Abstract
Epstein-Barr virus (EBV) has an accepted association with the epithelial malignancy nasopharyngeal carcinoma and has also been reported in other more controversial carcinoma settings. Evaluation of EBV association with epithelial carcinomas such as breast cancer would benefit from a better understanding of the outcome of EBV infection of these cells. Cell-free preparations of a green fluorescent protein-expressing virus, BX1, were used to infect breast cancer cell lines, which were then examined for EBV gene expression and viral genome copy number. Reverse transcription-PCR analyses revealed that the cells supported a mixture of latency II and lytic EBV gene expression. Lytic Zta and BMRF1 protein expression was detected by immunohistochemistry, and DNA PCR analyses estimated an EBV copy number of 300 to 600 genomes per infected cell. Evidence for lytic EBV expression was also found in breast tissue, where reverse transcription-PCR analyses detected lytic Zta transcripts in 7 of 10 breast carcinoma tissues and 4 of 10 normal tissues from the same patients. Scattered cells immunoreactive for Zta protein were also detectable in breast carcinoma. Quantitative real-time PCR analysis of EBV-positive breast carcinoma tissues suggested that less than 0.1% of the cells contained viral genomes. We suggest that sporadic lytic EBV infection may contribute to PCR-based detection of EBV in traditionally nonvirally associated epithelial malignancies.
Epstein-Barr virus (EBV) infects 90% of the population, and primary infection in young adulthood may result in infectious mononucleosis. In the majority of individuals, the virus persists for life in the memory B-cell pool (2) without recognized health consequences. However, EBV is associated with a growing list of malignancies of both lymphoid and epithelial origin, including Burkitt's lymphoma, posttransplant lymphoproliferative disease, B-cell lymphoma in the immunocompromised, Hodgkin's lymphoma, NK/T-cell lymphoma, nasopharyngeal carcinoma, leiomyosarcoma in AIDS patients, and a subset of gastric carcinomas (13, 43, 48). In addition, there have been reports linking EBV to carcinomas in sites such as the breast (4, 16, 30, 33), lung, and prostate (7, 24, 53).
In different studies with DNA PCR, 19 of 21 (30) and 15 of 28 (33) breast cancer samples from Britain were found to be EBV positive, as were 51 of 100 breast carcinoma samples from France (4), 161 of 509 cases from Europe and North Africa (16), and 19 of 92 samples from the United Kingdom (39). While EBV DNA has been found in breast cancer with some frequency, this has not correlated with an equivalent detection of viral gene expression or viral proteins. In situ hybridization probing for the highly abundant and stable small RNA genes (EBERs) has been negative (10, 15, 20) or has detected only focal expression (11).
Immunohistochemical analyses to detect EBV latency proteins have also been largely negative (11, 15). Positive staining for EBNA1 has been reported in some studies (4, 24), but the specificity of the EBNA1 reagents in clinical material has been questioned (6) and the EBNA1 2B4-1 antibody has recently been shown to cross-react with a nonviral tumor antigen (39). Thus, a question remains regarding the basis for the positive detection of EBV DNA in the face of negative data for EBV latency gene products.
To address this issue, we evaluated the outcome of EBV infection of breast carcinoma cell lines with an in vitro infection model. We demonstrate that these cells can support progression into the viral lytic cycle and suggest that sporadic lytic infection of epithelial cells by EBV may contribute to the detection of EBV DNA in clinical studies reliant on DNA PCR technology.
MATERIALS AND METHODS
Cell lines and EBV infection.
Breast epithelial tumor cell lines were obtained from the American Type Culture Collection and cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The green fluorescent protein (GFP)-positive Akata cell line BX1 (38), Raji, and Namalwa were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. BX1 cells were treated with anti-immunoglobulin G (IgG) at a concentration of 50 μg/ml for 5 days to induce virus production. The supernatant was collected, passed through a 0.45-μm filter, and centrifuged at 15,000 rpm at 4°C for 1 h. The concentrated cell-free virus was then added to the breast epithelial cell cultures, and cells were collected for analysis 48 h later. To select a BX1-converted MDA-MB468 cell line, G418 was added to the culture medium at a concentration of 400 μg/ml 48 h after infection. The medium was changed every 7 days until G418-resistant clones emerged (3 to 5 weeks).
Tissue samples.
Snap-frozen late-stage invasive breast carcinoma, normal breast tissues from the same patients, and fixed paraffin-embedded specimens were obtained from the Department of Pathology at Johns Hopkins Hospital and kept at −70°C until analysis.
RT-PCR for EBV transcripts.
The sequence of the reverse transcription (RT)-PCR primers used are listed in Table 1. mRNA was extracted from virus-infected breast epithelial cell lines and frozen tissues with a GeneElute Direct mRNA miniprep kit (Sigma, St. Louis, Mo.). cDNA was synthesized with random hexamers and avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). Both primary and secondary PCRs involved an initial denaturation at 95°C for 5 min, followed by 40 cycles consisting of 95°C for 30 s, optimal annealing temperature for 1 min, 72°C for 1 min, and a final extension at 72°C for 10 min. The RT-PCR products were electrophoresed on a 1.2% agarose gel and visualized by ethidium bromide staining.
TABLE 1.
