Abstract
In the title compound, C24H19NO2S, the mean plane of the benzo[b]carbazole ring system makes a dihedral angle of 79.26 (5)° with the phenyl ring. The S atom is in a distorted tetrahedral configuration. The crystal structure exhibits weak C—H⋯O and C—H⋯π interactions.
Related literature
For related literature, see: Allen et al. (1987 ▶); Chakkaravarthi et al. (2007 ▶, 2008 ▶); Diaz et al. (2002 ▶); Etter et al. (1990 ▶); Friend et al. (1999 ▶); Govindasamy et al. (1998 ▶); Hökelek et al. (1998 ▶); Hosomi et al. (2000 ▶); Itoigawa et al. (2000 ▶); Ramsewak et al. (1999 ▶); Rodriguez et al. (1995 ▶); Sankaranarayanan et al. (2000 ▶); Tachibana et al. (2001 ▶); Zhang et al. (2004 ▶).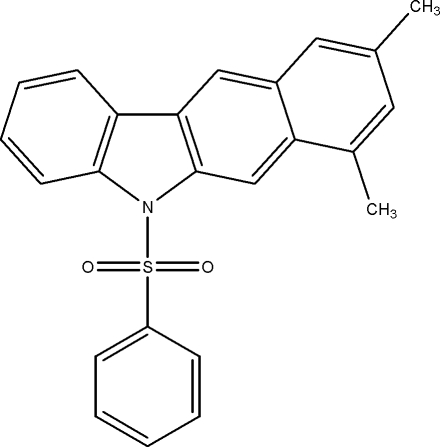
Experimental
Crystal data
C24H19NO2S
M r = 385.46
Orthorhombic,

a = 13.9260 (6) Å
b = 10.1995 (5) Å
c = 27.3014 (14) Å
V = 3877.8 (3) Å3
Z = 8
Mo Kα radiation
μ = 0.19 mm−1
T = 295 (2) K
0.30 × 0.20 × 0.16 mm
Data collection
Bruker Kappa APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.940, T max = 0.971
26413 measured reflections
5626 independent reflections
3490 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.157
S = 1.04
5626 reflections
255 parameters
H-atom parameters constrained
Δρmax = 0.68 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808024380/bt2759sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808024380/bt2759Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C8—H8⋯O1 | 0.93 | 2.37 | 2.945 (3) | 120 |
| C21—H21⋯O2 | 0.93 | 2.34 | 2.935 (2) | 122 |
| C8—H8⋯Cg1i | 0.93 | 3.00 | 3.859 | 155 |
| C11—H11⋯Cg2ii | 0.93 | 2.90 | 3.693 | 144 |
Symmetry codes: (i)  ; (ii)
; (ii)  . Cg1 and Cg2 are the centroids of the C7–C12 and C15–C20 rings, respectively.
. Cg1 and Cg2 are the centroids of the C7–C12 and C15–C20 rings, respectively.
Acknowledgments
The authors acknowledge the Sophisticated Analytical Instrument Facility, Indian Institute of Technology, Madras, for the data collection.
supplementary crystallographic information
Comment
Carbazole and its derivatives have become quite attractive compounds owing to their applications in pharmacy and molecular electronics. It has been reported that carbazole derivatives possess various biological activities, such as antitumor (Itoigawa et al., 2000), antioxidative (Tachibana et al., 2001), anti-inflammatory and antimutagenic (Ramsewak et al., 1999). Carbazole derivatives also exhibit electroactivity and luminescence properties and are considered to be potential candidates for electronic such as colour displays, organic semiconductor lasers and solar cells (Friend et al., 1999). These compounds are thermally and photochemically stable, which makes them useful materials for technological applications. For instance, the carbazole ring is easily funtionalized and covalently linked to other molecules (Diaz et al., 2002). This enables its use as a convenient building block for the design and synthesis of molecular glasses, which are widely studied as components of electroactive and photoactive materials (Zhang et al., 2004).
Against this background, and in order to obtain detailed information on molecular conformations in the solid state, X-ray studies of the title compounds (I) have been carried out. X-Ray analysis confirms the molecular structure and atom connectivity for (I), as illustrated in Fig. 1. The benzocarbazole ring is planar, with bond distances and angles comparable to those reported similiar structures (Hökelek et al., 1998; Hosomi et al., 2000). The mean planes of the benzo(b)carbazole and phenyl rings form a dihedral angle of 79.26 (5)°. The N1—S1—C1 plane is almost orthogonal to carbazole ring [dihedral angle 87.20 (7)°] and phenyl ring [dihedral angle 88.44 (7)°]. The best plane of pyrrole ring N1/C7/C12/C13/C22 subtends a dihedral angle of 28.11 (11)° with sulfonyl group.
