Abstract
Barrier-to-autointegration factor (BAF) is a conserved human chromatin protein exploited by retroviruses. Previous investigators showed that BAF binds double-stranded DNA nonspecifically and is a host component of preintegration complexes (PICs) isolated from cells infected with human immunodeficiency virus type 1 (HIV-1) or Moloney murine leukemia virus. BAF protects PIC structure and stimulates the integration of salt-stripped PICs into target DNA in vitro. PICs are thought to acquire BAF from the cytoplasm during infection. However, we identified two human tissues (of 16 tested) in which BAF mRNA was not detected: thymus and peripheral blood leukocytes, which are enriched in CD4+ T lymphocytes and macrophage precursors, respectively. BAF protein was detected in activated but not resting CD4+ T lymphocytes; thus, if BAF were essential for PIC function, we hypothesized that virions might “bring their own BAF.” Supporting this model, BAF copurified with HIV-1 virions that were digested with subtilisin to remove microvesicle contaminants, and BAF was present in approximately zero to three copies per virion. In three independent assays, BAF bound directly to both p55 Gag (the structural precursor of HIV-1 virions) and its cleaved product, matrix. Using lysates from cells overexpressing Gag, endogenous BAF and Gag were coimmunoprecipitated by antibodies against Gag. Purified recombinant BAF had low micromolar affinities (1.1 to 1.4 μM) for recombinant Gag and matrix. We conclude that BAF is present at low levels in incoming virions, in addition to being acquired from the cytoplasm of newly infected cells. We further conclude that BAF might contribute to the assembly or activity of HIV-1 PICs through direct binding to matrix, as well as DNA.
Human immunodeficiency virus type 1 (HIV-1) is a retrovirus that is transmitted through sexual contact, contaminated blood, or other body fluids (54).Primary targets for HIV-1 infection are CD4+ (helper) T lymphocytes and macrophages (1, 33, 65). The virus infects cells that express the CD4 surface receptor plus chemokine receptors, including CCR5 or CXCR4 (2). After the virus fuses with the cell membrane, the virus coat is removed, revealing the reverse transcription complex. This complex contains two positive-strand copies of the viral RNA genome, tRNALys primer, reverse transcriptase (RT), integrase (IN), nucleocapsid (NC), viral protein R (Vpr [26]), and host proteins. RT then completes the reverse transcription of viral RNA into double-stranded DNA, which is assembled into preintegration complexes (PICs). Mature PICs are large (∼28-nm-diameter) structures that include HIV-encoded matrix (MA) and NC proteins, plus IN, RT, Vpr, host proteins HMGa1 and BAF, and 3 μm of retroviral DNA (25, 26). The structure and composition of the PIC appear to change over time and are incompletely understood (34). The PIC translocates rapidly toward the nucleus by engaging microtubule-dependent motors (46). In nondividing or G1-phase cells, several viral proteins including IN, MA, and Vpr are proposed to mediate PIC entry into the nucleus through the nuclear pore complexes (5, 15, 23).
Once inside the nucleus, the PIC must integrate the viral DNA into a host chromosome to establish a productive infection (41). HIV-1 integration favors regions of chromosomes with active genes, which have more “open” chromatin structure (56). It is not known if this bias for expressed chromatin is trivial (easier access) or deliberate. In contrast, the mechanics of the DNA end processing and joining events for HIV-1 are well characterized (29) and are mediated by IN (17, 59).
PICs isolated from the cytoplasm of cells infected with either Moloney murine leukemia virus (MoMLV) or HIV-1 can fully and efficiently integrate into target DNA in vitro (16, 19). Interestingly, PICs that are first extracted with 1 M KCl contain IN but fail to integrate (12, 38, 39), suggesting that PICs contain salt-extractable factors required for integration. Salt-extracted PICs lose a special structure, termed the intasome, normally present at each end of the viral DNA (13, 63). A host factor purified from the cytoplasm of uninfected NIH 3T3 cells was found to restore intasome structure (12, 28) when added to salt-extracted HIV-1 PICs. This factor was a small (10-kDa) human protein, barrier-to-autointegration factor (BAF), dimers of which bind directly but nonspecifically to double-stranded DNA (8, 38, 67). Purified BAF protein also protects salt-extracted MoMLV PICs against suicidal autointegration (hence, barrier-to-autointegration factor [38]). These findings suggest that BAF has both protective and positive roles early in HIV-1 infection. As evidence for direct roles, BAF was recently shown to be a bona fide component of HIV-1 and MoMLV PICs (43, 60). A different host protein named HMGa1 (formerly known as HMG I/Y) is also present in PICs and promotes integration in vitro but is ∼500-fold less active than BAF in vitro (12, 42).
BAF is an evolutionarily conserved, essential chromatin protein in metazoans (64; M. Segura-Totten and K. L. Wilson, unpublished results). When incubated with DNA, BAF dimers oligomerize in groups of ca. six to form higher-order nucleoprotein complexes in vitro (67). BAF also interacts with LAP2β, a nuclear inner membrane protein (22), and can form complexes with both LAP2 and DNA in vitro (58), suggesting that BAF might link chromatin to the nuclear envelope. BAF recognizes a conserved 40-residue motif, termed the LEM domain (named LEM for LAP2, emerin, and MAN1), which defines a family of nuclear proteins including LAP2, emerin, and MAN1 (7, 61). A subset of BAF resides in the cytoplasm of mammalian cells (30, 43, 57), consistent with its original purification from NIH 3T3 cells (38). A significant fraction of nuclear BAF concentrates near the nuclear envelope in vertebrate cells, where LEM domain proteins are enriched (31, 57, 64). In Caenorhabditis elegans, BAF enrichment near the nuclear envelope requires emerin and MAN1 (44). During mitosis, BAF localizes to chromatin and appears to have a structural role in recruiting emerin during nuclear envelope assembly (27). BAF can influence higher-order chromatin structure either positively (enhanced chromatin decondensation) or negatively (compressing chromatin) in Xenopus nuclear assembly extracts (57). Interestingly, BAF also appears to have direct roles in gene regulation (30, 62). The mechanisms of BAF's functions in healthy uninfected cells are not yet understood.