Oligonucleotide primers used in RT-PCR analysis
| Transcript | Oligonucleotide sequence | 3′ coordinatesa | cDNA products (bp) |
|---|---|---|---|
| EBER1 | |||
| E1 | 5′-AGGACCTACGCTGCCCTAGAG-3′ | 6,649 | |
| E1 nested | 5′-AGAGGTTTTGCTAGGGAGG-3′ | 6,664 | |
| E2 | 5′-AAAACATGCGGACCACCAGC-3′ | 6,776 | |
| E2 nested | 5′-GACCACCAGCTGGTACTTG-3′ | 6,767 | 140 |
| BARF0 | |||
| P3 | 5′-GTGAGGGAAATAACCAGGATC-3′ | 157,086 | |
| P3 nested | 5′-CAGGACCAGAATGAGCATGC-3′ | 157,110 | |
| P4 | 5′-GCTTTCCTTTCCGAGTCTGC-3′ | 159,171 | |
| P4 nested | 5′-CTTCTCCTCGGACATCCAGTG-3′ | 159,121 | 237 |
| RPMS1 | |||
| P1 | 5′-CACGATGTCCTGGTCAGAGTG-3′ | 10,248* | |
| P1 nested | 5′-GGCTTGAGGAATACCTCGTTG-3′ | 10,282* | |
| P2 | 5′-TGGCCTTCGATATCGAGTGTC-3′ | 155,847 | |
| P2 nested | 5′-ACCAACGAGGCTGACCTGATC-3′ | 155,801 | 296 |
| EBNA1 (Q-Kexon) | |||
| Qp | 5′-GCGGGATAGCGTGCGCTA-3′ | 62,447 | |
| Qp nested | 5′-GTGCGCTACCGGATGGCG-3′ | 62,457 | |
| K1 | 5′-CTCTTCTTTGAGGTCCACTG-3′ | 108,032 | |
| K1 nested | 5′-CTTCTGGTCCAGATGTGT-3′ | 108,002 | 264 |
| Wp/Cp (Y-K exon) | |||
| Y2 | 5′-ATTAGAGACCACTTTGAGCC-3′ | 47,921 | |
| Y2 nested | 5′-TGGCGTGTGACGTGGTGTAA-3′ | 48,416 | |
| K1 | 5′-CTCTTCTTTGAGGTCCACTG-3′ | 108,032 | |
| K1 nested | 5′-CTTCTGGTCCAGATGTGT-3′ | 108,002 | NA |
| LMP1 | |||
| L1 | 5′-CTGAGATCTATGGAACACGACCTTGAG-3′ | 169,455 | |
| L1 nested | 5′-CTAGGCCTTGCTCTCCTTCTC-3′ | 169,382 | |
| L2 | 5′-GCAGAGCATCTCCAATAAGTAG-3′ | 168,949 | |
| L2 nested | 5′-GGAACAATGCCTGTCCGTG-3′ | 169,096 | 249 |
| ZTA | |||
| Z1 | 5′-AGCAGACATTTGGTGTTCCAC-3′ | 102,716 | |
| Z1 nested | 5′-ACGCACGGAAACCACAAC-3′ | 102,666 | |
| Z2 | 5′-ACATCTGCTTCAACAGGAGG-3′ | 102,303 | |
| Z2 nested | 5′-GCGCAGCCTGTCATTTTCAG-3′ | 102,323 | 174 |
Sequences from B95-8 or Raji (*).
Real-time PCR for EBV copy number.
Genomic DNA from infected MDA-MB468 cells, breast carcinoma, and normal breast tissues was extracted by digesting with proteinase K at 50°C overnight, followed by phenol-chloroform extraction and ethanol precipitation. PCRs were set up in a volume of 50 μl with a TaqMan PCR core reagent kit (Perkin-Elmer Corp., Branchburg, N.J.). Fluorescent probes were custom synthesized by Perkin-Elmer Applied Biosystems. PCR primers were synthesized by Gibco BRL (Frederick, Md.). Each reaction contained 5 μl of 10× buffer A (4 mM MgC12), amplification primers (300 nM), fluorescent probe (25 nM), dATP, dCTP, and dGTP (200 μM each), dUTP (400 uM), 1.25 U of AmpliTaq Gold, and 0.5 U of AmpErase uracil N-glycosylase. Extracted genomic DNA was used for amplification. DNA amplifications were carried out in a 96-well reaction plate in a Perkin-Elmer Applied Biosystems 7700 sequence detector. Each sample was analyzed in duplicate, and multiple negative water blanks were included in each analysis. A calibration curve was run in parallel and in duplicate with each analysis, with DNA extracted from the EBV-positive cell line Namalwa as a standard.
Immunohistochemistry and immunofluorescence.
For immunohistochemistry, infected breast epithelial cells were fixed with ice-cold methanol for 5 min. Paraffin-embedded sections were deparaffinized and pretreated with 1 mM citric acid buffer (pH 6.0) for 20 min in a microwave oven. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide. After three washes with phosphate-buffered saline, slides were incubated in 10% normal goat serum for 10 min and then with monoclonal antibodies at a dilution of 1:200 in phosphate-buffered saline at room temperature for 1 h. Bound antibody was detected by use of biotinylated anti-mouse IgG at a dilution of 1:100. A standard immunoperoxidase staining protocol with StreptABC Complex/HRP Duet, Mouse and Rabbit (Dako, Carpinteria, Calif.) was used for signal detection. Hematoxylin was used for counterstaining. Mouse monoclonal anti-Zta antibody was purchased from Argene (North Massapequa, N.Y.). Mouse monoclonal anti-EA-D antibody recognizing the protein product of EBV BMRF1 was purchased from Advanced Biotechnologies (Gaithersburg, Md.). Anti-LMP-1 monoclonal antibody was obtained from the culture supernatant of the S12 hybridoma cell line (34).
For immunofluorescent analysis, MDA-MB468-BX1 cells were fixed in 2% paraformaldehyde and permeabilized with 0.1% Triton X-100. Zta was visualized with mouse monoclonal anti-Zta antibody and rhodamine-conjugated secondary antibody.
RESULTS
In vitro infection of breast cancer cell lines.
