the retro-orbital at the indicated times postinjection. Total DNA, including the Ad vector DNA, was isolated from the blood, and slot-blot analysis was then performed as described in Materials and Methods. All data represent the mean ± SD of four mice.
Since the systemically injected conventional Ad vectors are delivered to the liver (14, 48), we next determined the amounts of Ad/ΔFΔP-S35-L2 DNA in the liver in comparison with Ad-L2 DNA by slot-blot analysis. The amounts of viral DNA were measured 1 and 48 h after injection (Fig. 6). Eighty-one percent of the input Ad-L2 DNA had accumulated in the liver 1 h after injection. More than 60% of Ad/ΔFΔP-S35-L2 DNA was also detected in the liver 1 h after injection. In contrast, a clear difference in the amount of Ad vector DNA in the liver was observed at 48 h postadministration. Twenty-one percent of the input Ad-L2 DNA was detected in the liver, while Ad/ΔFΔP-S35-L2 DNA was not detected 48 h after injection. These data suggest that both Ad-L2 and Ad/ΔFΔP-S35-L2 are predominantly delivered to the liver after intravenous administration and that the Ad/ΔFΔP-S35-L2 DNA degrades more rapidly in the liver than Ad-L2 DNA.
FIG. 6.
Liver accumulation of Ad-L2 and Ad/ΔFΔP-S35-L2 1 and 48 h after intravenous administration into mice. (A) Fluorimager image for determination of accumulation in the liver of Ad-L2 and Ad/ΔFΔP-S35-L2. (B) Liver accumulation of Ad-L2 and Ad/ΔFΔP-S35-L2 after intravenous administration into mice. Livers were recovered from mice 1 or 48 h after intravenous injection of Ad-L2 or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP/mouse). Total DNA, including the Ad vector DNA, was isolated from the liver, and slot-blot analysis was then performed as described in Materials and Methods. All data represent the means ± SD for four mice. n.d., not detectable.
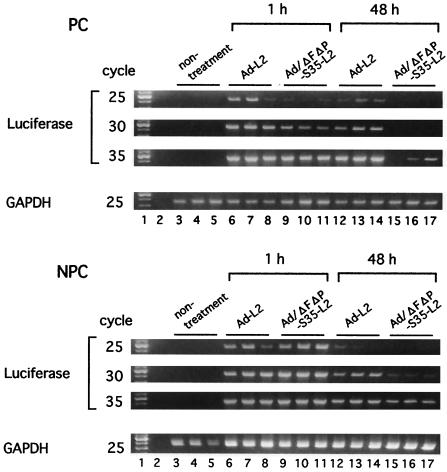
Next, to determine why there is a big difference between luciferase production and viral DNA accumulation in the liver (48 h after injection) in the case of Ad/ΔFΔP-S35-L2 and why Ad/ΔFΔP-S35-L2 DNA is more rapidly degraded in the liver than Ad-L2 DNA, the cellular distributions of Ad-L2 and Ad/ΔFΔP-S35-L2 in the liver were examined. The amounts of Ad-L2 and Ad/ΔFΔP-S35-L2 delivered to the PC (hepatocytes) and NPC (Kupffer cells and endothelial cells) 1 and 48 h after injection were examined by semiquantitative PCR analysis (Fig. 7). Similar amounts of Ad-L2 DNA were detected in the PC and NPC at 1 and 48 h postinjection, although the amounts of Ad-L2 DNA at 48 h were less than those at 1 h. Ad-L2 is likely to be equally distributed to the PC and NPC after injection at a dose of 3.0 × 1010 VP/mouse. Ad-L2 DNA in the NPC was more susceptible to degradation than that in the PC. In contrast, Ad/ΔFΔP-S35-L2 DNA accumulated more in the NPC at both 1 and 48 h postinjection than in the PC, and the amount of Ad/ΔFΔP-S35-L2 DNA at 48 h postinjection was much less than that at 1 h in both PC and NPC. These results suggest that the NPC contribute more to the hepatic uptake of Ad/ΔFΔP-S35-L2 than the PC and that Ad/ΔFΔP-S35-L2 DNA delivered to the NPC is easily degraded.
FIG. 7.