Previous reports suggested that BAF was expressed in all cell types (67), consistent with its proposed essential roles in cell division (27, 57, 67). BAF was not detected in virions in previous studies (38) and was therefore hypothesized to be acquired by newly formed PICs from the cytoplasm (38, 43). However, we found that BAF mRNA and protein were low or not detected in both thymus tissue and resting CD4+ T lymphocytes. Given its proposed role in protecting HIV-1 PICs, we hypothesized that HIV-1 virions might “bring their own BAF.” Our results support this hypothesis and further show that BAF binds to both p55 Gag and mature recombinant MA with low micromolar affinities. Direct binding between BAF and MA has important implications for PIC assembly and structure, which are discussed.
MATERIALS AND METHODS
Semiquantitative PCR.
Primers specific for the 5′ and 3′ coding sequences of BAF cDNA (57) were used to PCR amplify a 273-bp BAF fragment from a human cDNA panel of 16 tissues (BD Biosciences, Clontech, Palo Alto, Calif.). For PCR, samples were treated as follows: (i) an initial denaturation step of 60 s at 94°C, (ii) 38 cycles, with each cycle consisting of 60 s at 94°C, 2 min at 60°C, and 1 min at 72°C, and (iii) a final extension step of 5 min at 72°C. Control samples amplified using primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) verified the presence of equal amounts of total cDNA in all tissues.
Purification of resting and activated CD4+ T lymphocytes. (i) Purification of resting CD4+ T lymphocytes.
Human peripheral blood mononuclear cells (PBMCs) were purified by Hypaque-Ficoll gradient centrifugation followed by monocyte depletion via adherence. A highly purified population of resting CD4+ HLA-DR− T lymphocytes was obtained by bead depletion of unwanted cells and subsequent sorting as described previously (14). Purity of the resulting CD4+ HLA-DR− T lymphocytes was determined by fluorescence-activated cell sorting (FACS) analysis using phycoerythrin (PE)-conjugated anti-CD4 antibodies and fluorescein isothiocyanate (FITC)-conjugated HLA-DR antibodies (BD Biosciences Pharmingen, San Diego, Calif.).
(ii) Purification of activated CD4+ T lymphocytes.
PBMCs were purified by Hypaque-Ficoll gradient centrifugation and activated by adding 5 μg of phytohemagglutinin (PHA) per ml and culturing for 3 days in medium containing interleukin-2 and cytokine-rich supernatant from activated T lymphocytes. On day four, CD8+ T lymphocytes were removed by magnetic bead depletion (Dynabeads M-450 CD8; Dynal Biotech), and CD4+ T lymphocytes were positively selected using magnetic beads followed by bead detachment (CD4 positive isolation kit; Dynal Biotech) per the manufacturer's instructions. The purity and activation status of CD4+ T lymphocytes were determined by FACS analysis using FITC-conjugated anti-CD4 antibodies (Caltag, Burlingame, Calif.), PE-conjugated anti-CD25 antibodies (Coulter-Immunotech, Brea, Calif.), PE-conjugated anti-CD69 antibodies (Pharmingen, San Diego, Calif.), and PE-conjugated HLA-DR antibodies (Coulter-Immunotech).
Preparation and purification of HIV-1 virions and microvesicles.
Viruses were purified as described previously (4) from clarified cell culture supernatants by two successive rounds of ultracentrifugation in sucrose density gradients (double banded). Virus-containing fractions were identified by absorption with UV light at 280- and 254-nm wavelengths. Peak UV-absorbing fractions were pooled, diluted to less than 20% sucrose in TNE buffer (10 mM Tris-HCl [pH 7.2], 100 mM NaCl, and 1 mM EDTA in deionized water), pelleted by ultracentrifugation at 100,000 × g, and resuspended in TNE buffer. Samples were stored at −70°C. Microvesicles were isolated from the culture supernatant of uninfected H9 cells as described previously (3). The H9 cell line was obtained from the American Type Culture Collection (Rockville, Md.) and maintained in complete RPMI 1640 medium (GIBCO-BRL, Life Technologies, Gaithersburg, Md.) containing 10% fetal calf serum (HyClone, Logan, Utah) and 10 mM HEPES. The T-tropic virus used in this work was identified according to the virus strain and cell line in which it was propagated (the AIDS Vaccine Program [AVP], Frederick, Md.) as HIV-1MN/H9 and represents a single-cell clone produced by limiting dilutions (52).
Western blotting for BAF, CD45, and p24 (capsid [CA]).
Before loading on gels, HIV-1 virion samples and purified BAF protein were heated to 60°C for 5 to 10 min in sodium dodecyl sulfate (SDS) sample buffer supplemented with 5% β-mercaptoethanol. We then loaded 106 resting or activated CD4+ T lymphocytes per lane on NuPAGE gels (Invitrogen Corp., Carlsbad, Calif.) with SDS and 4 to 12% polyacrylamide. After electrophoresis, proteins were transferred to nitrocellulose filters at 100 V for 45 min in transfer buffer (50 mM Tris [pH 7.5], 380 mM glycine, 0.1% SDS, and 20% methanol). After blocking with phosphate-buffered saline (PBS) containing 5% nonfat powdered milk (Safeway) and 0.1% Tween 20, filters were incubated at 4°C overnight with anti-BAF rabbit serum 3273 diluted 1:1,000 (57). Blots were then washed three times (15 min each time), incubated with horseradish peroxidase-conjugated goat anti-rabbit antibodies (1:5,000 dilution; Pierce, Rockford, Ill.), and washed three times in PBS containing Tween 20 for 15 min each. Proteins were visualized by enhanced chemiluminescence and exposure to Hyperfilm MP (Amersham Biosciences, Piscataway, N.J.).