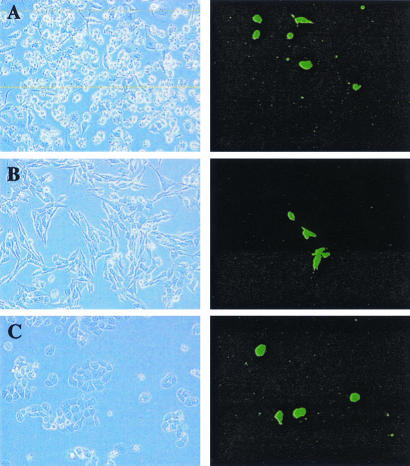
To better understand the outcome of EBV infection of breast cancer cells, an in vitro infection protocol was established. In vitro infection of epithelial cells by EBV occurs at very low frequency compared to the efficient infection seen with B cells, and it has been reported that cell-cell contact is required for epithelial cell infection (9, 46). However, a problem with cocultivation is that the donor B cells stick avidly to the epithelial cells, making it difficult to subsequently separate the two cell populations for analysis. We therefore prepared cell-free virus from BX1 cells, an Akata strain carrying the GFP open reading frame integrated within the EBV genome at BXLF1 (38). BX1 cells were stimulated to enter the lytic cycle by incubation with IgG, and virus in the culture supernatant was prepared by passage of the culture supernatant through a 0.45-μm filter and concentration by centrifugation at 15,000 rpm for 1 h. This EBV preparation was able to infect breast carcinoma cell lines, as indicated by the appearance of GFP-positive cells in infected MDA-MB231, MDA-MB435, and MDA-MB468 cultures (Fig. 1). Approximately 0.5 to 1.0% of the cultured cells were infected, based on GFP expression.
FIG. 1.
In vitro EBV infection of breast cell lines. Breast cancer cell lines were incubated with concentrated, cell-free BX1 virus and monitored for GFP expression 48 h after infection. (A) MDA-MB231; (B) MDA-MB435; (C) MDA-MB468. Left panels, phase contrast. Right panels, immunofluorescence.
EBV gene expression in infected MDA-MB468 cells.
In posttransplant lymphoproliferative disease, as in latently infected lymphoblastoid B-cell lines in culture, the full spectrum of EBV latency genes are expressed; EBNAs 1, 2, 3A, 3B, 3C, and LP, the differentially spliced BARTs (RPMS, RK-BARFO, and A73), the membrane proteins LMP1 and LMP2A, and the polymerase III-transcribed EBERs. This pattern of gene expression has been termed latency III. EBV-associated tumors arising in the immunocompetent host exhibit a more tightly regulated latency, latency I (Burkitt's lymphoma) or latency II (nasopharyngeal carcinoma and Hodgkin's lymphoma), in which viral gene expression is restricted to EBNA1, the BARTs, and the EBERs in latency I or these genes plus the LMPs in latency II (28).
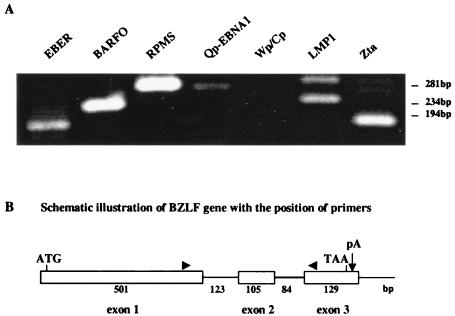
Nested RT-PCR assays were performed to determine the pattern of EBV gene expression in infected MDA-MB468 cells. The latency-associated BARTs (the primers used detected either BARFO or RPMS splices), Qp-initiated EBNA1 and LMP1 transcripts, and the polymerase III EBER RNAs were detected, but no Wp/Cp-initiated RNAs were found (Fig. 2A). This pattern of EBV latency gene expression is typical of latency II. However, transcripts for the BZLF1 open reading frame encoding the lytic transactivator Zta were also detected (Fig. 2A), suggesting a mixture of latent and lytic infection. The BZLF1 open reading frame contains two introns (Fig. 2B), and the sizes of the minor RT-PCR bands are consistent with products missing one or the other of the introns. DNA sequencing confirmed the partially spliced nature of the minor RT-PCR products. These splice variants were also detected in RT-PCR analyses of the BX1 converted MDA-MB468 cell line described below (data not shown) and have been described in analyses of nasopharyngeal carcinoma tumor cells (12).
FIG. 2.
Latent and lytic EBV gene expression in infected MDA-MB468 cells. (A) Ethidium bromide-stained gels showing the DNA products amplified by nested RT-PCR with the specific primers listed in Table 1. mRNA was isolated from MDA-MB468 cells 48 h after infection with cell-free BX1 virus. The upper band present in the LMP1 RT-PCR migrates at the position expected for an unspliced LMP1 transcript. The minor bands in the Zta amplification represent partially spliced transcripts that retain either the first or second intron. Sizes indicated at the right were determined from the migration of φX174 DNA. (B) Diagram of the Zta coding region, showing the relative sizes (in base pairs) of the three exons and two introns and the locations of the primer (arrowheads) used in the RT-PCR analyses.
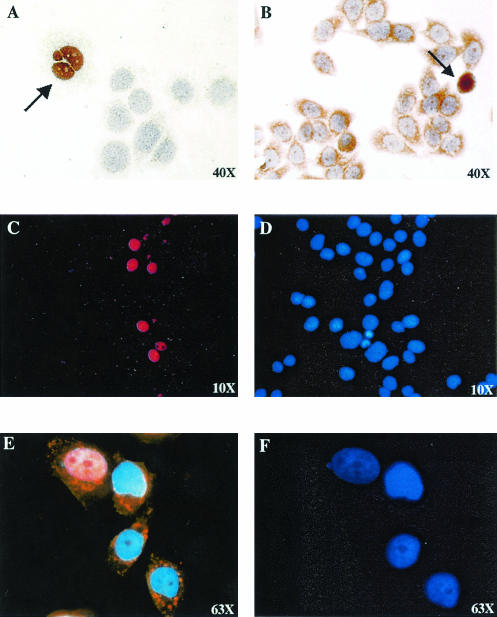
The expression of lytic cycle gene products after infection of MDA-MB468 cells was further examined by immunohistochemistry and staining with horseradish peroxidase. Expression of both the immediate-early Zta protein (Fig. 3A) and the early BMRF1 (EA-D) protein (Fig. 3B) was detected. We noted that the nuclei of Zta-expressing cells were frequently abnormal in shape. This may relate to a mitotic defect in chromosome condensation that has been described in adenovirus Zta-infected cells (35).