Semiquantitative PCR analysis for Ad-L2 and Ad/ΔFΔP-S35-L2 in liver PC and NPC. Mice were intravenously administered Ad-L2 or Ad/ΔFΔP-S35-L2 (3.0 × 1010 VP/mouse). Collagenase perfusion was performed 1 or 48 h after Ad vector injection to separate the cells. Total DNA, including the Ad vector DNA, was isolated from the cells, and semiquantitative PCR was then performed as described in Materials and Methods. Lane 1, 100-bp ladder; lanes 3 to 5, nontreatment cells; lanes 6 to 11, collagenase perfusion 1 h after Ad vector injection to separate the cells; lanes 12 to 17, collagenase perfusion 48 h after Ad vector injection to separate the cells.
DISCUSSION
For the development of targeted Ad vectors, the construction of vectors that abolish (or reduce) natural viral tropism is a first step. Identification and incorporation of a foreign ligand (i.e., peptide) that has high affinity with the specific cellular receptor into the capsid of Ad vectors that no longer infect cells are the next steps. This study was undertaken to develop vectors that would be functional for the first step. The triple-mutant Ad vector containing the fiber knob with CAR binding ablated, the fiber shaft of Ad type 35, and the penton base with a deletion of the RGD motif is the outcome of our study to reduce natural viral tropism. This vector mediated levels of liver transduction that were >30,000-fold lower than a conventional Ad vector when it was systemically injected into mice (Fig. 4). This vector showed more restricted liver transduction than the vectors reported by Einfeld et al. (8) and Nakamura et al. (34), which mutated two domains each of the fiber knob and shaft and the RGD motif of the penton base, respectively. Einfeld et al. have developed Ad vectors with ablation of both CAR and αv integrin binding from the Ad type 5 fiber shaft, which shows approximately 700-fold lower liver transduction (8), while Nakamura et al. have developed vectors containing the Ad type 40 short fiber (hypothesized not to bind to any receptors) with an intact penton base, which shows 64-fold lower liver transduction (34).
The Ad type 5-based vector delivers the foreign gene predominantly in the liver after intravenous injection into mice (14, 48). This Ad tropism to the liver is considered to be involved in both the interaction of viral components and cellular receptors (e.g., the fiber CAR and the RGD motif of the penton base αv integrin) (8) and the anatomical properties of the liver sinusoid (9). Several groups, including us, have reported that Ad vectors with CAR binding ablated, which mutate the AB, DE, or FG loop of the fiber knob, do not change systemic gene transfer properties (1, 24, 32, 40), although Einfeld et al. have shown that Ad vectors with CAR binding ablated containing a mutation of the AB loop of the fiber knob exhibited a 10-fold decrease in liver transduction compared to CAR-binding Ad vectors (8). Ad vectors with αv integrin binding ablated also show similar or only slightly decreased liver transduction compared with conventional Ad vectors (32). However, the present study shows that Ad vectors with ablation of both CAR and αv integrin binding mediate approximately 270-fold less liver transduction than conventional Ad vectors, which is consistent with the report of Einfeld et al. (8). Furthermore, because the length of the fiber shaft (34, 39, 43) and the KKTK motif of the Ad type 5 fiber shaft (41) have been reported to influence the gene transfer of Ad vectors, we also replaced the Ad type 5 fiber shaft with the Ad type 35 fiber shaft, which is short (Ad type 35 fiber shaft, 6 β-repeats; Ad type 5 fiber shaft, 22 β-repeats) and does not have the KKTK motif, in the Ad vectors with ablation of both CAR and αv integrin binding. As a result, we succeeded in developing a triple-mutant vector (Ad/ΔFΔP-S35-L2) which drastically reduces natural viral tropism to the mouse liver. Double-mutant Ad vectors (both Ad/ΔFΔP-L2 and Ad/ΔF-S35-L2), which had mutations of two domains of the fiber knob, the fiber shaft, and the penton base, showed intermediate liver transduction between that of the conventional Ad vector (Ad-L2) and that of the triple-mutant Ad vector (Ad/ΔFΔP-S35-L2) (Fig. 4). Ad tropism is determined by at least three factors: the fiber knob, the fiber shaft, and the RGD motif of the penton base. Interestingly, an in vitro transduction experiment showed that Ad/ΔFΔP-L2 and Ad/ΔFΔP-S35-L2 mediate similar levels of transduction in both CAR-positive and -negative cells (Fig. 3). The fiber shaft might have a minimal effect on the efficiency of in vitro gene transfer.
