Abstract
The gammaherpesviruses characteristically drive the proliferation of latently infected lymphocytes. The murine gammaherpesvirus 68 (MHV-68) MK3 protein contributes to this process in vivo by evading CD8+-T-cell recognition during latency, as well as during lytic infection. We analyzed some of the molecular mechanisms that control MK3 expression. No dedicated MK3 mRNA was detected. Instead, the MK3 open reading frame (ORF) was transcribed as part of a bicistronic mRNA, downstream of a previously unidentified ORF, 13M. The 13M/MK3 promoter appeared to extend approximately 1 kb 5′ of the transcription start site and included elements both dependent on and independent of the ORF50 lytic transactivator. MK3 was translated from the bicistronic transcript by virtue of an internal ribosome entry site (IRES) element. RNA structure mapping identified two stem-loops between 13M and MK3 that were sufficient for IRES activity in a bicistronic reporter plasmid and a third stem-loop just within the MK3 coding sequence, with a subtler, perhaps regulatory role. Overall, translation of the MHV-68 MK3 bore a striking resemblance to that of the Kaposi's sarcoma-associated herpesvirus vFLIP, suggesting that IRES elements are a common theme of latent gammaherpesvirus immune evasion in proliferating cells.
The murine gammaherpesvirus 68 (MHV-68) is a natural parasite of mice (4, 5) that is related to the Kaposi's sarcoma-associated herpesvirus (KSHV). Thus, we can learn from MHV-68 something of how KSHV persists in immunocompetent hosts and causes disease. Some 90% of MHV-68 genes have clear position or sequence homologs in KSHV (42). However, the homology is greatest for the genes encoding structural virion components and essential lytic replication enzymes; there is much less conservation of host interaction genes such as those concerned with immune evasion. Of course, it is precisely these functions that are difficult to define in vitro and about which MHV-68 can be most informative. Thus, much of the utility of MHV-68 as a model for human disease mechanisms depends on identifying how the host interaction functions of each virus—assumed to have a greater commonality than is apparent from DNA sequence alignments—are distributed among their more variable genes.
Immune evasion is a case in point. The list of immune evasion genes for either MHV-68 or KSHV genes is far from complete, but already the general impression is that those of each virus have evolved as a coordinated set, with the acquisition of a new gene leading to modified functions for the others. KSHV has two lytic cycle genes that downregulate major histocompatibility complex (MHC) class I expression, K3 and K5, while MHV-68 has just one, MK3 (10, 17, 37). An MHV-68 chemokine binding protein, M3, also mediates CD8+-T-cell evasion (7, 32) and may compensate for the lack of a K5, although exactly where it fits into in vivo pathogenesis remains controversial (41). M3 may also overlap in function with the KSHV vMIPs (26). In addition to its lytic cycle repertoire, KSHV has a latency gene, vFLIP (11, 36), that blocks death domain receptor signaling (40) and can protect a B-cell tumor against immune elimination (12). MHV-68 has no vFLIP. However, the MHV-68 MK3 is transcribed in latently infected germinal center B cells as well as in the viral lytic cycle, and a major feature of the MK3-deficient MHV-68 phenotype is a defect in viral latency amplification (38). Thus, MHV-68 may have evolved broader MK3 expression rather than a vFLIP to protect latent genomes against T cells. Understanding the control of MK3 expression is thus essential to interpreting its function and relating this to immune evasion by other gammaherpesviruses. In particular, we wish to know how MK3 might be adapted to function in proliferating cells.
CD8+-T-cell evasion by MHV-68 was originally localized to MK3 by transfecting genomic library clones plus the ORF50 viral transactivator into target cells presenting an MHC class I-restricted T-cell epitope (37). This approach also established that MK3 transcription in fibroblasts depends on ORF50-responsive promoter elements sited more than 500 bp upstream of the start of the MK3 open reading frame (ORF). While there is a consensus polyadenylation site just 3′ of the MK3 ORF, close to ORF11, the 5′ end of its transcript is unknown. Furthermore, the 5′ end of the MK3 ORF abuts a 1.5-kb genomic region of unknown function. In order to identify the MK3 promoter and to understand more about the control of its expression, we mapped the MK3 transcript and investigated its translation.
MATERIALS AND METHODS
Retroviral expression plasmids.
The 13M/MK3 transcript, from the start of the 13M ORF to the end of the MK3 ORF, was amplified from MHV-68 genomic DNA by PCR (Hi-Fidelity; Roche Diagnostics, Lewes, United Kingdom) including a 5′ MfeI site and a 3′ XhoI site in the respective primers. The PCR product was cloned into the EcoRI and XhoI sites of pMSCV-IRES-GFP, which expresses green fluorescent protein (GFP) from an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) 3′ of the XhoI site (29). The 13M ORF by itself and 13M/MK3-CT, which has an inactivating C-terminal truncation of MK3 (17), were similarly cloned into pMSCV-IRES-GFP. pMSCV-MK3-IRES-GFP has been described previously (37). For some experiments, the 13M/MK3 was cloned into pMSCV-IRES-PURO, in which the GFP gene was replaced with puromycin acetyltransferase. To replace the 13M ORF with part of the MHV-68 thymidine kinase (TK), the entire TK ORF (genomic coordinates 32879 to 34813) was first amplified by PCR and was cloned into pMSCV-IRES-GFP by using EcoRI and XhoI sites introduced at the 5′ and 3′ ends of TK, respectively. The 3′ end of TK was then removed by digestion with BglII (cuts at genomic coordinate 32999) and XhoI. The 13M/MK3 PCR product (see above) was digested with BamHI (cuts at genomic coordinate 25494, near the 3′ end of the 13M ORF) and XhoI and was ligated into the BglII and XhoI sites of pMSCV-TK-IRES-GFP. The TK/MK3 construct thus comprised the first 367 amino acids of TK, a stop codon in the 3′ fragment of the 13M ORF, the 13M/MK3 intergenic region, and the MK3 ORF.
Viruses and cell lines.
All cells were maintained in Dulbecco's modified Eagle's medium (Sigma Chemical Co., Poole, United Kingdom), supplemented with penicillin (100 U/ml, streptomycin (100 μg/ml), glutamine (2 mM), and 10% fetal bovine serum (PAA Laboratories, Linz, Austria). NIH 3T3 cells (American Type Culture Collection CRL-1658), BALB/c-3T3 cells (CCL-163), 293T cells (CRL-11268), and BHK-21 cells (CCL-10) have been described previously (37). A20 cells (TIB-208) were obtained from F. K. Stevenson (Tenovus Research Laboratories, Southampton, United Kingdom). Replication-defective retroviruses were generated by transfecting 293T cells with the relevant pMSCV-based retroviral expression plasmid plus pEQPAM3 (29), using Fugene-6 (Roche) as previously described (6). Supernatants were collected 48 and 72 h after transfection, filtered (0.45-μm pore size), and were used to transduce cells in the presence of 6 μg of hexadimethrine bromide (Sigma)/ml. Cells transduced with pMSCV-13M/MK3-IRES-PURO were selected with 30 μg of puromycin/ml. MHV-68 was grown and titered in BHK-21 cells as previously described (9).
RACE analysis.