FIG. 3.
Detection of lytic Zta and BMRF1 protein expression in in vitro-infected MDA-MB468 cells. (A and B) Immunoperoxidase staining of MDA-MB468 cells fixed 48 h after infection with cell-free BX1 virus. (A) Anti-Zta primary antibody. (B) Anti-BMRF1 (EA-D) primary antibody. Positive-staining cells are indicated with arrows. (C and D) Immunofluorescence assay showing Zta expression in a converted MDA-MB468-BX1 cell line established by BX1 infection followed by selection for neomycin resistance. (C) Cells were stained with anti-Zta primary antibody and rhodamine-conjugated secondary antibody. (D) 4′,6′-Diamidino-2-phenylindole (DAPI) staining of cell nuclei. (E and F) MDA-MB468-BX1 cells triple stained for LMP1 expression (indocarbocyanine, orange), nuclear Zta expression (rhodamine, red), and DAPI (nuclear, blue) (E) or stained with DAPI alone (F).
The low level of infectivity in the MDA-MB468 cultures made it difficult to evaluate the prevalence of lytic gene expression. To address this point, MDA-MB468 cells were infected with BX1 virus, which carries a neomycin resistance gene in addition to GFP, and a BX1-converted MDA-MB468 cell line was established by G418 selection. Immunofluorescence staining revealed that approximately 10% of the MDA-MB468-BX1 cells constitutively expressed Zta protein (Fig. 3C and D). Double staining of MDA-MB468 converted cells for LMP1 and Zta expression was also performed with rhodamine (red)- and indocarbocyanine (orange)-conjugated secondary antibodies. (Expression of GFP from the endogenous EBV genomes complicated the use of fluorescein isothiocyanate as an alternative fluorophor.) Greater than 90% of the cells were LMP1 expressing, in contrast to approximately 10% Zta-positive cells (Fig. 3E and F).
EBV genome copy number in infected MDA-MB468 cells.
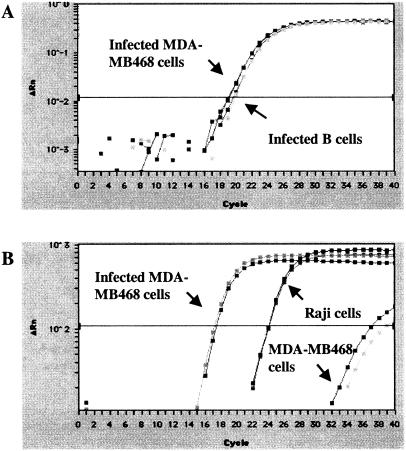
The detection of Zta transcripts and protein in newly infected MDA-MB468 cells indicated that a proportion of the cells were entering the lytic cycle. To investigate whether the lytic cycle was progressing to the stage of viral DNA replication, real-time quantitative PCR was carried out to determine the EBV copy number in the infected MDA-MB468 cells. The PCR primers used amplified a region of the EBV BamHI-W fragment, and a calibration curve was run in parallel with each analysis, with DNA extracted from the EBV-positive Namalwa B-cell line. Namalwa carries two integrated copies of the EBV genome per cell (31).
The EBV genome copy number in MDA-MB468 cells 2 days postinfection with BX1 virus was compared with that in EBV-negative Akata B cells 2 days postinfection with BX1 virus (Fig. 4A). In a second analysis, the EBV copy number in uninfected MDA-MB468 cells, EBV-positive Raji B cells, and BX1-infected MDA-MB468 cells was measured (Fig. 4B). The percentage of infected MDA-MB468 cells and Akata cells was estimated by counting the number of GFP-positive cells per 100 cells and found to be 0.5 to 1.0% for the breast cancer cells and 30% for the Akata B cells. Taking the infection rate into consideration, the EBV genome number in the infected MDA-MB468 cells was estimated from an average of the two assays to be 311 to 622 copies per cell, while the number of copies in newly infected Akata B cells was 10 copies per cell. The higher number of genomes present in the infected breast cancer cells is consistent with a proportion of the cells' supporting lytic viral DNA replication. The estimated copy number in Raji cells was 54 genomes per cell, which is in line with previous reports (1).
FIG. 4.
Estimation of EBV genome copy number in MDA-MB468 cells infected in vitro. Real-time quantitative PCR was performed in duplicate with primers specific for the EBV BamHI-W fragment. (A) Amplification plots of the fluorescence intensity against PCR cycle number are shown to compare the EBV genome load in MDA-MB468 and virus-negative Akata (−) B cells 48 h after infection with cell-free BX1 virus. (B) EBV genome load in uninfected and infected MDA-MB468 cells versus EBV-positive Raji B cells. x axis, PCR cycle number. y axis, fluorescence intensity.
Detection of lytic EBV gene expression in human breast tissues.