Several groups have reported that the length of the fiber shaft influences Ad-mediated gene transfer (34, 39, 43). Two groups have speculated that electrostatic interference, by which the short-shafted vectors would have a more charge-dependent repulsion between Ad type 5 hexon (highly negatively charged) and acidic cell surface proteins, might prevent efficient transduction (34, 39). On the other hand, Vigne et al. have proposed that shaft shortening may induce steric hindrance between the fiber knob and the RGD motif of the penton base, preventing each other from efficiently interacting with CAR and αv integrin, respectively (43). In the triple-mutant Ad vector developed in the present study, the participation of charge-dependent repulsion and steric hindrance on the less efficient liver transduction remains unclear. However, the shortened fiber shaft might reduce the remaining weak affinities of the mutated fiber knob and penton base for their cognate receptors and other interactions between the triple-mutant Ad vector and the cell by means of the charge-dependent repulsion and/or steric hindrance, resulting in much less efficient liver transduction. Components other than the fiber shaft length, such as certain sequences containing the Ad type 5 fiber shaft, but not the Ad type 35 fiber shaft, may be involved in reduced infectivity in vivo.
To support the propagation of mutant Ad vectors with ablation of both CAR and αv integrin binding, we generated new packaging cell lines. In the previously developed 293 cells expressing Ad type 5 fiber protein (Fiber-293), the tripartite leader sequence (44) or rabbit β-globin splicing signals (23) were inserted in order to enhance fiber expression. In contrast, a fiber expression plasmid (pCMVmfiber-Hyg) transfected into 293 cells in the present study contains the sequence of the CMV promoter-enhancer, intron A, SV40 poly(A), and the SV40 enhancer, which is much more efficient than the conventional vector containing the sequence of the CMV promoter-enhancer and SV40 or bovine growth hormone poly(A) (50). Successful construction of Fiber-293 cells would be achieved by the addition of these optimized transcriptional regulatory sequences. Importantly, the triple-mutant Ad vector (Ad/ΔFΔP-S35-L2) was generated to a particle titer similar to that of conventional Ad vectors (see Materials and Methods).
Several groups have used packaging cell lines that express an artificial receptor molecule, which should not have any natural analogs and should be completely artificial, such as anti-His scFv and anti-hemagglutinin (HA) scFv, for the amplification of mutated Ad vectors (7, 37). In the case of cell lines expressing anti-His scFv, the His tag sequence has been introduced in the C-terminal region of the fiber knob in the Ad vectors (7), while in the case of cell lines expressing anti-HA scFv, the HA tag sequence has been introduced in the HI loop of the fiber knob or the penton base instead of the RGD motif (37). When the Fiber-293 cells are used for a packaging cell line, either the HI loop or the C-terminal region of the fiber knob as well as the penton base can be used for displaying a pseudoligand in the vectors, making them advantageous over the cell lines expressing anti-His scFv or anti-HA scFv.
For the development of targeted Ad vectors, incorporation of a foreign ligand (i.e., peptide) that has a high affinity for the specific cellular receptor into the capsid of Ad vectors is also required. There are several possible locations for displaying a foreign ligand, including the HI loop or C terminus of the fiber knob and the region of the RGD motif of the penton base. In our triple-mutant Ad vector, both the HI loop and C-terminal coding region of the fiber knob and the region of the RGD motif of the penton base were designed to have unique restriction sites (Csp45I, ClaI, and XbaI, respectively) (18, 32, 33). Therefore, the targeting ligands can easily be displayed in the capsids of the vectors by cloning genes into their regions by a simple in vitro ligation.