To probe for BAF protein in purified HIV-1 virions, equal amounts of protein were loaded per lane on NuPAGE gels (Invitrogen Corp.), resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to filters, and probed with antibodies against BAF (as described above), CA protein (1:7,000 dilution) (AVP), or CD45 (BD Biosciences, Palo Alto, Calif.) (1:5,000 dilution). All primary antibodies were diluted in PBS containing 0.1% Tween 20 and 5% milk and processed as described above. Fresh and recently frozen virions gave the best BAF signal; we speculate that BAF is slowly degraded in frozen samples.
Blot overlays.
Purified proteins (p55 Gag, IN, and RT) were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. MA and CA were also produced recombinantly in Escherichia coli transformed with plasmids obtained from the NIH AIDS Research and Reference Reagent Program. Proteins or crude bacterial lysates were separated on SDS-10% polyacrylamide gels, transferred to nitrocellulose membranes (Schleicher and Schuell Bioscience, Keene, N.H.), and blocked for 1 h in PBS containing Tween 20 and 5% nonfat dry milk. Blots were then washed twice in blot rinse buffer (BRB) (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20) for 5 min at 22 to 24°C and incubated overnight with 20 μCi of 35S-labeled BAF diluted 1:200 into BRB containing 0.1% fetal calf serum (final volume, 3 ml). Probe 35S-labeled BAF was synthesized in eukaryotic transcription and translation extracts as described previously (37). Blots were washed twice in BRB, dried, and exposed to Hyperfilm MP.
Binding assays. (i) Microtiter well binding assay.
Purified p55 Gag was obtained from the NIH AIDS Research and Reference Reagent Program. Recombinant BAF was synthesized in E. coli and purified as described previously (57); for a detailed protocol, contact K. L. Wilson. Known amounts of protein, namely, p55 Gag or BAF (2 μg per well) or bovine serum albumin (BSA) (as negative control) in binding buffer (20 mM HEPES [pH 7.4], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA), were adsorbed to microtiter wells, and then 3% BSA was added to block nonspecific binding sites. In fact, 1.8 pmol of Gag and 10 pmol of BAF dimer actually bound to the well. Wells were not allowed to dry. Increasing concentrations of soluble 35S-labeled BAF or 35S-labeled MA, transcribed and translated in rabbit reticulocyte lysates, were incubated with immobilized p55 Gag or BAF, respectively, or the corresponding BSA-coated control wells. After the wells were washed three times with binding buffer, bound proteins were eluted with 5% SDS and quantified by scintillation counting as described previously (30). 35S-labeled BAF and 35S-labeled MA did not bind significantly to BSA controls (data not shown).
(ii) Coimmunoprecipitation in vitro.
Different amounts of purified proteins were mixed and incubated for 30 min at 22 to 25°C to allow binding. We then added 100 μl of immunoprecipitation (IP) buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 10 mM EDTA, 2 mM EGTA, 0.1% Nonidet P-40 [NP-40], 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 μg of leupeptin per ml) to each sample. MA was immunoprecipitated using polyclonal serum (AVP) and incubated 1 h at 4°C. Washed protein A Sepharose beads (50 μl per sample; Amersham/Pharmacia Biotech, Piscataway, N.J.) were added, and samples were incubated overnight at 4°C and centrifuged at 2,000 rpm (Eppendorf 5415C) for 5 min to pellet beads. Pellets were washed four times with IP buffer. Bound proteins were eluted by boiling in 30 μl of 2× SDS sample buffer, resolved by SDS-PAGE, and immunoblotted as described above.
(iii) Immunoprecipitation from cell lysates.
Transfected HeLa cells were rinsed twice with PBS, incubated with 200 μl of lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40, 1 mM PMSF, 20 μg of leupeptin per ml) and collected by scraping. The entire lysate was transferred into a 1.5-ml tube and centrifuged at 14,000 rpm for 1 min (22 to 25°C) to remove cellular debris. Each 200-μl cell lysate was precleared by incubation with protein A Sepharose beads (20 μl) at 4°C for 30 min. The supernatant (precleared lysate) was then divided into aliquots. Each IP reaction mixture consisted of 10 μl of precleared lysate plus 4 μl of MA antibody, which were incubated overnight at 4°C and then supplemented with 20 μl of protein A Sepharose beads (Amersham/Pharmacia Biotech), incubated for 2 h at 4°C, and centrifuged at 2,000 rpm (Eppendorf centrifuge 5415C) for 5 min. Pelleted beads were washed four times with lysis buffer. Bound proteins were extracted by boiling in 30 μl of 2× SDS sample buffer, resolved by SDS-PAGE, and immunoblotted as described above.
RESULTS
BAF protein is present at very low levels in resting CD4+ T lymphocytes.
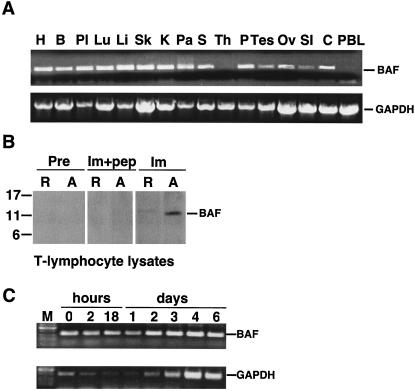
This work originated from a control experiment, which was expected to verify that BAF is expressed ubiquitously in human tissues. A first-strand cDNA panel of 16 human tissues was assayed for BAF mRNA by quantitative PCR analysis. To our surprise, BAF mRNA was not detected in 2 of 16 tissues (Fig. 1A). All tissues had intact mRNA, which was shown by using primers specific for the housekeeping enzyme GAPDH (Fig. 1A). Furthermore, the two tissues in which BAF mRNA was not detected were the thymus (site of T-lymphocyte development) and peripheral blood leukocytes (enriched for quiescent lymphocytes and monocytes; Fig. 1A). Activated CD4+ T lymphocytes and macrophages are the principal targets for HIV-1 replication in vivo, whereas resting CD4+ T lymphocytes provide a latent reservoir for the virus.