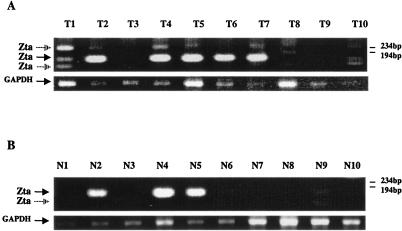
The finding of lytic EBV gene expression in the in vitro-infected breast cancer cell lines led us to test for expression of the BZLF1 open reading frame encoding Zta in snap-frozen samples of 10 late-stage, invasive breast carcinomas as well as in 10 control normal breast tissues from the same patients. Positive RT-PCR signals for BZLF1 expression were observed in both tumor and normal samples (Fig. 5). The frequency of detection of positive RT-PCR for Zta in the normal tissues may be influenced by the fact that these tissues were from the same patients as the tumor samples. Multiple ethidium bromide-stained bands were observed in the Zta-primed RT-PCR amplifications of most of the PCR-positive tumor samples. Individual amplified bands were isolated, cloned, and sequenced to confirm their identity. The major band in the majority of the samples (solid arrow) represented a fully spliced Zta cDNA. The smaller minor band, which was particularly abundant in tumor sample T1, represented a cDNA lacking Zta exon 2, and the larger band, which was also abundant in T1, represented a cDNA in which the first intron was retained.
FIG. 5.
RT-PCR detection of EBV Zta transcripts in breast carcinoma and normal breast tissues. Ethidium bromide-stained gels showing the DNA products amplified by nested RT-PCR with the EBV Zta and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers described in Table 1. (A) Ten snap-frozen breast carcinoma samples. (B) Ten snap-frozen samples of normal breast tissue from the same patients. The major (closed arrow) and minor (open arrow) spliced forms of Zta are indicated. GAPDH amplification was used as a control for RNA integrity. Sizes indicated at the right were determined from the migration of φX174 DNA.
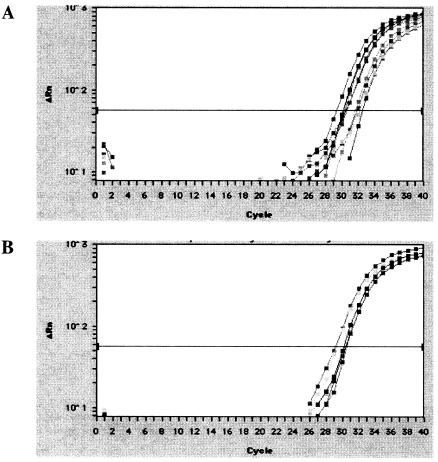
To evaluate the relative EBV genome load in the positive breast carcinoma tissues versus the positive normal breast tissues, quantitative PCR was performed on the five tumor samples (T1, T4, T5, T6, and T7) and three normal samples (N2, N4, and N5) that gave the strongest positive RT-PCR signals (Fig. 6). No significant difference was found between the two groups, with the breast carcinoma tissues (Fig. 6A) calculated to contain 1,187 ± 673 EBV genomes per 106 cells and the normal tissues (Fig. 6B) 1,643 ± 907 EBV genomes per 106 cells. If one assumes the lowest infectivity rate of 1 EBV genome per cell, then the data would indicate that less than 0.1% of the cells in either the normal or the breast cell carcinoma tissues were EBV positive.
FIG. 6.
Comparison of the EBV genome load in EBV-positive breast carcinoma and normal breast tissues. Real-time quantitative PCR was performed with primers specific for the EBV BamHI-W fragment. Duplicate analysis of (A) five breast carcinoma samples (T2, T4, T5, T6, and T7) and (B) three normal breast samples (N2, N4, and N5) that were positive by RT-PCR for Zta expression. x axis, PCR cycle number. y axis, fluorescence intensity.
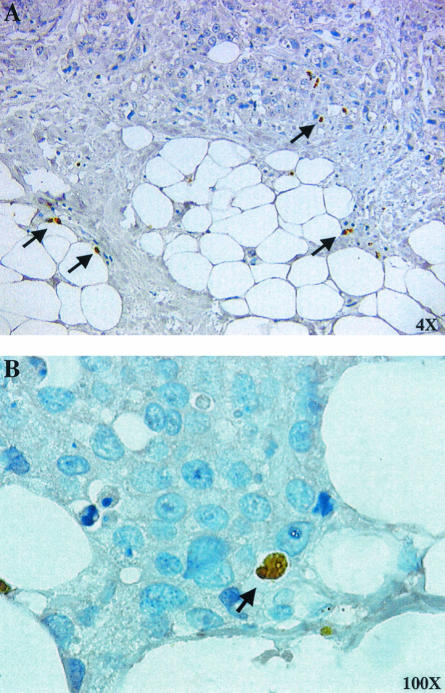
In an attempt to determine whether there was evidence for EBV lytic protein expression in breast tissues, immunohistochemistry for Zta was performed on formalin-fixed, paraffin-embedded breast tissue samples. Occasional Zta-positive cells were observed within the breast carcinoma tissues (Fig. 7A and B). We were unable to visualize Zta protein expression in normal breast samples (data not shown).
FIG. 7.
Detection of Zta protein expression in breast carcinoma tissue. Immunohistochemical staining of paraffin-embedded breast carcinoma tissue with anti-Zta primary antibody. Rare Zta-positive cells are indicated by arrows. Magnifications are indicated.
DISCUSSION
EBV has a recognized association with epithelial cancers. Nasopharyngeal carcinoma is 100% EBV positive (43), and gastric carcinomas are 10% positive worldwide, with up to 18% positivity in some regions (48). A number of rarer carcinomas, such as lymphoepithelioma-like carcinomas of the salivary gland, lung, and thymus, are also EBV associated (21). The presence of EBV DNA in breast cancer was first reported in 1995 by Labrecque et al. (30). This report has been followed by additional positive findings plus a large number of papers reporting a lack of association (6, 15, 20, 37). The positive findings of EBV association in breast cancer were based on detection of EBV DNA by PCR amplification, while the negative data came predominantly from studies examining EBV gene expression.