RNA was extracted from NIH 3T3 cells 18 h after infection with MHV-68 (5 PFU/cell) by using RNAzol B (Tel-Test, Friendswood, Tex.). The ends of the MK3-specific mRNA were identified by using the 5′/3′ rapid-amplification-of-cDNA ends (RACE) kit (Roche Diagnostics). Briefly, for the 5′ end total RNA was reverse transcribed by using a primer corresponding to genomic coordinates 25129 to 25154 of the MK3 ORF (24730 to 25335) (42). A 5′ poly(A) tail was added to the cDNA by using terminal transferase, and the product was amplified by PCR, by using a 5′ poly(A)-specific primer that also included 5′ restriction sites and a 3′ primer corresponding to genomic coordinates 25165 to 25189. For the 3′ end, cDNA synthesis was primed with the poly(A)-specific primer and was then amplified with a 5′ primer matching the start of the 13M ORF (genomic coordinates 25717 to 25701) and a 3′ primer identical to the 5′ part of the poly(A)-specific primer but lacking its oligo-dT tract. The PCR products were gel purified (Qiaquick gel extraction kit; Qiagen Ltd., Crawley, United Kingdom) and were sequenced directly. The 5′ RACE product was also cloned into pSP73 (Promega Corporation, Southampton, United Kingdom) by using a ClaI site in the poly(A)-specific primer and an AatII site in MK3, and three independent clones were sequenced. In vivo 13M/MK3 transcripts were amplified by PCR with primers corresponding to genomic coordinates 25219 to 25244 and 25717 to 25701.
Northern blotting.
RNA was purified from uninfected or MHV-68-infected NIH 3T3 cells (5 PFU/cell, 18 h) by using RNAzol-B, electrophoresed (20 μg/lane) on a 0.8% formaldehyde agarose gel, and blotted overnight onto uncharged nylon membranes (Roche Diagnostics). Probe templates for β-actin, ORF MK3 (genomic coordinates 24730 to 25335), and ORF 13M (genomic coordinates 25500 to 25717) were generated by PCR of either cellular cDNA or MHV-68 BAC DNA. These were gel purified and random prime labeled (Qbiogene, Nottingham, United Kingdom) with [32P]dCTP (Amersham Biosciences, Little Chalfont, United Kingdom). Blots were hybridized with probe overnight at 45°C in 50% formamide, 5× Denhardt's solution, 6× SSC, 0.1% sodium dodecyl sulfate (SDS), and 100 μg of sonicated salmon sperm DNA buffer/ml, followed by washing (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 65°C) and exposure to X-ray film.
RNase protection assay.
Genomic coordinates 25275 to 25773 of the MHV-68 genome were amplified by PCR, including HindIII and XbaI sites as 5′ extensions in the respective 5′ and 3′ primers. The PCR product was cloned into the HindIII and XbaI sites of pSP73. The plasmid was linearized with XbaI, and an antisense riboprobe was generated with T7 RNA polymerase (Ambion Europe Ltd., Huntingdon, United Kingdom), labeling with [32P]UTP (Amersham Biosciences) according to the manufacturer's instructions. The probe was hybridized to mRNA from MHV-68-infected NIH 3T3 cells (5 PFU/cell, 18 h) using the Direct Protect lysate RPA kit (Ambion). Following RNase A/RNase T1 treatment, protected fragments were purified and separated on 8 M urea/6% polyacrylamide gels.
Flow cytometry.
All antibodies were obtained from BD-Pharmingen (Oxford, United Kingdom). Cells were washed in phosphate-buffered saline-bovine serum albumin (0.1%)-azide (0.01%), incubated for 15 min on ice with 10% mouse serum plus anti-CD16/32 monoclonal antibody (2.4G2), and stained for 1 h on ice by using biotinylated anti-H-2Kd (SF1-1.1) or biotinylated anti-H-2Dd (34-2-12), followed by phycoerythrin-conjugated streptavidin. After two washes in ice-cold phosphate-buffered saline, cells were analyzed on a FACSCalibur by using Cellquest (BD Pharmingen). Data were plotted with FCSPress v1.3 (http://www.fcspress.com).
RNA structure mapping.
The 3′ end of the 13M ORF, the 13M/K3 intergenic region and the 5′ end of the MK3 ORF (genomic coordinates 25501 to 25300) were amplified by PCR from viral DNA, including 5′ and 3′ SacI and EcoRI restriction sites in the respective primers. The PCR product was cloned into pBluescript II KS(+) (Stratagene, La Jolla, Calif.). RNA was transcribed from EcoRI-cleaved plasmid (100 μg/ml) by using T7 polymerase (New England Biolabs, Hitchin, United Kingdom) (2,500 U/ml) in 40 mM Tris-Cl (pH = 8) with 15 mM MgCl2, 5 mM dithiothreitol, and 2.5 mM nucleoside triphosphates for 3 h at 37°C. Following phenol-chloroform extraction and ethanol precipitation, DNA template was removed by digestion with RQ1 RNase-free DNase (Promega) and Sephadex G-50 column chromatography. RNA was then concentrated by ethanol precipitation and dephosphorylated by using Puccinellia borealis alkaline phosphatase, followed by heat inactivation at 68°C with 0.5% SDS, phenol-chloroform extraction, and ethanol precipitation. The RNA was then 5′ end labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-33P]ATP (110 TBq/mmol; APBiotech, Amersham, United Kingdom) according to the manufacturer's instructions, followed by purification in a 15% acrylamide-7 M urea gel and ethanol precipitation. Mapping reactions (2.5 × 107 cpm/ml of labeled RNA + 10 μg of unlabeled baker's yeast tRNA as a carrier) followed previously described protocols (23). RNase T1 digestions (Roche), which cut preferentially at single-stranded G residues, were for 20 min at 4°C in 50 mM sodium cacodylate (pH = 7) and 2 mM MgCl2 with 0 to 4 U of enzyme/ml. RNase CV1 (Ambion Ltd.), which cuts at double-stranded or stacked RNA bases, was used (0 to 1 U/ml) according to the manufacturer's instructions. RNase U2 digestions (APBiotech), which cut preferentially at single-stranded A residues, were for 20 min at 4°C in 20 mM NaAc (pH = 4.8), 2 mM MgCl2, 100 mM KCl with 0 to 4 U of enzyme/ml. All reactions were stopped by adding 3 volumes of ethanol. Lead probing was for 5 min at 25°C in 20 mM HEPES (pH = 7.5), 5 mM MgAc, and 50 mM KAc with 2 to 4 mM PbAc. Reactions were stopped with EDTA (final concentration, 33 mM), and RNA was precipitated with ethanol. For imidazole probing, labeled RNA was dried in a desiccator and was redissolved in 40 mM NaCl, 10 mM MgCl2, and 2 M imidazole (pH = 7). After 2 to 4 h at 37°C, the reaction was stopped by adding 1 volume of 2% lithium perchlorate in acetone. All RNA was recovered by centrifugation (13,000 × g, 15 min, 4°C), washed in acetone, dried, and resuspended in water. As a size marker, labeled RNA was subjected to partial alkaline hydrolysis in 22.5 mM NaHCO3, 2.5 mM Na2CO3 for 1 min at 100°C. All samples were mixed with an equal volume of formamide gel loading buffer (95% [wt/vol] formamide, 10 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol), boiled for 2 min, and resolved in 6% polyacrylamide-7 M urea gels. Gels were dried and exposed to X-ray film.
Luciferase assays.