The average S—O, S—C, and S—N distances are 1.418, 1.760 and 1.664 Å, respectively, in (I); these are comparable as observed in similiar structures (Chakkaravarthi et al., 2007; Sankaranarayanan et al., 2000). The N—C bond lengths, namely N1—C7 and N1—C22 [1.430 (2) & 1.434 (2) Å in (I)] deviate slightly from the normal mean value reported in the literature (Allen et al., 1987). This indicates that the substitution of the phenylsulfonyl group at atom N1 results in an lengthening of the C—N bond lengths. This may be due to the electron-withdrawing character of the phenylsulfonyl group (Govindasamy et al., 1998). The S atom exhibits significant deviation from that of a regular tetrahedron, with the largest deviations being seen for the O—S—O [O1—S1—O2 119.66 (7)°] and O—S—N angles [O1—S—N1 106.78 (7)°]. The widening of the angles may be due to repulsive interactions between the two short S=O bonds, similar to what is observed in related structures (Chakkaravarthi et al., 2008; Rodriguez et al., 1995).
The sum of the bond angles around N1 [350.99°] indicate the sp2 hybridized state of the atom N, in the molecule. The benzene ring C15—C20 is almost coplanar with methyl groups [torsion angles C24—C19—C20—C21 and C15—C16—C17—C23 (0.17 (30)° and -179.18 (22)°, respectively] The torsion angles O1—S1—N1—C7 and O1—S1—C1—C6 [43.11 (16)° and 22.47 (20)°, respectively] describe the syn conformation of the phenylsulfonyl group with respect to benzocarbazole ring system. This conformation is influenced by the intramolecular C—H··· O hydrogen bonds (Table 1), C8—H8··· O1 and C21—H21···O2, involving sulfonyl atoms O1 and O2.
In addition, intramolecular C8—H8···O1 and C21—H21···O2 hydrogen bonds form six-membered rings, both with a graph-set motif of S(6) (Etter et al., 1990). The crystal packing exhibits weak intermolecular C—H···π interactions, involving the C7—C12 (centroid Cg1) and C15—C20 (centroid Cg2) rings (Table 1) [Fig. 2].
Experimental
To a solution of diethyl 2-((2-(bromomethyl)-1-(phenylsulfonyl)-1H-indol-3-yl)methylene) malonate (0.57 mmol) in dry 1,2-DCE (10 ml), ZnBr2 (1.15 mmol) and m-xylene (1.15 mmol) were added. The reaction mixture was then refluxed for 1 h under N2 atmosphere. It was then poured over ice-water (30 ml) containing 1 ml of concentrated HCl, extracted with chloroform (2 X 10 ml) and dried (Na2SO4). The solvent was removed under vacuo, then crude products was purified by flash column chromatography (silica gel, 230–420 mesh, n-hexane/ethyl acetate 99:1) afforded the compound (I), suitable for X-ray analysis.
Refinement
H atoms were positioned geometrically and refined using riding model with C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C) for aromatic H atoms and C—H = 0.96 Å and Uiso(H) = 1.5Ueq(C) for methyl H atoms.
Figures
Fig. 1.
The molecular structure of (I), with atom labeling scheme. Displacement ellipsoids are drawn at 50% probability level. H atoms are presented as a small spheres of arbitrary radius. Intramolecular H-bonds are shown as dashed lines.
Fig. 2.
The crystal structure of (I), viewed down the b axis. C—H···π interactions are shown as dashed lines For the sake of clarity, H atoms not involved in interaction have been omitted.
Crystal data
| C24H19NO2S | F000 = 1616 |
| Mr = 385.46 | Dx = 1.320 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 5907 reflections |
| a = 13.9260 (6) Å | θ = 2.6–28.4º |
| b = 10.1995 (5) Å | µ = 0.19 mm−1 |
| c = 27.3014 (14) Å | T = 295 (2) K |
| V = 3877.8 (3) Å3 | Block, colourless |
| Z = 8 | 0.30 × 0.20 × 0.16 mm |
Data collection