FIG. 1.
Expression of BAF mRNA in human tissues. (A) Agarose gel analysis of semiquantitative RT-PCR experiments done using two multiple tissue panels of first-strand cDNAs. Similar amounts of BAF cDNA (273 bp) were amplified from heart (H), brain (B), placenta (Pl), lung (Lu), liver (Li), skeletal muscle (Sk), kidney (K), pancreas (Pa), spleen (S), prostate (P), testis (Tes), ovary (Ov), small intestine (SI), and colon (C). BAF mRNA was not detected in thymus (Th) or peripheral blood leukocytes (PBL). Control experiments using primers specific to housekeeping enzyme GAPDH verified that all tissues amplified similar amounts of the 800-bp GAPDH fragment, confirming RNA integrity in these samples. (B) Western blot of protein lysates from resting (R) and day 4 in vitro-activated (A) CD4+ T lymphocytes probed with preimmune (Pre) or immune (Im) rabbit serum against human BAF. Monomeric BAF migrates at 11 kDa on gels (57). Recognition of BAF was specific, because no signal was obtained when immune antibodies were pretreated with peptide antigen (Im+pep). (C) Agarose gel analysis of semiquantitative RT-PCR experiments performed using RNA purified from CD4+ T lymphocytes at the indicated times after activation. M, molecular size markers.
To independently determine whether BAF was present in resting T lymphocytes, we isolated and purified resting CD4+ T lymphocytes from uninfected individuals. These resting cells were at least 99% pure. Alternatively, we activated PBMCs in vitro by culturing in the presence of interleukin-2 and cytokine-rich supernatant from activated T lymphocytes prior to purification (see Materials and Methods). More than 60% of CD4+ T lymphocytes became activated by day 4, based on the expression of CD69 and CD25 markers (data not shown). Whole-cell lysates from each population were resolved by SDS-PAGE and probed with antibodies against human BAF. No BAF signal was detected by preimmune antibodies in either resting or activated CD4+ T lymphocytes (Fig. 1B). Using immune antibodies, BAF protein was detected at very low levels in resting CD4+ T cells; this low signal may arise from the <1% contaminating cells (which could include activated T lymphocytes). However, BAF protein was abundant in activated CD4+ T lymphocytes (Fig. 1B). Recognition of BAF was specific, because it was competed by pretreating antibodies with the antigenic peptide (Fig. 1B).
We next used RT-PCR to assay BAF and GAPDH mRNA levels in purified CD4+ T cells as a function of time after activation (Fig. 1C). Low mRNA levels were detected for BAF and GAPDH at time zero (Fig. 1C); these low signals, which might be due to contaminating cells, decreased during the first 24 h after activation but then increased by day 2. By day 4, mRNA levels increased almost twofold for BAF and sixfold for GAPDH compared to the levels at time zero (Fig. 1C). A previous study of cyclin A expression (36) suggested that isolated resting T lymphocytes enter G1 phase of the cell cycle 2 to 3 days after activation. Thus, by both criteria (mRNA and protein), BAF expression was low in resting CD4+ T-lymphocyte populations and increased when cells became metabolically active and reentered the cell cycle.
BAF is present in HIV-1 virions.
Since BAF is hypothesized to be essential for the integrity of retroviral PICs, its apparent absence from thymus tissue and very low levels in resting CD4+ T lymphocytes suggested two possibilities: either HIV-1 can newly infect only activated CD4+ T lymphocytes, which express BAF protein, or BAF is preincorporated into HIV-1 virions. To revisit the latter possibility, we probed immunoblots of sucrose gradient-purified HIV-1 virions (HIVMN) obtained from the culture medium of infected H9 cells. Our first experimental results suggested that BAF was abundant in HIV-1 virions (data not shown). However, virion preparations can include many contaminating host proteins present in microvesicles, which are shed from cells and copurify with virions (3).
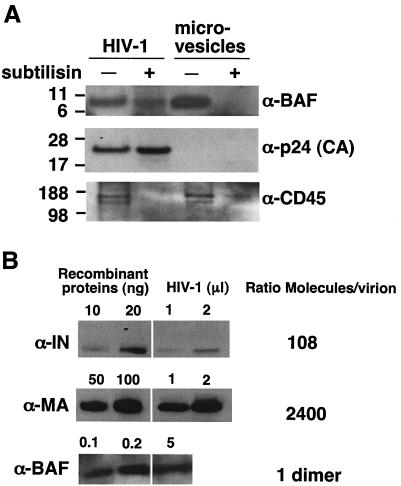
To rigorously determine whether BAF was virion associated, we isolated virus particles from infected H9 cells, and in parallel, we isolated microvesicles from uninfected H9 cells. Equal amounts of protein from each preparation were either left untreated or digested for 14 h with subtilisin, a nonspecific protease (52). Virion core particles are shielded from digestion by the virus membrane. In contrast, protease digestion removes >95% of contaminating nonviral cellular debris and makes microvesicles lighter, allowing them to be removed by centrifugation (51). We therefore centrifuged each sample through 20% sucrose to separate virions from proteolyzed debris and microvesicles. The pellets were resolved by SDS-PAGE and immunoblotted using antibodies against human BAF, virus-encoded p24 (CA) protein, and microvesicle marker protein CD45 (18, 48, 51). These markers verified the identity of each fraction, confirmed that viral protein p24 (CA) was quantitatively protected from proteolysis and showed that the exposed microvesicle protein CD45 was sensitive to proteolysis, as expected (Fig. 2A). Importantly, most of the BAF in HIV-1 virions was protected from proteolysis and copurified with CA, whereas microvesicle-associated BAF failed to pellet after proteolysis (Fig. 2A). On SDS-polyacrylamide gels, virion-associated BAF migrated predominantly as a 10-kDa protein, consistent with its monomeric mass. These results demonstrated for the first time that BAF is present in HIV-1 virions.