Portions of the 13M/MK3 transcript were amplified from viral DNA by PCR (Hi-Fidelity; Roche), including SacI and SalI sites in the respective 5′ and 3′ primers. The EMCV IRES was amplified from pMSCV-IRES-GFP. All PCR products were cloned into the bicistronic reporter plasmid p2LUC (13). Transcription from this plasmid is driven by a simian virus 40 (SV40) promoter. There is a 5′ Renilla luciferase gene (RLUC), lacking a stop codon, and a 3′ firefly luciferase gene (FLUC), lacking a start codon. Unique SacI and SalI sites separate the two luciferase coding sequences. A stop codon for the upstream Renilla luciferase was included in each 5′ PCR primer. The genomic coordinates of the 13M/MK3 inserts were as follows: clone 1, 25352 to 25261; clone 2, 25715 to 25105; clone 3, 25715 to 25330; clone 4, 25494 to 25105; clone 5, 25442 to 25105; clone 6, 25352 to 25105; and 22-nucleotide (nt) insert, 25352 to 25330. 293T cells were transfected with p2LUC derivatives by using Fugene-6 (Roche). Forty to 48 h later, cells were harvested and assayed for luciferase activity by using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
CAT assays.
MHV-68 genomic fragments upstream of the 13M/MK3 transcription start site were amplified from viral DNA by PCR (Hi-Fidelity; Roche), including PstI and HindIII sites in the respective 5′ and 3′ primers. The invariant 3′ primer corresponded to genomic coordinates 25710 to 25726. The upstream primers started at genomic coordinates 25898 (clone P1), 26093 (clone P2), 26228 (clone P3), and 26721 (clone P4). All PCR products were cloned into the PstI and HindIII sites of pCAT (Promega). The positive control vector has an SV40 early promoter upstream of chloramphenicol acetyltransferase (CAT) (pCAT-SV40). An ORF50 expression plasmid, comprising a genomic fragment spanning the entire ORF50 coding sequence (21) cloned into pcDNA3 (Invitrogen, Paisley, United Kingdom), has been described previously (37). 293T cells or NIH 3T3 cells were transfected with the relevant 13M/MK3 promoter construct plus either pcDNA3-ORF50 or empty pcDNA3 vector by using Fugene-6 (Roche). In some experiments, NIH 3T3 cells were infected with MHV-68 (5 PFU/cell) 24 h after transfection. Cells were harvested 40 to 48 h after transfection and were processed for CAT enzyme-linked immunosrbent assay (Roche) according to the manufacturer's instructions.
RESULTS
Identification of a bicistronic transcript incorporating MK3.
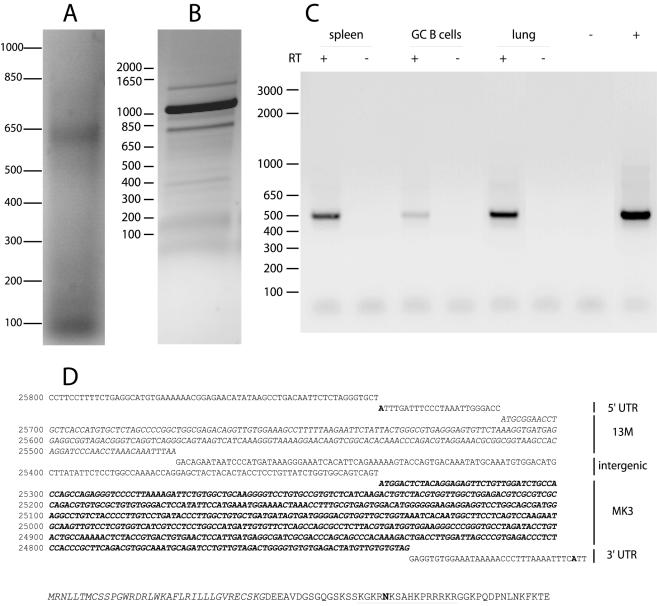
We located the 5′ and 3′ ends of the MK3 transcript in MHV-68-infected NIH 3T3 cells by RACE (Fig. 1A). No 5′ RACE product of the size expected for a dedicated MK3 mRNA (200 to 300 bp) was observed. Instead, there was a single band corresponding to an mRNA starting at least—depending on splicing—400 bp upstream of the 5′ end of the MK3 ORF (genomic coordinate 25335). The MK3 ORF differs from the KSHV K3 and K5 ORFs in having no consensus splice acceptor site near its 5′ end, and DNA sequencing of the 5′ RACE product revealed no evidence of splicing. The transcription start site, identified by a poly(A) tail added during the RACE procedure, was at genomic coordinate 25735 (Fig. 1D). The 3′ end of the mRNA was then also mapped by RACE, with a 3′ primer matching that used for oligo-dT-based reverse transcription and a 5′ primer matching the 5′ end of the MK3 mRNA (Fig. 1B). There was a predominant 1,050-bp product. DNA sequence analysis confirmed that this covered the entire MK3 ORF without splicing and terminated in a poly(A) tail at genomic coordinate 24697, just after the consensus polyadenylation site 3′ of the MK3 ORF (Fig. 1D). We used primers spanning the 5′ RACE-mapped transcription start site and the conserved MK3 RING finger to identify in in vivo samples an MK3 mRNA extending at least 400 nt upstream of the ORF. The mRNA was found both in the lungs of mice infected 3 days earlier with MHV-68 and in the spleens and germinal center B cells of mice infected 14 days earlier (Fig. 1C). We have previously shown germinal center B cells to be a site of latent MK3 expression (38). Thus, there was evidence of MK3 transcription initiating at least 400 bp upstream of the MK3 ORF in both lytically and latently infected tissues.
FIG. 1.
Identification of a bicistronic MK3 transcript. (A) 5′ RACE analysis of MHV-68-infected NIH 3T3 cells gave a single band of approximately 600 bp but gave no band corresponding to a transcription start site close to the 5′ end of the MK3 ORF (200 to 300 bp). The sub-100-bp band corresponds to paired PCR primers. An MK3 mRNA of this length would lack the RING domain that is essential for MK3 function (6). Equivalent data were obtained in two separate experiments. (B) 3′ RACE analysis starting from the identified 5′ end gave a predominant band of approximately 1,050 bp. This was then sequenced from its 5′ end (D). (C) In order to identify the putative bicistronic mRNA by reverse transcriptase PCR of in vivo samples, we used one primer matching the 5′ end of the RACE product (25717 to 25701) and another in the conserved RING finger coding region of MK3 (25219 to 25244). A single band corresponding to an unspliced mRNA was identified in poly(A)-primed cDNA from lungs 3 days after intranasal MHV-68 infection, spleens 14 days after intranasal MHV-68 infection (five mice pooled), and germinal center (GC) B cells (CD19+ PNAhi), purified from the same spleens by flow cytometric sorting. We have previously characterized germinal center B cells as a site of latency-associated MK3 transcription (38). The negative control (−) was without cDNA, and the positive control (+) was MHV-68 genomic DNA. RT = reverse transcriptase. (D) DNA sequence analysis starting within the MK3 ORF identified the MK3 transcription start site in both total and cloned 5′ RACE products as genomic coordinate 25735 (shown in boldface). The most 5′ AUG codon in this transcript (25711) would give a short ORF ending at genomic coordinate 25476 that we have called 13M (shown in italics) and then a 140-nt intergenic region before the start of the MK3 ORF (boldfaced italics). The 3′ RACE product was also unspliced and was terminated in a poly(A) tail at genomic coordinate 24697 (shown in boldface), 22 nt after a consensus polyadenylation site (underlined). The inferred 13M protein coding sequence is shown below, including a possible 35-amino-acid signal sequence (italics), a consensus N-linked glycosylation site (boldface), and a positively charged domain (underlined).