EBV is a ubiquitous virus that establishes a life-long infection in its host. In such a setting, the detection of EBV DNA by sensitive PCR technology in nonlymphoid cancers that have not traditionally been considered virally associated is difficult to evaluate (52). EBV has been detected in cells shed into human breast milk (27), and in an animal model system, murine gammaherpesvirus 72 has been found to replicate in mammary gland epithelial cells and to be shed into breast milk (42). One interpretation of the positive PCR data in breast cancer has been that the signal arises from infiltrating EBV-positive lymphocytes. We wish to offer an alternative explanation, that lytic infection of a very small number of breast epithelial cells could produce a disproportionately strong DNA PCR signal.
EBV infection of B cells leads to the establishment of a latent infection, with lytic viral gene expression occurring spontaneously only within particular type III latency cell lines and only in a small percentage of the cell population. However, other cell types have been found to provide a more permissive environment. In the case of hairy leukoplakia, a lesion associated with the differentiated layers of the tongue epithelium, there is aggressive EBV lytic replication with extensive virion production along with concurrent expression of EBV latency genes (22, 50, 51). In vitro infection of both nonepithelial cells, for example, primary monocytes and explanted tissue from leiomyosarcoma (26, 40, 45), as well as epithelial cells derived from gastric carcinoma and nasopharyngeal carcinoma (29, 55, 56), has also resulted in cultures in which both latency gene expression and lytic BZLF1 gene expression have been detected.
Our observations with in vitro-infected breast carcinoma cells suggest that these cells are similarly expressing a mixture of latency II and lytic EBV genes. Both BZLF1 transcripts and Zta protein expression were detected, and 10% of the EBV-converted MDA-MB468-BX1 cells exhibited spontaneous Zta expression in immunofluorescence assays. Furthermore, the assessment of viral genome copy number per infected cell measured in primary infections gave a number (300 to 600) that is consistent with the viral lytic cycle progressing at least as far as productive viral DNA replication.
Zta transcripts were detected by RT-PCR in breast carcinoma tissues and rare examples of Zta-positive cells were also observed by immunohistochemistry. While occasional Zta-positive cells were observed in immunohistochemical analyses of breast cancer tissues, we were unable to detect positive cells in the normal breast tissue samples from the same patients even although the quantitative PCR data indicated that comparable numbers of viral genomes were present in PCR-positive normal and breast cancer samples. This apparent difference in the ability to detect Zta may derive in part from an increased resistance to cell death in the cancer cells.
EBV infection of B cells utilizes CR2 (CD21) as the receptor and HLA class II as a coreceptor (17, 25, 32, 41). Engagement of CD21 by gp350/220 on the incoming virus is believed to stimulate signaling that provides an initial environment favorable to establishment of viral infection. Activation of CR2 either by binding of gp350/220 or through cross-linking has been found to stimulate interleukin-6 expression (14, 49) and phosphatidylinositol-3 kinase activity (3), both of which are associated with cell survival and proliferative signaling. Epithelial cells do not express CR2, and infection is mediated by the viral glycoprotein gH (38). For epithelial cells, where the B-cell-like prosurvival signals are not initiated during receptor binding, cell death may be more aggressive. Constitutive expression of prosurvival genes such as STAT3 and STAT5a/b, which are commonly activated in breast cancer cells and cell lines (5, 18), could provide additional protection that enhances immunohistochemical detection of lytically infected cells.
The presence of cells supporting the EBV lytic cycle in these tissues has implications for assays based on EBV DNA detection. A single lytically infected cell containing 600 copies of the EBV genome would give a DNA PCR signal indistinguishable from that arising from 600 cells each containing a single copy of the genome. Real-time PCR analysis of EBV-positive breast carcinoma samples indicated that less than 0.1% of the cells in the biopsy samples were positive at the 1 EBV genome copy per cell level. Taking into account the information on Zta positivity and hence the likelihood that some of the cells were undergoing lytic EBV DNA replication, the percentage of EBV-infected carcinoma cells is likely to be considerably less than 0.1%.
The data suggesting that only a very small proportion of the breast carcinoma cells are EBV infected and the indication of lytic cycle progression may account in part for discrepancies between studies in which PCR analysis was used to evaluate EBV association with breast cancer and those in which immunohistochemistry for latency proteins or EBER in situ hybridization was employed. In the case of the EBERs, these polymerase III RNAs are expressed at a reduced level in cells undergoing lytic viral replication (19, 23). Although growth stimulation is usually thought of as being a property of EBV latency genes, the EBV lytic cycle Rta protein can induce E2F-1 genes, which are associated with cell cycle progression (47), and Zta, which normally is associated with a G1 or G2/M cell cycle block (8, 44, 54), has also been described as stimulating E2F-1 genes in certain cell types (36). However, the very small number of EBV-infected cells estimated by quantitative RT-PCR to be present in the PCR-positive breast cancer samples makes it unlikely that EBV latent or lytic infection could be contributing directly to tumor growth. Our study focused on EBV infection of breast cancer cells, but the observations are likely to be applicable to other settings in which EBV association with epithelial malignancies is being evaluated.
Acknowledgments
We thank Pedram Argani and Donte Trusty for breast tissue acquisition and Yvette Tanhehco and Leslie Meszler for help with real-time PCR and microscopy.