The elucidation of the different fates of systemically administered triple-mutant Ad vector and conventional Ad vectors should also provide valuable insight into the development of targeted Ad vectors. The clearance kinetics from the circulation were similar for both vectors, having a half-life of <2 min (Fig. 5). Also, >60% of both vectors were detected in the liver 1 h after injection (Fig. 6). These observations suggest that sequestration into the liver is responsible for the rapid clearance of both vectors from the circulation. However, 48 h after injection, triple-mutant Ad vector DNA could not be detected in the liver, while 21% of the conventional Ad vector DNA remained. This large discrepancy could explain why the transgene expression levels of both vectors in the liver differed so tremendously (Fig. 4). For further clarification of what led to this discrepancy, we found that the triple-mutant Ad vector was preferentially sequestered into NPC (Kupffer and endothelial cells) of the liver, while the conventional Ad vector was sequestered (delivered) into both NPC cells and PC (Fig. 7). It has been shown previously that Kupffer cells play a central role in eliminating the input Ad vectors within the first 24 h after intravenous administration (11, 49). We suppose that phagocytosis by the liver's Kupffer cells might be the leading cause of the rapid degradation of the triple-mutant Ad virus as well as of conventional Ad vectors. This Kupffer cell-mediated clearance might present an obstacle in the next step for targeted Ad vectors, which is incorporation of a foreign ligand into the viral capsid. Selective depletion or blockade of Kupffer cells by treatment with clodronate liposome (21, 22, 38), gadolinium chloride (25), or another drug might be required.
In summary, the fiber knob, fiber shaft, and RGD motif of the penton base each plays an important role in Ad vector-mediated transduction to the mouse liver. The triple-mutant Ad vector, which contains the fiber knob with CAR binding ablated, the fiber shaft of Ad type 35, and the penton base with a deletion of the RGD motif, exhibits little tropism for any organs and should be a fundamental vector for targeted gene delivery.
Acknowledgments
We thank Zhi-Li Xu for discussions. We also thank Momoko Ariki and Yuko Ohtsuka for their technical assistance.
This work was supported by grants from the Ministry of Health, Labour and Welfare of Japan and a grant-in-aid for Scientific Research on Priority Areas of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
REFERENCES
- 1.Alemany, R., and D. T. Curiel. 2001. CAR-binding ablation does not change biodistribution and toxicity of adenoviral vectors. Gene Ther. 8:1347-1353. [DOI] [PubMed] [Google Scholar]
- 2.Alemany, R., K. Suzuki, and D. T. Curiel. 2000. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 81:2605-2609. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, K., S. Furuhata, K. Hatanaka, M. Maeda, J. S. Remy, J. P. Behr, M. Terada, and T. Yoshida. 2001. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Ther. 8:508-514. [DOI] [PubMed] [Google Scholar]
- 4.Asaoka, K., M. Tada, Y. Sawamura, J. Ikeda, and H. Abe. 2000. Dependence of efficient adenoviral gene delivery in malignant glioma cells on the expression levels of the coxsackievirus and adenovirus receptor. J. Neurosurg. 92:1002-1008. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 6.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 7.Douglas, J. T., C. R. Miller, M. Kim, I. Dmitriev, G. Mikheeva, V. Krasnykh, and D. T. Curiel. 1999. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat. Biotechnol. 17:470-475. [DOI] [PubMed] [Google Scholar]
- 8.Einfeld, D. A., R. Schroeder, P. W. Roelvink, A. Lizonova, C. R. King, I. Kovesdi, and T. J. Wickham. 2001. Reducing the native tropism of adenovirus vectors requires removal of both CAR and integrin interactions. J. Virol. 75:11284-11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fechner, H., A. Haack, H. Wang, X. Wang, K. Eizema, M. Pauschinger, R. Schoemaker, R. Veghel, A. Houtsmuller, H. P. Schultheiss, J. Lamers, and W. Poller. 1999. Expression of coxsackie adenovirus receptor and alphav-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 6:1520-1535. [DOI] [PubMed] [Google Scholar]