Translation from the most 5′ AUG of the MK3 mRNA predicted an 81-amino-acid protein, terminating at a stop codon 141 bp upstream of the MK3 ORF. DNA sequence analysis of the upstream ORF revealed a potential leader sequence, a consensus N-linked glycosylation site, and highly positively charged C terminus, although no homology with other sequenced herpesvirus genes (Fig. 1D) was revealed. This putative gene product we have called 13M.
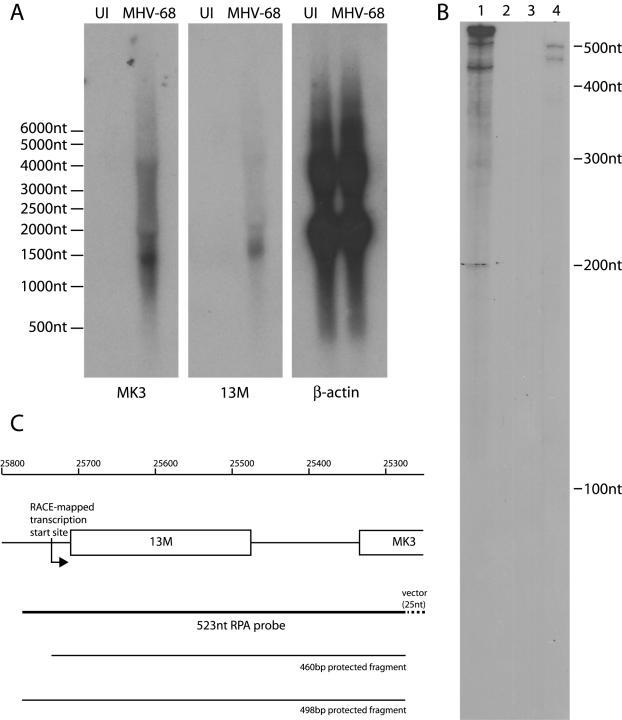
Northern blots (Fig. 2A) identified a predominant 1.4-kb MK3 mRNA, consistent with the 1,050-nt transcript predicted by RACE mapping plus a poly(A) tail. There was no band corresponding to the predicted size of a monocistronic MK3 mRNA (approximately 1 kb). Furthermore, identical patterns of Northern blot hybridization were obtained with a 13M ORF probe and the MK3 ORF probe. Thus, MK3 and 13M appeared to be transcribed as a single unit.
FIG. 2.
Northern blot and RNase protection assay of the 13M/MK3 mRNA. (A) RNA from NIH 3T3 cells either uninfected (UI) or infected overnight with MHV-68 at 5 PFU/cell (MHV-68) was probed for β-actin, the MK3 ORF and the 13M ORF as indicated. The positions of RNA size markers (Ambion Europe Ltd.) are shown. Equivalent results were obtained in two experiments. (B) A 523-nt 32P-labeled RNA probe, comprising 25 nt of vector sequence and 498 nt of MHV-68 sequence (genomic coordinates 25275 to 25773) antisense to the MK3 coding strand, was used for RNase protection. Protected fragments were separated by polyacrylamide gel electrophoresis and were exposed to X-ray film. Lane 1 = probe only, no RNase; lane 2 = probe only, plus RNase; lane 3 = probe hybridized to uninfected NIH 3T3 cell lysate, plus RNase; and lane 4 = probe hybridized to MHV-68-infected NIH 3T3 cell lysate, plus RNase. The positions of RNA size markers are shown. The data are representative of two independent experiments. (C) Diagram of the 13M/MK3 transcription start site relative to the RNase protection probe used in panel B. Genomic coordinates are shown, together with the locations of the 13M ORF, the 5′ end of the MK3 ORF, and the probable ends of the protected fragments.
RNase protection assays (Fig. 2B) provided additional evidence that the predominant MK3 transcription start site lies upstream of the 13M ORF. Two bands were consistently observed. The smaller (460 bp) band corresponded to the predicted overlap between the labeled probe and the RACE-mapped 5′ end of the 13M/MK3 mRNA (Fig. 2C). The larger (498 bp) band corresponded to the full length of the MHV-68-specific sequence in the probe. While this band may have reflected an additional, upstream transcription start site, it seemed likelier either that it was either an RNA/DNA hybrid, resulting from the very large number of viral genomes in lytically infected cell lysates (the 13M/MK3 mRNA was of relatively low abundance) or that it represented aberrant late transcripts from further upstream that failed to terminate normally. The key point was that there was no smaller band (approximately 100 bp) that would have been expected for a monocistronic MK3 mRNA.
MK3 expression from the 13M/MK3 transcript.
As MK3 was transcribed only downstream of 13M, an immediate question was how this mRNA, which contained six AUG codons before the start of the MK3 ORF, produced MK3 protein (Fig. 1D). Several of the upstream AUG codons were in a reasonable context for translation initiation. The 13M ORF was out of frame with MK3 and was followed by stop codons in all three reading frames, making a fusion of the two unlikely. And the presence of four AUG codons between the end of 13M and the start of MK3 argued against scanning ribosomes translating 13M and then reinitiating at MK3. MK3 translation seemed likeliest to depend on an IRES.
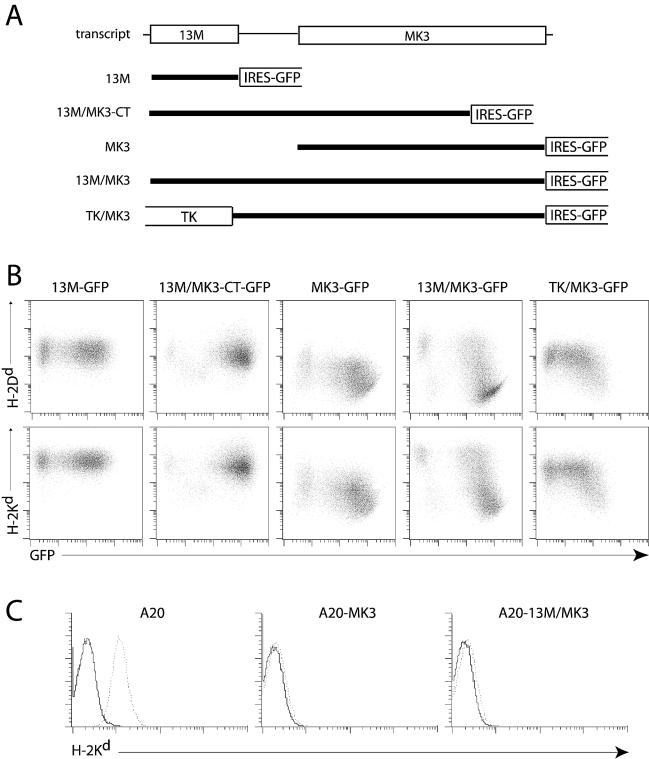
In order to establish whether MK3 was translated from the putative bicistronic mRNA, DNA corresponding to the complete 13M/MK3 transcript—from the start of 13M to the end of MK3—was cloned into pMSCV-IRES-GFP (Fig. 3A). We used MHC class I downregulation as a marker of MK3 protein expression (37). The level of GFP fluorescence in cells transduced with cell pMSCV-IRES-GFP derivatives generally reflects retroviral promoter activity and is thus proportional to the transcription of any upstream genes. MK3 translation therefore manifested itself as a loss of MHC class I staining on GFP+ cells. Neither 13M by itself nor 13M plus a truncated form of MK3 that does not bind MHC class I heavy chains (6) caused MHC class I downregulation (Fig. 3B). In contrast, 13M/MK3 was highly effective and comparable to MK3 alone. Thus, MK3-dependent MHC class I downregulation was mediated by the 13M/MK3 mRNA.