This work was funded by Public Health Service grants RO1 CA 30356 from the National Cancer Institute to S.D.H. and AI 20662 to L.H.-F. and the SPORE in Breast Cancer, CA 88893.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, G. L., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Barel, M., M. Le Romancer, and R. Frade. 2001. Activation of the EBV/C3d receptor (CR2, CD21) on human B lymphocyte surface triggers tyrosine phosphorylation of the 95-kDa nucleolin and its interaction with phosphatidylinositol 3 kinase. J. Immunol. 166:3167-3173. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 91:1376-1381. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 6.Brink, A. A., A. J. van Den Brule, P. van Diest, and C. J. Meijer. 2000. Re: detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 92:655-656. [DOI] [PubMed] [Google Scholar]
- 7.Castro, C. Y., M. L. Ostrowski, R. Barrios, L. K. Green, H. H. Popper, S. Powell, P. T. Cagle, and J. Y. Ro. 2001. Relationship between Epstein-Barr virus and lymphoepithelioma-like carcinoma of the lung: a clinicopathologic study of 6 cases and review of the literature. Hum. Pathol. 32:863-872. [DOI] [PubMed] [Google Scholar]
- 8.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., C.-H. Tung, Y.-T. Huang, J. Lu, J.-Y. Chen, and C.-H. Tsai. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J. Virol. 73:8857-8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, J. S., C. C. Chen, and K. J. Chang. 1998. In situ detection of Epstein-Barr virus in breast cancer. Cancer Lett. 124:53-57. [DOI] [PubMed] [Google Scholar]
- 11.Chu, P. G., K. L. Chang, Y. Y. Chen, W. G. Chen, and L. M. Weiss. 2001. No significant association of Epstein-Barr virus infection with invasive breast carcinoma. Am. J. Pathol. 159:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochet, C., D. Martel-Renoir, V. Grunewald, J. Bosq, G. Cochet, G. Schwaab, J. F. Bernaudin, and I. Joab. 1993. Expression of the Epstein-Barr virus immediate early gene, BZLF1, in nasopharyngeal carcinoma tumor cells. Virology 197:358-365. [DOI] [PubMed] [Google Scholar]
- 13.Crawford, D. H. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Addario, M., T. A. Libermann, J. Xu, A. Ahmad, and J. Menezes. 2001. Epstein-Barr Virus and its glycoprotein-350 upregulate IL-6 in human B-lymphocytes via CD21, involving activation of NF-kappaB and different signaling pathways. J. Mol. Biol. 308:501-514. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande, C. G., S. Badve, N. Kidwai, and R. Longnecker. 2002. Lack of expression of the Epstein-Barr Virus (EBV) gene products, EBERs, EBNA1, LMP1, and LMP2A, in breast cancer cells. Lab. Investig. 82:1193-1199. [DOI] [PubMed] [Google Scholar]
- 16.Fina, F., S. Romain, L. Ouafik, J. Palmari, F. Ben Ayed, S. Benharkat, P. Bonnier, F. Spyratos, J. A. Foekens, C. Rose, M. Buisson, H. Gerard, M. O. Reymond, J. M. Seigneurin, and P. M. Martin. 2001. Frequency and genome load of Epstein-Barr virus in 509 breast cancers from different geographical areas. Br. J. Cancer 84:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fingeroth, J. D., J. J. Weis, T. F. Tedder, J. L. Strominger, P. A. Biro, and D. T. Fearon. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 81:4510-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, R., T. L. Bowman, G. Niu, H. Yu, S. Minton, C. A. Muro-Cacho, C. E. Cox, R. Falcone, R. Fairclough, S. Parsons, A. Laudano, A. Gazit, A. Levitzki, A. Kraker, and R. Jove. 2001. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20:2499-2513. [DOI] [PubMed] [Google Scholar]
- 19.Gilligan, K., P. Rajadurai, L. Resnick, and N. Raab-Traub. 1990. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc. Natl. Acad. Sci. USA 87:8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser, S. L., R. F. Ambinder, J. A. DiGiuseppe, P. L. Horn-Ross, and J. L. Hsu. 1998. Absence of Epstein-Barr virus EBER-1 transcripts in an epidemiologically diverse group of breast cancers. Int. J. Cancer 75:555-558. [DOI] [PubMed] [Google Scholar]
- 21.Goldenberg, D., A. Golz, A. Netzer, E. Rosenblatt, A. Rachmiel, R. F. Goldenberg, and H. Z. Joachims. 2001. Epstein-Barr virus and cancers of the head and neck. Am. J. Otolaryngol. 22:197-205. [DOI] [PubMed] [Google Scholar]
- 22.Greenspan, J. S., D. Greenspan, E. T. Lennette, D. I. Abrams, M. A. Conant, V. Petersen, and U. K. Freese. 1985. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N. Engl. J. Med. 313:1564-1571. [DOI] [PubMed] [Google Scholar]
- 23.Greifenegger, N., M. Jager, L. A. Kunz-Schughart, H. Wolf, and F. Schwarzmann. 1998. Epstein-Barr virus small RNA (EBER) genes: differential regulation during lytic viral replication. J. Virol. 72:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grinstein, S., M. V. Preciado, P. Gattuso, P. A. Chabay, W. H. Warren, E. De Matteo, and V. E. Gould. 2002. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer Res. 62:4876-4878. [PubMed] [Google Scholar]
- 25.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenson, H. B., E. A. Montalvo, K. L. McClain, Y. Ench, P. Heard, B. A. Christy, P. J. Dewalt-Hagan, and M. P. Moyer. 1999. Characterization of natural Epstein-Barr virus infection and replication in smooth muscle cells from a leiomyosarcoma. J. Med. Virol. 57:36-46. [PubMed] [Google Scholar]
- 27.Junker, A. K., E. E. Thomas, A. Radcliffe, R. B. Forsyth, A. G. Davidson, and L. Rymo. 1991. Epstein-Barr virus shedding in breast milk. Am. J. Med. Sci. 302:220-223. [DOI] [PubMed] [Google Scholar]