- 10.Heffelfinger, S. C., H. H. Hawkins, J. Barrish, L. Taylor, and G. J. Darlington. 1992. SK HEP-1: a human cell line of endothelial origin. In Vitro Cell Dev. Biol. 28A:136-142. [DOI] [PubMed] [Google Scholar]
- 11.Hegenbarth, S., R. Gerolami, U. Protzer, P. L. Tran, C. Brechot, G. Gerken, and P. A. Knolle. 2000. Liver sinusoidal endothelial cells are not permissive for adenovirus type 5. Hum. Gene Ther. 11:481-486. [DOI] [PubMed] [Google Scholar]
- 12.Henry, L. J., D. Xia, M. E. Wilke, J. Deisenhofer, and R. D. Gerard. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J. Virol. 68:5239-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong, S. S., M. K. Magnusson, P. Henning, L. Lindholm, and P. A. Boulanger. 2003. Adenovirus stripping: a versatile method to generate adenovirus vectors with new cell target specificity. Mol. Ther. 7:692-699. [DOI] [PubMed] [Google Scholar]
- 14.Huard, J., H. Lochmuller, G. Acsadi, A. Jani, B. Massie, and G. Karpati. 1995. The route of administration is a major determinant of the transduction efficiency of rat tissues by adenoviral recombinants. Gene Ther. 2:107-115. [PubMed] [Google Scholar]
- 15.Kanerva, A., M. Wang, G. J. Bauerschmitz, J. T. Lam, R. A. Desmond, S. M. Bhoola, M. N. Barnes, R. D. Alvarez, G. P. Siegal, D. T. Curiel, and A. Hemminki. 2002. Gene transfer to ovarian cancer versus normal tissues with fiber-modified adenoviruses. Mol. Ther. 5:695-704. [DOI] [PubMed] [Google Scholar]
- 16.Kirby, I., E. Davison, A. J. Beavil, C. P. C. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. J. Sutton, and G. Santis. 1999. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J. Virol. 73:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koizumi, N., H. Mizuguchi, T. Hosono, A. Ishii-Watabe, E. Uchida, N. Utoguchi, Y. Watanabe, and T. Hayakawa. 2001. Efficient gene transfer by fiber-mutant adenoviral vectors containing RGD peptide. Biochim. Biophys. Acta 1568:13-20. [DOI] [PubMed] [Google Scholar]
- 18.Koizumi, N., H. Mizuguchi, N. Utoguchi, Y. Watanabe, and T. Hayakawa. 2003. Generation of fiber-modified adenovirus vectors containing heterologous peptides in both the HI loop and C terminus of the fiber knob. J. Gene Med. 5:267-276. [DOI] [PubMed] [Google Scholar]
- 19.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 75:4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasnykh, V. N., J. T. Douglas, and V. W. van Beusechem. 2000. Genetic targeting of adenoviral vectors. Mol. Ther. 1:391-405. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmin, A., M. Finegold, and R. Eisensmith. 1997. Macrophage depletion increases the safety, efficacy and persistence of adenovirus-mediated gene transfer in vivo. Gene Ther. 4:309-316. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmin, A. I., O. Galenko, and R. C. Eisensmith. 2001. An immunomodulatory procedure that stabilizes transgene expression and permits readministration of E1-deleted adenovirus vectors. Mol. Ther. 3:293-301. [DOI] [PubMed] [Google Scholar]
- 23.Legrand, V., D. Spehner, Y. Schlesinger, N. Settelen, A. Pavirani, and M. Mehtali. 1999. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J. Virol. 73:907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leissner, P., V. Legrand, Y. Schlesinger, D. A. Hadji, M. van Raaij, S. Cusack, A. Pavirani, and M. Mehtali. 2001. Influence of adenoviral fiber mutations on viral encapsidation, infectivity and in vivo tropism. Gene Ther. 8:49-57. [DOI] [PubMed] [Google Scholar]
- 25.Lieber, A., C. He, L. Meuse, D. Schowalter, I. Kirillova, B. Winther, and M. Kay. 1997. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J. Virol. 71:8798-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnusson, M. K., S. S. Hong, P. Henning, P. Boulanger, and L. Lindholm. 2002. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J. Gene Med. 4:356-370. [DOI] [PubMed] [Google Scholar]
- 27.Maizel, J. V. J., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115-125. [DOI] [PubMed] [Google Scholar]
- 28.Mizuguchi, H., and T. Hayakawa. 2002. Adenovirus vectors containing chimeric type 5 and type 35 fiber proteins exhibit altered and expanded tropism and increase the size limit of foreign genes. Gene 285:69-77. [DOI] [PubMed] [Google Scholar]