FIG. 3.
MK3 translation from the 13M/MK3 transcript. (A) Portions of the 13M/MK3 transcript were cloned into the retroviral expression vector pMSCV-IRES-GFP, which translates GFP from the EMCV IRES. Thus, the retroviral transcript was bicistronic or tricistronic depending on the insert. MK3-CT has been previously characterized as a nonfunctional MK3 mutant (6). In TK/MK3, the 13M coding sequence was replaced by that of the first 367 amino acids of the MHV-68 TK. (B) BALB/c-3T3 cells exposed to replication-defective retrovirus were analyzed for MHC class I downregulation—a characteristic of MK3—in transduced (GFP+) cells. The data shown are from one of three equivalent experiments. (C) The MK3 ORF or the 13M/MK3 bicistronic construct was cloned into pMSCV-IRES-PURO and was used to transduce the A20 B-cell line. After selection with puromycin, the cells were stained for H-2Kd (dotted line) or secondary antibody only (solid line).
One possibility was that the 13M ORF represented simply the 5′ untranslated region of a dedicated MK3 transcript, with the MK3 AUG being the one first accessible to cap-dependent ribosomes. To rule this out, we replaced the 13M ORF with a large, known coding sequence, the N-terminal 1.1 kb of the MHV-68 TK. The 13M/MK3 intergenic sequence was retained, with TK translation ending 10 nt upstream of the 13M stop codon. TK/MK3 still downregulated MHC class I expression in GFP+ cells, though to a lesser degree, possibly reflecting a loss of part of an MK3 IRES. However, large upstream ORFs tend generally to reduce downstream gene expression in this retroviral system, as seen by the lower GFP expression from the EMCV IRES in the same mRNA. The quantitative relationship between GFP fluorescence intensity and the degree of MHC class I downregulation was maintained. Thus, these data supported the idea of a 13M/MK3 intergenic IRES element being responsible for translation from the MK3 AUG.
Since the 13M/MK3 transcript was also identified in latently infected B cells (Fig. 1C), we looked for evidence of IRES activity in a B-cell line (Fig. 3C). A20 cells were transduced with retrovirus expressing either the 13M/MK3 bicistronic mRNA or MK3 alone. We used a retroviral vector encoding puromycin acetyltransferase after the EMCV IRES rather than GFP and selected cells on the basis of puromycin resistance. The MHC class I downregulation achieved by 13M/MK3 was equivalent to that with MK3 alone. Thus, the MK3 IRES was functional in a B cell line.
RNA structure in the 13M/MK3 intergenic region.
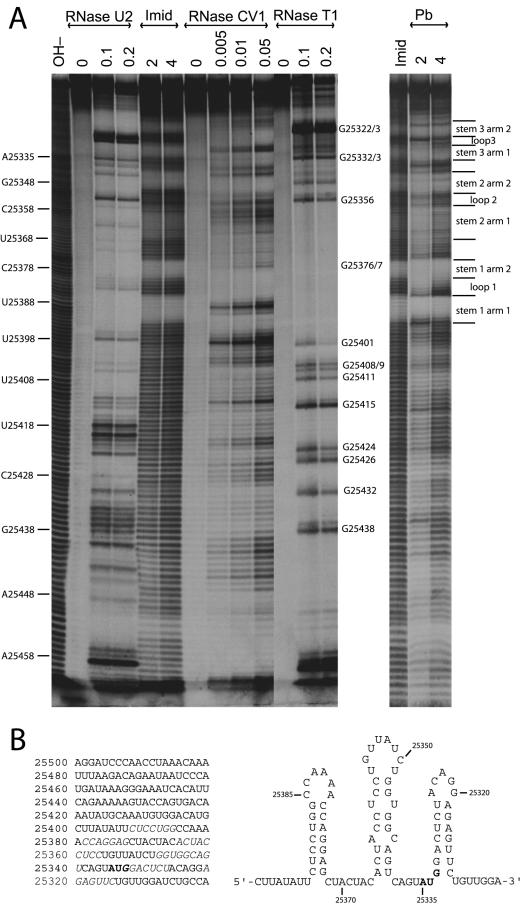
IRES activity generally requires RNA secondary structure 5′ of the relevant AUG codon that can replace a 5′ 7-methyl-guanosine mRNA cap and direct ribosome assembly. Since the 13M ORF appeared not to be essential for MK3 translation (Fig. 3), we looked for evidence of base pairing in an RNA corresponding to genomic coordinates 25501 to 25300—the entire 13M/MK3 intergenic region plus the start of the MK3 ORF (Fig. 4). Imidazole and lead treatments unambiguously identified six regions of stably base-paired RNA near the 5′ end of the MK3 ORF. Regions of less marked protection against cleavage by lead were also apparent further upstream and showed some correlation with susceptibility to cleavage by RNase CV1, for example, nucleotides 25426 to 25430 and 25442 to 25448. However, with the possible exception of nucleotides 25450 to 25453, there was little protection against imidazole, suggesting that these regions were not incorporated into a stable secondary structure at 37°C.
FIG.4.
RNA structure mapping of the 13M/MK3 transcript. (A) The 13M/MK3 intergenic region and the start of the MK3 ORF (genomic coordinates 25501 to 25300) were transcribed as a 5′ end-labeled RNA, subjected to chemical or enzymatic partial degradation as outlined in Materials and Methods, and analyzed by polyacrylamide gel electrophoresis and autoradiography. The genomic coordinates of specific residues and the locations of inferred secondary structure features are shown. OH− = alkaline hydrolysis; Imid = imidazole; Pb = lead. Molarities are shown for imidazole and lead; enzyme units per 50-μl reaction are shown for the RNase treatments. Equivalent results were obtained in two independent experiments. (B) Nucleotide sequence of the transcribed RNA, with regions showing evidence of base-pairing in italics and the MK3 start codon in boldface. On the right is shown an interpretation of the base-pairing: three stem-loops.