FIG. 2.
BAF is present in HIV-1 virions at low levels. (A) HIV-1 virions produced from HIVMN-infected H9 cell lines were either mock treated (−) or digested with the nonspecific protease subtilisin (+). In parallel, microvesicles were prepared from uninfected H9 cells and either mock treated (−) or treated with subtilisin (+). Protein extracts from all samples were then analyzed by immunoblotting using antibodies to human BAF, CD45 (microvesicle marker), and p24 (HIV-1 capsid protein) (α-BAF, α-p24, and α-CD45, respectively). (B) Stoichiometry of BAF in the virion. Protein extracts from subtilisin-digested HIV-1 virions and known amounts of recombinant IN, MA, and BAF proteins were immunoblotted using antibodies specific for each protein. The number of molecules per unit volume of purified virions was calculated and expressed as a ratio relative to BAF.
How much BAF is present in virions?
We used semiquantitative immunoblot analysis to quantify BAF relative to MA and IN in mature HIV-1 virions. Titrated amounts of purified recombinant MA (45), IN (32), and BAF protein plus different volumes of subtilisin-digested virion samples were resolved on SDS-polyacrylamide gels, transferred to nitrocellulose filters, and probed with antibodies specific for BAF (57), MA, or IN. Each protein was quantified by densitometry, and we then estimated the number of BAF dimers per virion (Fig. 2B) relative to MA (a structural protein) and IN (an enzyme). This quantification suggested that HIV-1 virions contain a molar ratio of one BAF dimer per 108 copies of IN and 2,400 copies of MA. Our numbers for IN and MA are consistent with previously published estimates (∼100 copies of IN and ∼2,000 MA per virion [20]). We therefore conclude that BAF is present at very low copy number in virions, with at least one dimer per virion. Given the errors inherent in such estimates, we suggest that individual virions contain approximately zero to three BAF dimers.
BAF binds directly to p55 Gag.
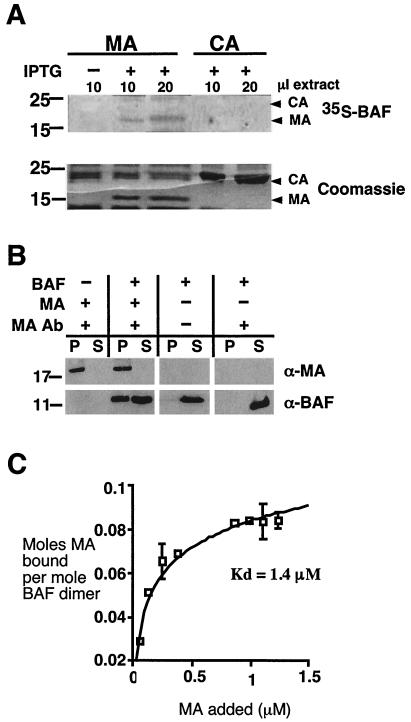
How is BAF recruited into HIV-1 virions? BAF is not known to bind RNA, and we found no evidence in database searches for a LEM domain in any HIV-1-encoded protein. Because BAF binds DNA and protects intasome structures in the PIC, we hypothesized that it might associate with either IN or RT, which are also DNA-associated components of the PIC. To test this model, different amounts of purified IN (32) and RT (40) proteins, plus the viral structural precursor p55 Gag as a control, were resolved by SDS-PAGE, transferred to filters, and probed with 35S-labeled BAF (Fig. 3A). BAF showed only background binding to bands containing as much as 1 μg of purified RT or IN (Fig. 3A). However, BAF gave a strong signal with the 55-kDa Gag polyprotein, even at the lowest level tested (125 ng [Fig. 3A]). We concluded that BAF binds directly to p55 Gag, a structural protein with key roles in virion assembly (21).
FIG. 3.
BAF binds directly to p55 Gag. (A) Blot overlay assay. Purified p55 protein (125, 250, and 500 ng), RT (250, 500, and 1,000 ng) and IN (250, 500, and 1,000 ng) proteins (prot.) were resolved by SDS-PAGE, transferred to filters, and probed with 35S-labeled BAF. The autoradiograph is shown. (B) In vitro coimmunoprecipitation assay. Recombinant p55 Gag protein (500 ng) was incubated with (+) or without (−) 200 ng of recombinant BAF and then immunoprecipitated using protein A beads with (+) or without (−) antibodies against BAF (BAF Ab). One-tenth of each pellet (P) and 10% of each corresponding supernatant (S) fraction were resolved by SDS-PAGE and immunoblotted with antibodies specific for either BAF or the MA domain of p55 Gag. (C) Binding affinity. The affinity of BAF for p55 Gag was determined in microtiter well assays. Increasing concentrations of 35S-labeled BAF were incubated with constant amounts of recombinant p55 Gag (1.8 pmol) immobilized in microtiter wells. Double-reciprocal plots (not shown) were used to determine the affinity constant as described previously (30). (D) Endogenous BAF coimmunoprecipitates with p55 Gag in human (HeLa) cell extract. Full-length Gag (pCiGagPRE) was expressed in HeLa cells, and protein lysates (L) from transfected cells were incubated with protein A beads alone (Protein A) or protein A beads plus antibodies against MA (α-MA Ab). A fraction (15%) of the supernatant (S) and 30% of each immunoprecipitate (P) were resolved by SDS-PAGE and immunoblotted for BAF. Transf., transfected.
Binding between BAF and p55 Gag was independently confirmed by both in vitro and in vivo coimmunoprecipitation assays. We first incubated 500 ng of recombinant p55 Gag protein for 30 min with (or without) purified recombinant human BAF and then added protein A Sepharose with or without antibodies against human BAF. Pelleted beads and supernatants were then Western blotted with antibodies specific for Gag or BAF (Fig. 3B). Controls showed that Gag did not spontaneously pellet and that Gag plus BAF remained soluble in the absence of antibody (Fig. 3B). The antibody against BAF did not immunoprecipitate Gag in the absence of BAF (data not shown). However when all three components were present, a majority of Gag coimmunoprecipitated with BAF (Fig. 3B). These results confirmed direct binding between BAF and Gag in solution.