- 28.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Knox, P. G., Q. X. Li, A. B. Rickinson, and L. S. Young. 1996. In vitro production of stable Epstein-Barr virus-positive epithelial cell clones which resemble the virus:cell interaction observed in nasopharyngeal carcinoma. Virology 215:40-50. [DOI] [PubMed] [Google Scholar]
- 30.Labrecque, L. G., D. M. Barnes, I. S. Fentiman, and B. E. Griffin. 1995. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 55:39-45. [PubMed] [Google Scholar]
- 31.Lawrence, J. B., C. A. Villnave, and R. H. Singer. 1988. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell 52:51-61. [DOI] [PubMed] [Google Scholar]
- 32.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luqmani, Y., and S. Shousha. 1995. Presence of Epstein-Barr virus in breast carcinoma. Int. J. Oncol. 6:899-903. [DOI] [PubMed] [Google Scholar]
- 34.Mann, K. P., D. Staunton, and D. A. Thorley-Lawson. 1985. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J. Virol. 55:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauser, A., E. Holley-Guthrie, D. Simpson, W. Kaufmann, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces both a G(2) and a mitotic block. J. Virol. 76:10030-10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauser, A., E. Holley-Guthrie, A. Zanation, W. Yarborough, W. Kaufmann, A. Klingelhutz, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J. Virol. 76:12543-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCall, S. A., J. H. Lichy, K. E. Bijwaard, N. S. Aguilera, W. S. Chu, and J. K. Taubenberger. 2001. Epstein-Barr virus detection in ductal carcinoma of the breast. J. Natl. Cancer Inst. 93:148-150. [DOI] [PubMed] [Google Scholar]
- 38.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray, P. G., D. Lissauer, J. Junying, G. Davies, S. Moore, A. Bell, J. Timms, D. Rowlands, C. McConkey, G. M. Reynolds, S. Ghataura, D. England, R. Caroll, and L. S. Young. 2003. Reactivity with a monoclonal antibody to Epstein-Barr virus (EBV) nuclear antigen 1 defines a subset of aggressive breast cancers in the absence of the EBV genome. Cancer Res. 63:2338-2343. [PubMed] [Google Scholar]
- 40.Nakamura, H., D. Iwakiri, Y. Ono, and S. Fujiwara. 1998. Epstein-Barr-virus-infected human T-cell line with a unique pattern of viral-gene expression. Int. J. Cancer 76:587-594. [DOI] [PubMed] [Google Scholar]
- 41.Nemerow, G. R., R. Wolfert, M. E. McNaughton, and N. R. Cooper. 1985. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2). J. Virol. 55:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raslova, H., M. Berebbi, J. Rajcani, A. Sarasin, J. Matis, and M. Kudelova. 2001. Susceptibility of mouse mammary glands to murine gammaherpesvirus 72 (MHV-72) infection: evidence of MHV-72 transmission via breast milk. Microb. Pathog. 31:47-58. [DOI] [PubMed] [Google Scholar]
- 43.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 44.Rodrigiuez, A., M. Armstrong, D. Dwyer, and E. Flemington. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 73:9029-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savard, M., C. Belanger, M. Tardif, P. Gourde, L. Flamand, and J. Gosselin. 2000. Infection of primary human monocytes by Epstein-Barr virus. J. Virol. 74:2612-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speck, P., and R. Longnecker. 2000. Infection of breast epithelial cells with Epstein-Barr virus via cell-to-cell contact. J. Natl. Cancer Inst. 92:1849-1851. [DOI] [PubMed] [Google Scholar]
- 47.Swenson, J. J., A. E. Mauser, W. K. Kaufmann, and S. C. Kenney. 1999. The Epstein-Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J. Virol. 73:6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takada, K. 2000. Epstein-Barr virus and gastric carcinoma. Mol. Pathol. 53:255-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanner, J. E., C. Alfieri, T. A. Chatila, and F. Diaz-Mitoma. 1996. Induction of interleukin-6 after stimulation of human B-cell CD21 by Epstein-Barr virus glycoproteins gp350 and gp220. J. Virol. 70:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walling, D. M., C. M. Flaitz, C. M. Nichols, S. D. Hudnall, and K. Adler-Storthz. 2001. Persistent productive Epstein-Barr virus replication in normal epithelial cells in vivo. J. Infect. Dis. 184:1499-1507. [DOI] [PubMed] [Google Scholar]
- 51.Webster-Cyriaque, J., J. Middeldorp, and N. Raab-Traub. 2000. Hairy leukoplakia: an unusual combination of transforming and permissive Epstein-Barr virus infections. J. Virol. 74:7610-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, M., J. S. Pagano, J. T. Schiller, S. S. Tevethia, N. Raab-Traub, and J. Gruber. 2002. New associations of human papillomavirus, Simian virus 40, and Epstein-Barr virus with human cancer. J. Natl. Cancer Inst. 94:1832-1836. [DOI] [PubMed] [Google Scholar]
- 53.Wong, M. P., L. P. Chung, S. T. Yuen, S. Y. Leung, S. Y. Chan, E. Wang, and K. H. Fu. 1995. In situ detection of Epstein-Barr virus in non-small cell lung carcinomas. J. Pathol. 177:233-240. [DOI] [PubMed] [Google Scholar]
- 54.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with Zta and mediates Zta-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanai, H., K. Takada, N. Shimizu, Y. Mizugaki, M. Tada, and K. Okita. 1997. Epstein-Barr virus infection in non-carcinomatous gastric epithelium. J. Pathol. 183:293-298. [DOI] [PubMed] [Google Scholar]
- 56.Yoshiyama, H., S. Imai, N. Shimizu, and K. Takada. 1997. Epstein-Barr virus infection of human gastric carcinoma cells: Implication of the existence of a new virus receptor different from CD21. J. Virol. 71:5688-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]