There was obvious nucleotide complementarity between the six adjacent regions protected against both imidazole and lead. Thus, the simplest explanation for the RNA mapping data was three stem-loops (Fig. 4B). Approaching the MK3 AUG in a 5′-to-3′ direction, the first stem would have a CUCCUGG arm 1, paired to a CCAGGAG arm 2, with an intervening CCAAAA loop. RNase U2 cleaved the 4 A residues in the loop but not the two A residues in arm 2; RNase T1 did not cleave the G residues in either arm 1 or arm 2; and RNase CV1 cleaved in both arm 1 and arm 2. The second stem would have ACUACCUCC paired with GGUGGCAGU, incorporating two mismatches, and a UGUUAUCU loop. The less marked protection against imidazole in stem 2 arm 1 was consistent with imperfect base pairing. RNase U2 also showed some cleavage at the mispaired A residue in arm 1 (genomic coordinate 25362) but not at the 2 other A residues (25342, 25365). RNase CV1 cleaved within each stem arm. RNase T1 showed weak or absent cleavage of the 5 G residues in arm 2 but cleaved in the loop at genomic coordinate 25337. The third stem would have GGACUCU paired with AGAGUUC. RNase T1 showed evident cleavage of the 2 G residues at the base of arm 1, implying that the G of the MK3 start codon (genomic coordinate 25333) contributed only weakly to stem-loop 3. There was very strong cleavage at the two G residues in the loop and protection from RNase T1 at the G residues in stem 3 arm 2 (genomic coordinates 25320 and 25318). RNase U2 failed to cleave at the A residues in either arm of stem 3 but did cleave strongly in the loop (genomic coordinates 25334 and 25336). RNase CV1 cleaved within stem 3 arm 1 and to a lesser degree within stem 3 arm 2. Stem-loop 3 was unusual in occupying the 5′ coding sequence of the dependent gene. An alternative translation start site for MK3 seemed unlikely, as there are no other in-frame AUG codons upstream of its conserved RING finger. The presumed unwinding of stem-loop 3 that would be required for the MK3 AUG to enter the ribosomal P site raised the question as to how it would contribute to IRES function.
IRES activity of the 13M/MK3 intergenic region.
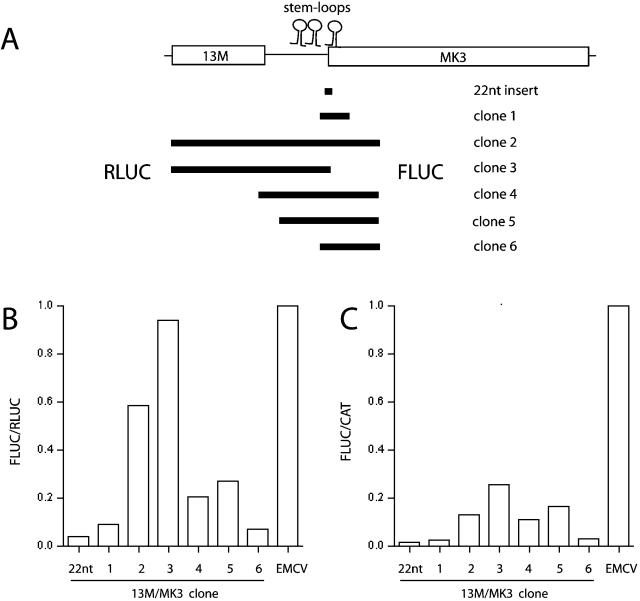
In order to correlate the RNA structure mapping data with putative IRES function, we cloned regions from the start of 13M up to the first transmembrane domain of MK3 (6) into the p2LUC bicistronic reporter plasmid, in which the relative efficiencies of cap-dependent and internal translation initiation read out as a ratio of upstream and downstream luciferase activities. Because IRES-dependent translation often depends on a fixed spacing of RNA secondary structure and the initiating AUG and because the 13M/MK3 stem-loop 3 incorporated the MK3 AUG, we retained this codon in its natural context as the start of the downstream luciferase gene. The reporter plasmid thus comprised a 5′ cap-dependent RLUC, a stop codon, the putative IRES sequence, and FLUC fused in frame to an N-terminal sequence derived from MK3 (Fig. 5A). Unmodified p2LUC has no AUG for FLUC, which is out of frame with RLUC, and no stop codon for RLUC, which consequently reads across the start of the FLUC coding sequence before terminating. Unsurprisingly, FLUC activity was undetectable in cells transfected with p2LUC (data not shown). As a more realistic assay background, we used a 22-nt insert from the 13M/MK3 transcript (genomic coordinates 25330 to 25352), which provided a stop codon for RLUC and the MK3 AUG (25333 to 25335) in frame with FLUC but none of the stem-loop structures. The low FLUC activity in cells transfected with p2LUC-22nt presumably reflected rare events such as translational reinitiation or transcription from cryptic promoter sequences within RLUC generating a capped FLUC mRNA. IRES activity was taken as a reproducible increase over this background. As a positive control, we used the well-characterized EMCV IRES (18).
FIG. 5.
Dual luciferase assays of 13M/MK3 IRES function. (A) Portions of the 13M/MK3 transcript were cloned between RLUC and FLUC in the bicistronic vector p2LUC. In each case there was an identical termination codon for RLUC and FLUC translation started at the MK3 AUG. The EMCV IRES was used as a positive control. The positions of the stem-loops identified by RNA structure mapping (Fig. 2) are shown. The 22-nt insert included none of these. (B and C) IRES activity was expressed as a ratio of FLUC/RLUC. Each transfection also included a fixed amount of pCAT-SV40 so as to normalize independently by CAT activity, shown as the ratio FLUC/CAT. The data shown are from one of five experiments. There was a consistent hierarchy of IRES activity between the 13M/MK3 clones, although maximal 13M/MK3 activity (clone 3) ranged from 20 to 100% of EMCV IRES activity for unknown reasons.
According to the FLUC/RLUC ratio (Fig. 5B), extending the 22-nt 13M/MK3 insert to include the entire 5′ region of the transcript (clone 3) increased IRES activity 26-fold. Clone 3 lacks stem-loop 3 but includes stem-loops 1 and 2, which thus appeared to be sufficient for IRES function. With stem-loop 3 intact, adding stem-loops 1 and 2 (clone 2 versus clone 6) increased IRES activity only ninefold, suggesting that stem-loop 3 inhibited MK3 translation. Although stem-loop 3 conferred a twofold increase in IRES activity in the absence of stem-loops 1 and 2 (22-nt insert versus clone 6), a caveat here is that the MK3 RING finger fused to FLUC in clone 6 could potentially alter its stability or activity: an intact RING finger generally confers stability on MK3 (unpublished data). With the 22-nt insert, FLUC contains just 2 amino acids (MD) from MK3. Truncating the RING finger (to MDSTGEFCWICH) while retaining stem-loop 3 made no difference to IRES activity (clones 1 and 6), but this fusion may still have been more favorable for FLUC activity than MD alone. A 52-bp truncation that removed the weak secondary structure at genomic coordinates 25450 to 25453 had no effect (clones 4 and 5), arguing that this was not functionally significant. Overall, the data pointed toward a primary role for stem-loops 1 and 2 in IRES function and a more complex, possibly regulatory role for stem-loop 3.
Truncating the 13M/MK3 insert at its 5′ end retained all three stem-loops but still reduced the FLUC/RLUC ratio twofold (clone 2 versus clone 4). One possibility is that additional RNA structure within the 13M ORF was required for full IRES function. However, removing the 13M ORF also increased RLUC activity twofold (data not shown), suggesting that 13M was reducing RLUC rather than contributing to IRES function. Even though the 13M ORF was downstream of an RLUC stop codon, it is possible that some 13M protein was still produced, as occurred with FLUC in the 22-nt insert clone, and that this affected RLUC. Removing the stop codon between RLUC and 13M to leave an in-frame fusion protein almost completely abolished RLUC activity (data not shown). As an independent method of normalization, we included a fixed amount of a CAT reporter plasmid in transfections and expressed IRES activity as a FLUC/CAT ratio (Fig. 5C). By this measure, stem-loops 1 and 2 were still essential (clones 5 and 6) and stem-loop 3 still appeared to be inhibitory (clones 2 and 3), but there was little effect of removing 13M. This was consistent with the fact that the 3 stem-loops without 13M were sufficient for MHC class I downregulation by the TK/MK3 construct depicted in Fig. 2.