- 29.Mizuguchi, H., and T. Hayakawa. 2002. Enhanced anti-tumor effect and reduced vector dissemination with fiber-modified adenovirus vectors expressing herpes simplex virus thymidine kinase. Cancer Gene Ther. 9:236-242. [DOI] [PubMed] [Google Scholar]
- 30.Mizuguchi, H., and M. A. Kay. 1998. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 9:2577-2583. [DOI] [PubMed] [Google Scholar]
- 31.Mizuguchi, H., and M. A. Kay. 1999. A simple method for constructing E1 and E1/E4 deleted recombinant adenovirus vector. Hum. Gene Ther. 10:2013-2017. [DOI] [PubMed] [Google Scholar]
- 32.Mizuguchi, H., N. Koizumi, T. Hosono, A. Ishii-Watabe, E. Uchida, N. Utoguchi, Y. Watanabe, and T. Hayakawa. 2002. CAR- or αv integrin-binding ablated adenovirus vectors, but not fiber-modified vectors containing RGD peptide, do not change the systemic gene transfer properties in mice. Gene Ther. 9:769-776. [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi, H., N. Koizumi, T. Hosono, N. Utoguchi, Y. Watanabe, M. A. Kay, and T. Hayakawa. 2001. A simplified system for constructing recombinant adenoviral vectors containing heterologous peptides in the HI loop of their fiber knob. Gene Ther. 8:730-735. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, T., K. Sato, and H. Hamada. 2003. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J. Virol. 77:2512-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishikawa, M., S. Takemura, Y. Takakura, and M. Hashida. 1998. Targeted delivery of plasmid DNA to hepatocytes in vivo: optimization of the pharmacokinetics of plasmid DNA/galactosylated poly(l-lysine) complexes by controlling their physicochemical properties. J. Pharm. Exp. Ther. 287:408-415. [PubMed] [Google Scholar]
- 36.Peng, K. W., L. Pham, H. Ye, R. Zufferey, D. Trono, F. L. Cosset, and S. J. Russell. 2001. Organ distribution of gene expression after intravenous infusion of targeted and untargeted lentiviral vectors. Gene Ther. 8:1456-1463. [DOI] [PubMed] [Google Scholar]
- 37.Roelvink, P. W., G. Mi Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 38.Schiedner, G., S. Hertel, M. Johnston, V. Dries, N. van Rooijen, and S. Kochanek. 2003. Selective depletion or blockade of Kupffer cells leads to enhanced and prolonged hepatic transgene expression using high-capacity adenoviral vectors. Mol. Ther. 7:35-43. [DOI] [PubMed] [Google Scholar]
- 39.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, T., N. Idamakanti, H. Kylefjord, M. Rollence, L. King, M. Kaloss, M. Kaleko, and S. C. Stevenson. 2002. In vivo hepatic adenoviral gene delivery occurs independently of the coxsackievirus-adenovirus receptor. Mol. Ther. 5:770-779. [DOI] [PubMed] [Google Scholar]
- 41.Smith, T. A. G., N. Idamakanti, M. L. Rollence, J. Marshall-Neff, J. Kim, K. Mulgrew, G. R. Nemerow, M. Kaleko, and S. C. Stevenson. 2003. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum. Gene Ther. 14:777-787. [DOI] [PubMed] [Google Scholar]
- 42.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vigne, E., J. F. Dedieu, A. Brie, A. Gillardeaux, D. Briot, K. Benihoud, M. Latta-Mahieu, P. Saulnier, M. Perricaudet, and P. Yeh. 2003. Genetic manipulations of adenovirus type 5 fiber resulting in liver tropism attenuation. Gene Ther. 10:153-162. [DOI] [PubMed] [Google Scholar]
- 44.Von Seggern, D. J., J. Kehler, R. I. Endo, and G. R. Nemerow. 1998. Complementation of a fibre mutant adenovirus by packaging cell lines stably expressing the adenovirus type 5 fibre protein. J. Gen. Virol. 79:1461-1468. [DOI] [PubMed] [Google Scholar]
- 45.Wickham, T. J. 2000. Targeting adenovirus. Gene Ther. 7:110-114. [DOI] [PubMed] [Google Scholar]
- 46.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 48.Wood, M., P. Perrotte, E. Onishi, M. E. Harper, C. Dinney, L. Pagliaro, and D. R. Wilson. 1999. Biodistribution of an adenoviral vector carrying the luciferase reporter gene following intravesical or intravenous administration to a mouse. Cancer Gene Ther. 6:367-372. [DOI] [PubMed] [Google Scholar]
- 49.Worgall, S., G. Wolff, E. Falck-Pedersen, and R. Crystal. 1997. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 8:37-44. [DOI] [PubMed] [Google Scholar]
- 50.Xu, Z.-L., H. Mizuguchi, A. Ishii-Watabe, E. Uchida, T. Mayumi, and T. Hayakawa. 2001. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene 272:149-156. [DOI] [PubMed] [Google Scholar]