The equilibrium binding affinity of BAF dimers for Gag was 1.1 μM (Fig. 3C), determined using a microtiter assay with BSA-coated wells as negative controls (30) (see Materials and Methods). This affinity was comparable to BAF's affinity for lamin A (1 μM) and about sixfold weaker than its affinity for emerin (200 nM [30]).
We next determined whether BAF and Gag interact in vivo. HeLa cells were transfected with pCiGagPRE (J. Wong and R. F. Siliciano, unpublished data), in which codons were optimized for efficient translation of Gag protein in mammalian cells. Cells were lysed 36 h after transfection, and whole-cell lysates were incubated with either protein A beads alone or with protein A beads plus antibodies against the MA domain of Gag. Beads were then pelleted, and the corresponding pellet and supernatant fractions, along with starting lysate were resolved by SDS-PAGE and immunoblotted for endogenous BAF (Fig. 3D). Controls showed that most BAF remained soluble in the absence of antibody (Fig. 3D), as expected. Interestingly, antibodies against MA quantitatively coimmunoprecipitated a slow-migrating (∼50-kDa) form of BAF from cell lysates (Fig. 3D; see Discussion). We concluded that Gag and BAF associate in vivo and that their interactions in vitro were therefore physiologically relevant.
BAF binds directly to mature MA.
The Gag polyprotein is responsible for building virions at the cell surface, and each immature virion contains ∼2,000 copies of Gag. After virions are released from cells, most Gag proteins are proteolyzed to generate four mature proteins: CA (24 kDa), MA (17 kDa), NC (7 kDa), and p6 (6 kDa) (reviewed in reference 21). cDNAs encoding mature CA and MA were available; to determine whether BAF binds directly to either CA or MA, these proteins were expressed in bacteria (45, 66), resolved by SDS-PAGE, and either transferred to filters and probed with 35S-labeled BAF, or stained with Coomassie blue (Fig. 4A). BAF bound specifically to MA, despite larger amounts of CA on the filter.
FIG. 4.
BAF binds directly to MA with low micromolar affinity. (A) BAF binds directly to MA, but not CA, in blot overlay assays. Different volumes of protein lysate from induced (+ IPTG) or uninduced bacteria (− IPTG) containing plasmids encoding either MA or CA were resolved by SDS-PAGE. Gels were either transferred to filters and probed with 35S-labeled BAF or stained with Coomassie blue to verify equal protein loads. (B) MA binds BAF in coimmunoprecipitation assays. Purified recombinant MA (200 ng) was incubated with (+) or without (−) 200 ng of recombinant BAF and then immunoprecipitated using protein A beads with or without antibodies against MA. A fraction (15%) of each pellet (P) and 30% of each supernatant (S) were resolved by SDS-PAGE and immunoblotted for BAF and MA, as indicated with antibodies to BAF (α-BAF) and MA (α-MA). (C) The equilibrium affinity of MA for BAF was determined in microtiter well assays by adding increasing concentrations of 35S-labeled MA to constant amounts of recombinant BAF immobilized in microtiter wells. Double-reciprocal plots (not shown) were used to determine the affinity constant as described previously (30).
To independently verify the BAF-MA interaction, we did coimmunoprecipitation experiments with recombinant purified MA and BAF (Fig. 4B). Negative controls confirmed that the MA antibody recognized MA, but not BAF, and that most BAF remained soluble in the absence of antibody (Fig. 4B). However when both proteins were present, the MA antibody coimmmunoprecipitated about half of the available BAF (Fig. 4B), confirming direct binding between BAF and MA in vitro. The equilibrium binding affinity between BAF and MA was determined in microtiter assays. Recombinant BAF dimers (10 pmol) were immobilized in microtiter wells, and different concentrations of 35S-labeled MA were added to each well. BSA-coated wells served as negative controls. The affinity of BAF for recombinant MA was 1.4 μM (Fig. 4C), slightly lower than BAF's affinity for full-length Gag. This biochemical analysis suggested a stoichiometry of 0.5 mol of MA per mol of BAF dimers (Fig. 4C) and further explained why less than half of the available BAF coprecipitated with MA in the earlier experiment (Fig. 4B), where the concentrations of BAF and MA were 0.5 and 0.6 μM, respectively. (These concentrations are close to the equilibrium affinity where, by definition, 50% of BAF would bind.) Because MA is a known component of the PIC, we concluded that protein-protein interactions between BAF and MA have the potential to contribute to the assembly, structure, and integration competence of the PIC.
DISCUSSION
This study produced three main results. First, BAF expression is very low or not detected in thymus and peripheral leukocytes, in contrast to all other tissues tested which express relatively uniform levels of BAF. Although unexpected, this finding emphasizes the nearly inactive metabolic state of resting T lymphocytes (6, 53). T lymphocytes have an unusual mechanism for organizing and regulating chromatin structure, which involves a three-dimensional scaffold or cage formed by SATB1 protein (9). We speculate that this structure might compensate for the absence of BAF. Our second finding was that BAF is present at low stoichiometry in purified HIV-1 virions, with the interesting caveat that in vivo, Gag appears to prefer a slower-migrating form (∼50 kDa) of BAF (see below). Third, we found that BAF binds directly to two HIV-encoded proteins, p55 Gag and MA, with low micromolar affinities. BAF thus joins a growing number of host proteins known to be incorporated into HIV-1 virions, including cyclophilin A, elongation factor 1α, actin, several actin-binding proteins (ezrin, moesin, and cofilin) and signaling proteins ERK2 and Lck (49). Host proteins are proposed to have roles in virus assembly or postentry functions (49). Our results for BAF favor the latter model; BAF is unlikely to be essential for HIV-1 virion assembly per se, because it is a minor component (zero to three dimers per virion) that might be absent from a subset of virions. We therefore hypothesize that HIV-1 virions incorporate BAF either (i) by accident, due to BAF's affinity for the MA domain of Gag, or (ii) on purpose, to promote PIC survival in resting CD4+ T lymphocytes (which lack BAF), or because BAF has a role in reverse transcription complexes or PIC assembly prior to the acquisition of cytoplasmic BAF.