A 5′ T7 promoter in p2LUC can be used to transcribe mRNA for in vitro translation. However, although we were able to translate FLUC from T7 transcripts in reticulocyte lysates by using the EMCV IRES, the 13M/MK3 IRES was ineffective, even when the reticulocyte lysate was supplemented with an S100 fraction derived from A20 cells. This presumably reflected a sensitivity of the MK3 IRES to intracellular conditions that were not reproduced in lysates. Such sensitivity is not uncommon in IRES elements.
Control of 13M/MK3 transcription.
Having established that the 13M/MK3 mRNA was responsible for producing MK3 protein, our next aim was to define the 13M/MK3 promoter. The reason for this was twofold: first to identify a basis for the regulation of 13M/MK3 transcription, and second to learn more about a poorly understood region of the MHV-68 genome between the 5′ end of the 13M/MK3 transcript and the left-hand set of internal repeats (genomic coordinates 25735 to 26778). Sequence analysis identified two ORFs here (42), but these are unlikely to encode proteins (27); KSHV has at least seven genes in the equivalent region (28). We had earlier used the transfection of MHV-68 genomic clones to define a functional transcription unit for reduced antigen presentation by MK3 (37). This was ORF50 dependent and required promoter elements sited between genomic coordinates 25889 and 26216. One prediction of MK3 translation from a bicistronic transcript was that the promoter elements responsible for inhibiting antigen presentation would also be responsible for 13M transcription. We tested this by using a set of 3′ coterminal promoter region clones to express CAT in place of 13M in NIH 3T3 and 293T cells (Fig. 6).
FIG. 6.
Analysis of the 13M/MK3 promoter. (A) MHV-68 genomic clones upstream of the 13M/MK3 transcription start site were used to drive CAT expression as shown. The limits of the HindIII, PvuII, and XhoI restricted genomic clones previously used to drive ORF50-dependent MK3 expression (37) are indicated. The DNA sequence of the relevant region is shown below, with the internal repeat region in italics. The limits of each genomic clone are shown in boldface. The 13M/MK3 transcript is in boldfaced italics. (B) 293T cells and NIH 3T3 cells were transfected with the CAT expression constructs, with or without an additional ORF50 expression vector. We also used MHV-68 infection (5 PFU/cell) to drive expression from transfected CAT plasmids in NIH 3T3 cells. Nil, untransfected; +, pCAT-SV40. CAT activity was expressed as mean ± standard deviation increase (n-fold) over untransfected or vector-only controls. Each graph is representative of two or three replicate experiments.
In NIH 3T3 cells, there was no ORF50-independent CAT activity, even with a 986-bp promoter fragment. ORF50-dependent CAT activity was absent with a 163-bp fragment and progressively increased with extension of the promoter to 359, 494, and 986 bp. Superinfecting NIH 3T3 cells with MHV-68 rather than cotransfecting ORF50 gave a similar hierarchy of activity. Thus, the CAT assays in NIH 3T3 cells (Fig. 6) agreed well with the previous antigen presentation-based assays in L929 cells, in which PvuII-restricted and HindIII-restricted clones produced MK3 when transfected with pcDNA3-ORF50, while an XhoI-restricted clone did not (37). In functional terms, 13M and MK3 appeared to rely on the same promoter elements.
In 293T cells, a low level of ORF50-independent promoter activity was seen with the 986-bp promoter fragment and perhaps also with the 494-bp fragment but not with the smaller fragments. Promoter upregulation by cotransfected ORF50 was low with the 163-bp fragment but reached a maximum with the 359-bp fragment. We also used the HindIII site in P4 to add in a HindIII/SacI genomic clone containing the adjacent internal repeat elements (genomic coordinates 26711 to 28336). This achieved no additional promoter activity with or without cotransfected ORF50 in either 293T or NIH 3T3 cells (data not shown). These repeats tend to be unstable in Escherichia coli (1, 42), and the plasmid used had 14 of the estimated 36 copies of the 40-bp repeat present in the intact virus. This limitation, plus possible additional features of latently infected murine germinal center B cells, left some uncertainty over the control of latent 13M/MK3 transcription. But it appeared that the lytic promoter, which also incorporated some ORF50-independent activity, extended approximately 1 kb upstream of the transcription start site.
DISCUSSION
The MHV-68 MK3 was found to be transcribed solely as the downstream ORF of a bicistronic mRNA. MK3 translation occurred by virtue of an IRES, analogous to that described for the KSHV vFLIP (3, 14, 22). The fact that the MHV-68 MK3 and probably also the KSHV vFLIP are involved in T-cell evasion during viral latency suggested that these IRES elements have independently evolved to maintain immune evasion during the G2 and M phases of rapidly cycling cells, when cap-dependent translation is reduced. They may also help to offset any reduction in translation initiation caused by virus-induced eIF2α phosphorylation. If T-cell evasion contributes to the survival of gammaherpesvirus-induced B-cell tumors, then attacking IRES function might be a possible general route to therapy. Along these lines, it is noteworthy that several Epstein-Barr virus latent mRNAs also include an IRES element (16).
Although downstream translation initiation from the 13M/MK3 intergenic region with the MHV-68 TK (Fig. 3) or RLUC (Fig. 5) as an upstream ORF implied IRES-driven MK3 translation, any demonstration of IRES function must address the alternative possibility of cap-dependent translation from rare mRNAs generated by cryptic splice sites or cryptic promoters (20). The lack of shorter RACE products argued against this with MK3. Any functional MK3 mRNA must have initiated 5′ of the gene-specific RACE primers used, since they were sited within the coding sequence of the essential MK3 RING finger (6). But, despite the strong bias of PCR towards amplifying short products, none was identified, even after an additional 30 cycles of seminested amplification (data not shown). Thus, any such transcripts were vanishingly rare. Northern blotting and RNase protection assays (Fig. 2) confirmed that the MK3 mRNA initiated 5′ of the 13M ORF. The failure to detect PCR products shorter than genomic controls when primers corresponding to the 13M start site and the MK3 RING finger (Fig. 1C) were used further argued against cryptic mRNA splicing. Meanwhile MHC class I downregulation by 13M/MK3 was comparable to that achieved by monocistronic MK3 (Fig. 3). The similar requirements for upstream promoter sequences to downregulate MHC class I via MK3 (37) and to transcribe 13M (Fig. 6) were also consistent with their sharing a common transcript.