BAF as a host component of HIV-1 virions.
Our findings suggest that BAF is a host component of HIV-1 virions, recruited (at least in part) through its affinity for the MA domain of Gag. In uninfected cultured cells, 30 to 50% of BAF is present in the cytoplasm (30, 43). However, an important subset of cells is deficient in BAF, namely, resting CD4+ T lymphocytes and blood monocytes, which comprise ∼5% of peripheral blood leukocytes. Thus, if HIV-1 enters a resting T lymphocyte, which is metabolically quiescent, the PIC must be able to survive low rates of reverse transcription and a potentially long latency period prior to cell activation and integration (53). Because BAF enters with the virus, we propose that BAF might also contribute to the earliest stages of PIC formation, by first interacting with MA and subsequently also interacting with retroviral DNA. In other words, BAF may facilitate the structural transition from reverse transcription complex to preintegration complex on the basis of its sequential interactions with MA and DNA.
BAF interactions with Gag in vitro versus in vivo.
Direct binding between BAF and Gag was shown by two methods (blot overlay and coimmunoprecipitation of purified proteins) and confirmed by coimmunoprecipitation of endogenous BAF from cells that overexpress p55 Gag. BAF binds Gag with an equilibrium binding affinity of 1.1 μM. We estimate that the concentration of BAF in HeLa cell cytosol is ∼7 nM (30). In cells, Gag proteins aggregate in groups of ∼2,000 at numerous sites on the cell surface, each of which can self-assemble into a retrovirus-like particle in cells that express no other HIV-encoded proteins. We therefore predict that Gag is sufficient to recruit BAF into assembling virions. The number of BAFs per virion will be dictated by two numbers: the concentration of BAF in cytoplasm (7 nM) and its affinity for Gag (1.1 μM). The cytoplasmic concentration of BAF is far too low for BAF to saturate Gag; the binding curve predicts that one or a few molecules of BAF will bind per ∼2,000 copies of Gag. Assuming that their affinity is the same in vivo, the concentration of BAF would have to be 157-fold higher (e.g., 1.1 μM) for there to be ∼1,000 copies of BAF per virion. Thus, our measured affinities are compatible with the experimentally determined low numbers (ca. zero to three) of BAF per virion.
Despite the agreement between our current in vitro and “in virion” results, our assumption that the BAF-Gag binding affinity is constant in vivo may need to be reexamined in future. We recently found that in HeLa cells, endogenous BAF is posttranslationally modified at several sites (L. Bengtsson and K. L. Wilson, unpublished data), potentially explaining the presence of several different slower-migrating forms of BAF in SDS-polyacrylamide gels (57). In cells, Gag bound preferentially to the most abundant (∼50-kDa) slow-migrating form of BAF, whereas HIV-1 virions contained the 10-kDa form of BAF. The modification status of slow-migrating forms of BAF are not yet understood. However, we speculate that BAF might be removed or modified by enzymes present in HIV-1 virions (11, 50).
Implications for PIC assembly and structure.
MA comprises the N-terminal domain of p55 Gag and is located close to the plasma membrane due to myristoylation of its N-terminal Gly residue. After proteolytic cleavage, most MA remains near the virion membrane. A missense mutation at a highly conserved residue (L20K) in MA disrupts an early event in infection, suggesting that MA might be important for the integrity or stability of the PIC (35). Interestingly, about 1% of MA molecules in the virion are phosphorylated on their C-terminal Tyr residue (Y132) by a membrane-associated kinase (10, 24). Tyrosine phosphorylation causes MA to bind IN (25), suggesting a mechanism by which phosphorylated MA might associate with IN, which is abundant in cytoplasmic reverse transcription complexes (47). Thus, binding to MA might be an effective way for BAF to associate with reverse transcription complexes prior to their maturation into PICs. However, it is important to note that HIV-1 can replicate under certain conditions in the absence of MA (55). Thus, MA-BAF interactions cannot be essential for the PIC under all conditions of infection.
Irrespective of the role of MA, future work will aim to determine whether virion-associated BAF is needed to establish an HIV-1 infection, since BAF's presence in the virion might be incidental. Nevertheless, the major conclusion from the present work is that BAF, regardless of its source (virion associated or cytoplasmically acquired), can bind protein components of the PIC, in addition to DNA. This may lead to new insights into PIC assembly and function.
Acknowledgments
We gratefully acknowledge the donors of the following reagents, which were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pWISP98-85 and pWISP93-93 cDNA encoding CA and MA, respectively, from Wes Sundquist; HIV-1 SF2 p55 Gag protein from Chiron Corporation and the Division of AIDS; HIV-1NL4-3 IN protein and IN antibody from Robert Craigie; and HIV-1 RT protein from Stuart Le Grice and Kathryn Howard. We thank James Holaska for biochemical advice, and members of the Wilson lab for stimulating discussions. We also acknowledge sharing of unpublished reagents and data by J. Wong, M. Segura-Totten, and L. Bengtsson.
This work was supported in part by NIH grant R01-GM48646 (to K.L.W.) and a pilot grant from the Johns Hopkins Center for AIDS Research (CFAR).