The presence of RNA secondary structure supported the idea of an IRES and revealed something of how it might function. Stem-loops 1 and 2 alone gave maximal IRES function, so the role of stem-loop 3 remained unclear. It is possible that other factors might interact with stem-loop 3 to enhance IRES activity in latently infected germinal center B cells, but it is tempting to speculate that stem-loop 3 must be unwound to accommodate a 14-nt downstream ribosomal footprint when the MK3 AUG enters the ribosomal P site. This would provide a potential mechanism for MHV-68 to modulate the relative proportions of 13M and MK3. The fairly simple structure of the 13M/MK3 IRES suggested that other functional elements remain to be identified. Complementarity to the 18S rRNA is a feature of several IRES elements (8); interestingly, 5 of the 7 single-stranded nucleotides between stem-loops 1 and 2 (ACUAC) were complementary to nucleotides 1069 to 1073 of the murine 18S rRNA, an unpaired region of helix 25 thought to be involved in translation initiation (25, 30). The adjacent sequence in arm 1 of stem-loop 2 contained 10 nucleotides (CACUACCUCC) also complementary to 18S rRNA but to a region that appears to be in a stable hairpin (25, 30). Obviously such matches must be confirmed functionally, but they illustrated the potential for intermolecular base pairing to supplement any interactions between ribosomal proteins and the MK3 stem-loops.
The extensive MK3 promoter underscored the importance to herpesviruses of gene regulation. Since ORF50 drives the viral lytic cycle (43), MK3 expression in latently infected germinal center B cells (24, 31, 38) implies that transcription can initiate from the 13M/MK3 promoter independently of ORF50. Not every aspect of regulated viral gene expression was likely to be captured in transfected fibroblasts—MHV-68 latency proteins or B cell-specific factors may activate the 13M/MK3 promoter—but the low level of ORF50-independent CAT activity seen in 293T cells possibly reflected such activity. In NIH 3T3 fibroblasts, the 13M/MK3 promoter was functionally dependent on ORF50 (Fig. 6), as it was in L929 cells (37). Consistent with this, although we could detect MK3 mRNA by reverse transcriptase PCR of cycloheximide-treated NIH 3T3 cells 6 h after MHV-68 infection, a much stronger signal was obtained without cycloheximide (data not shown). The progressive increase in CAT activity with increasing MK3 promoter length suggested that multiple binding sites for either ORF50 or ORF50-dependent proteins were distributed across it. Interesting in this regard were the multiple copies of a TTTTTNNNTGTTT consensus in the 13M/MK3 promoter: 1 in P1, 3 in P2, 4 in P3, and 6 in P4 (Fig. 6A). There are at least two copies of the same consensus upstream of the viral ORF6, ORF50, ORF57, and ORF61, among others. Thus, this is a candidate ORF50-responsive promoter element. The region between 13M and the left-hand internal repeats probably also incorporates an MHV-68 lytic replication origin. Genomic coordinates 26232 to 26374 are >70% identical to coordinates 101765 to 101624, just beyond the right-hand internal repeats (98980 to 101170). Both have A/T-rich and G/C-rich regions characteristic of herpesvirus lytic replication origins, and this function has been confirmed for the equivalent regions of KSHV (2).
What emerges from a comparison of MHV-68 and KSHV is that, while both viruses solve similar problems, their solutions do not necessarily show a simple equivalence. The KSHV K5 has a monocistronic spliced mRNA (33), and the KSHV K3, despite a complex transcription pattern (15), also has a 5′ splice acceptor site, consistent with cap-dependent translation. Both are lytic transcripts (19, 35, 39). Thus, in the viral life cycle, MK3 function probably overlaps with that of vFLIP as well as K3 and K5. The original equivalence of MK3 and K3 has perhaps been somewhat eroded by subsequent host/parasite coevolution. The use of MK3 rather than a vFLIP in MHV-68 latency perhaps reflects immunological differences between the respective hosts: mice have more restricted MHC class II expression than humans, so CD4+ T cells, which kill via death domain receptors, are probably a less significant component of antiviral cytotoxicity than are perforin/granzyme-dependent CD8+ T cells (34). CD8+-T-cell recognition may for MHV-68 be a more suitable immune evasion target than Fas.
Acknowledgments
We thank Sofia Marques and Pedro Simas for providing cDNA samples, Ann Kaminski for helpful discussion, Simon Talbot for providing plasmids used in preliminary experiments, and Anita Hancock for help with figures.
This work was funded by the Medical Research Council (United Kingdom) (project grant G9901295 of cooperative group G9800943) and the Wellcome Trust (project grant 059601). P.G.S. is an Academy of Medical Sciences/MRC Clinician Scientist (fellowship G108/462).
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 5.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 6.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 7.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chappell, S. A., G. M. Edelman, and V. P. Mauro. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 97:1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, H. M., B. de Lima, V. Morton, and P. G. Stevenson. 2003. Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J. Virol. 77:2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djerbi, M., V. Screpanti, A. I. Catrina, B. Bogen, P. Biberfeld, and A. Grandien. 1999. The inhibitor of death receptor signaling, FLICE-inhibitory protein, defines a new class of tumor progression factors. J. Exp. Med. 190:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grentzmann, G., J. A. Ingram, P. J. Kelly, R. F. Gesteland, and J. F. Atkins. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479-486. [PMC free article] [PubMed] [Google Scholar]
- 14.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaksson, A., M. Berggren, and A. Ricksten. 2003. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene 22:572-581. [DOI] [PubMed] [Google Scholar]
- 17.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 19.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, M. 2001. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 21:1899-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, S., I. V. Pavlova, H. W. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low, W., M. Harries, H. Ye, M. Q. Du, C. Boshoff, and M. Collins. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 75:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marczinke, B., R. Fisher, M. Vidakovic, A. J. Bloys, and I. Brierley. 1998. Secondary structure and mutational analysis of the ribosomal frameshift signal of rous sarcoma virus. J. Mol. Biol. 284:205-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques, S., S. Efstathiou, K. G. Smith, M. Haury, and J. P. Simas. 2003. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J. Virol. 77:7308-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melander, Y., L. Holmberg, and O. Nygard. 1997. Structure of 18 S ribosomal RNA in native 40 S ribosomal subunits. J. Biol. Chem. 272:3254-3258. [DOI] [PubMed] [Google Scholar]
- 26.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 27.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persons, D. A., M. G. Mehaffey, M. Kaleko, A. W. Nienhuis, and E. F. Vanin. 1998. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol. Dis. 24:167-182. [DOI] [PubMed] [Google Scholar]
- 30.Rairkar, A., H. M. Rubino, and R. E. Lockard. 1988. Chemical probing of adenine residues within the secondary structure of rabbit 18S ribosomal RNA. Biochemistry 27:582-592. [DOI] [PubMed] [Google Scholar]
- 31.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, J., B. de Lima, F. K. Stevenson, and P. G. Stevenson. 2002. A gamma-herpesvirus immune evasion gene allows tumor cells in vivo to escape attack by cytotoxic T cells specific for a tumor epitope. Eur. J. Immunol. 32:3481-3487. [DOI] [PubMed] [Google Scholar]
- 33.Rimessi, P., A. Bonaccorsi, M. Sturzl, M. Fabris, E. Brocca-Cofano, A. Caputo, G. Melucci-Vigo, M. Falchi, A. Cafaro, E. Cassai, B. Ensoli, and P. Monini. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell, J. H., and T. J. Ley. 2002. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20:323-370. [DOI] [PubMed] [Google Scholar]
- 35.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 39.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 41.van Berkel, V., B. Levine, S. B. Kapadia, J. E. Goldman, S. H. Speck, and H. W. Virgin. 2002. Critical role for a high-affinity chemokine-binding protein in gamma-herpesvirus-induced lethal meningitis. J. Clin. Investig. 109:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, T.-T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]