REFERENCES
- 1.Aquaro, S., P. Bagnarelli, T. Guenci, A. De Luca, M. Clementi, E. Balestra, R. Calio, and C. F. Perno. 2002. Long-term survival and virus production in human primary macrophages infected by human immunodeficiency virus. J. Med. Virol. 68:479-488. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 4.Bess, J. W., Jr., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J. Virol. 66:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, M., Y. Huang, R. Ghirlando, K. L. Wilson, R. Craigie, and G. M. Clore. 2001. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20:4399-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, M., Y. Huang, R. Zheng, S. Q. Wei, R. Ghirlando, M. S. Lee, R. Craigie, A. M. Gronenborn, and G. M. Clore. 1998. Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat. Struct. Biol. 5:903-909. [DOI] [PubMed] [Google Scholar]
- 9.Cai, S., H. J. Han, and T. Kohwi-Shigematsu. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34:42-51. [DOI] [PubMed] [Google Scholar]
- 10.Camaur, D., P. Gallay, S. Swingler, and D. Trono. 1997. Human immunodeficiency virus matrix tyrosine phosphorylation: characterization of the kinase and its substrate requirements. J. Virol. 71:6834-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartier, C., M. Deckert, C. Grangeasse, R. Trauger, F. Jensen, A. Bernard, A. Cozzone, C. Desgranges, and V. Boyer. 1997. Association of ERK2 mitogen-activated protein kinase with human immunodeficiency virus particles. J. Virol. 71:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., S. Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 14.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 15.de Noronha, C. M., M. P. Sherman, H. W. Lin, M. V. Cavrois, R. D. Moir, R. D. Goldman, and W. C. Greene. 2001. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294:1105-1108. [DOI] [PubMed] [Google Scholar]
- 16.Ellison, V., H. Abrams, T. Roe, J. Lifson, and P. Brown. 1990. Human immunodeficiency virus integration in a cell-free system. J. Virol. 64:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211-1221. [DOI] [PubMed] [Google Scholar]
- 18.Esser, M. T., D. R. Graham, L. V. Coren, C. M. Trubey, J. W. Bess, Jr., L. O. Arthur, D. E. Ott, and J. D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75:6173-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 21.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485-2492. [DOI] [PubMed] [Google Scholar]
- 23.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 25.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 26.Greene, W. C., and B. M. Peterlin. 2002. Charting HIV's remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 8:673-680. [DOI] [PubMed] [Google Scholar]
- 27.Haraguchi, T., T. Koujin, M. Segura-Totten, K. K. Lee, Y. Matsuoka, Y. Yoneda, K. L. Wilson, and Y. Hiraoka. 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114:4575-4585. [DOI] [PubMed] [Google Scholar]
- 28.Harris, D., and A. Engelman. 2000. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J. Biol. Chem. 275:39671-39677. [DOI] [PubMed] [Google Scholar]
- 29.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holaska, J. M., K. K. Lee, A. K. Kowalski, and K. L. Wilson. 2003. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 278:6969-6975. [DOI] [PubMed] [Google Scholar]
- 31.Holaska, J. M., K. L. Wilson, and M. Mansharamani. 2002. The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 14:357-364. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 33.Kedzierska, K., and S. M. Crowe. 2002. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr. Med. Chem. 9:1893-1903. [DOI] [PubMed] [Google Scholar]
- 34.Khiytani, D. K., and N. J. Dimmock. 2002. Characterization of a human immunodeficiency virus type 1 pre-integration complex in which the majority of the cDNA is resistant to DNase I digestion. J. Gen. Virol. 83:2523-2532. [DOI] [PubMed] [Google Scholar]
- 35.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacroix, I., C. Lipcey, J. Imbert, and B. Kahn-Perles. 2002. Sp1 transcriptional activity is up-regulated by phosphatase 2A in dividing T lymphocytes. J. Biol. Chem. 277:9598-9605. [DOI] [PubMed] [Google Scholar]
- 37.Lee, K. K., T. Haraguchi, R. S. Lee, T. Koujin, Y. Hiraoka, and K. L. Wilson. 2001. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114:4567-4573. [DOI] [PubMed] [Google Scholar]
- 38.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, M. S., and R. Craigie. 1994. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl. Acad. Sci. USA 91:9823-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Grice, S. F., C. E. Cameron, and S. J. Benkovic. 1995. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130-144. [DOI] [PubMed] [Google Scholar]
- 41.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, L., C. M. Farnet, W. F. Anderson, and F. D. Bushman. 1998. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J. Virol. 72:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, J., K. K. Lee, M. Segura-Totten, E. Neufeld, K. L. Wilson, and Y. Gruenbaum. 2003. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 100:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 46.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nermut, M. V., and A. Fassati. 2003. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J. Virol. 77:8196-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 50.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 51.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder II, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott, D. E., S. M. Nigida, Jr., L. E. Henderson, and L. O. Arthur. 1995. The majority of cells are superinfected in a cloned cell line that produces high levels of human immunodeficiency virus type 1 strain MN. J. Virol. 69:2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinn, T. C. 1996. Global burden of the HIV pandemic. Lancet 348:99-106. [DOI] [PubMed] [Google Scholar]
- 55.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 57.Segura-Totten, M., A. K. Kowalski, R. Craigie, and K. L. Wilson. 2002. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158:475-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shumaker, D. K., K. K. Lee, Y. C. Tanhehco, R. Craigie, and K. L. Wilson. 2001. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853-860. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 76:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umland, T. C., S.-Q. Wei, R. Craigie, and D. R. Davies. 2000. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39:9130-9138. [DOI] [PubMed] [Google Scholar]
- 62.Wang, X., S. Xu, C. Rivolta, L. Y. Li, G. H. Peng, P. K. Swain, C. H. Sung, A. Swaroop, E. L. Berson, T. P. Dryja, and S. Chen. 2002. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J. Biol. Chem. 277:43288-43300. [DOI] [PubMed] [Google Scholar]
- 63.Wei, S. Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson, K. L. 2000. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 10:125-129. [DOI] [PubMed] [Google Scholar]
- 65.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 66.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, R., R. Ghirlando, M. S. Lee, K. Mizuuchi, M. Krause, and R. Craigie. 2000. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. USA 97:8997-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]