| Bruker Kappa APEXII diffractometer | 5626 independent reflections |
| Radiation source: fine-focus sealed tube | 3490 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.038 |
| T = 295(2) K | θmax = 30.0º |
| ω and φ scans | θmin = 1.5º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −11→19 |
| Tmin = 0.940, Tmax = 0.971 | k = −14→11 |
| 26413 measured reflections | l = −37→38 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters constrained |
| wR(F2) = 0.157 | w = 1/[σ2(Fo2) + (0.0716P)2 + 1.0437P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 5626 reflections | Δρmax = 0.68 e Å−3 |
| 255 parameters | Δρmin = −0.31 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.53043 (3) | 0.01070 (5) | 0.179632 (16) | 0.04540 (15) | |
| O1 | 0.57705 (11) | 0.10159 (16) | 0.21087 (5) | 0.0640 (4) | |
| O2 | 0.58196 (10) | −0.09761 (15) | 0.16030 (5) | 0.0577 (4) | |
| N1 | 0.49029 (11) | 0.09650 (16) | 0.13202 (5) | 0.0426 (4) | |
| C1 | 0.42667 (14) | −0.0487 (2) | 0.20902 (7) | 0.0477 (5) | |
| C2 | 0.38759 (17) | −0.1665 (2) | 0.19457 (8) | 0.0591 (5) | |
| H2 | 0.4160 | −0.2151 | 0.1696 | 0.071* | |
| C3 | 0.30584 (19) | −0.2116 (3) | 0.21752 (10) | 0.0772 (7) | |
| H3 | 0.2787 | −0.2909 | 0.2079 | 0.093* | |
| C4 | 0.2648 (2) | −0.1411 (3) | 0.25400 (11) | 0.0809 (8) | |
| H4 | 0.2099 | −0.1727 | 0.2694 | 0.097* | |
| C5 | 0.30312 (19) | −0.0243 (3) | 0.26847 (10) | 0.0785 (8) | |
| H5 | 0.2741 | 0.0228 | 0.2936 | 0.094* | |
| C6 | 0.38543 (18) | 0.0250 (2) | 0.24591 (8) | 0.0627 (6) | |
| H6 | 0.4117 | 0.1049 | 0.2554 | 0.075* | |
| C7 | 0.44478 (13) | 0.22122 (19) | 0.13769 (7) | 0.0454 (4) | |
| C8 | 0.46434 (16) | 0.3208 (2) | 0.17055 (8) | 0.0581 (5) | |
| H8 | 0.5103 | 0.3106 | 0.1950 | 0.070* | |
| C9 | 0.4132 (2) | 0.4357 (3) | 0.16576 (9) | 0.0717 (7) | |
| H9 | 0.4250 | 0.5042 | 0.1874 | 0.086* | |
| C10 | 0.3448 (2) | 0.4517 (3) | 0.12961 (10) | 0.0745 (7) | |
| H10 | 0.3115 | 0.5304 | 0.1273 | 0.089* | |
| C11 | 0.32554 (17) | 0.3529 (2) | 0.09714 (9) | 0.0643 (6) | |
| H11 | 0.2794 | 0.3642 | 0.0729 | 0.077* | |
| C12 | 0.37554 (14) | 0.2355 (2) | 0.10081 (7) | 0.0486 (5) | |
| C13 | 0.37626 (13) | 0.1181 (2) | 0.07144 (6) | 0.0446 (4) | |
| C14 | 0.32703 (14) | 0.0819 (2) | 0.02981 (7) | 0.0497 (5) | |
| H14 | 0.2800 | 0.1367 | 0.0169 | 0.060* | |
| C15 | 0.34814 (13) | −0.0379 (2) | 0.00694 (7) | 0.0491 (5) | |
| C16 | 0.29952 (16) | −0.0775 (3) | −0.03650 (7) | 0.0600 (6) | |
| H16 | 0.2513 | −0.0243 | −0.0492 | 0.072* | |
| C17 | 0.32182 (18) | −0.1911 (3) | −0.05996 (8) | 0.0651 (7) | |
| C18 | 0.39480 (18) | −0.2716 (2) | −0.04030 (8) | 0.0648 (6) | |
| H18 | 0.4103 | −0.3488 | −0.0566 | 0.078* | |
| C19 | 0.44376 (15) | −0.2408 (2) | 0.00171 (7) | 0.0546 (5) | |
| C20 | 0.42069 (14) | −0.12157 (19) | 0.02648 (7) | 0.0459 (4) | |
| C21 | 0.47023 (13) | −0.08330 (19) | 0.06929 (6) | 0.0435 (4) | |
| H21 | 0.5176 | −0.1366 | 0.0827 | 0.052* | |
| C22 | 0.44720 (12) | 0.03323 (19) | 0.09060 (6) | 0.0410 (4) | |
| C23 | 0.2711 (2) | −0.2323 (3) | −0.10647 (9) | 0.0899 (9) | |
| H23A | 0.2321 | −0.1613 | −0.1182 | 0.135* | |
| H23B | 0.2312 | −0.3070 | −0.0999 | 0.135* | |
| H23C | 0.3179 | −0.2549 | −0.1309 | 0.135* | |
| C24 | 0.5218 (2) | −0.3292 (2) | 0.02088 (10) | 0.0711 (7) | |
| H24A | 0.5012 | −0.3687 | 0.0510 | 0.107* | |
| H24B | 0.5789 | −0.2788 | 0.0266 | 0.107* | |
| H24C | 0.5350 | −0.3966 | −0.0027 | 0.107* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0375 (2) | 0.0617 (3) | 0.0370 (2) | 0.0086 (2) | −0.00775 (18) | 0.0004 (2) |
| O1 | 0.0572 (9) | 0.0790 (11) | 0.0557 (9) | 0.0017 (8) | −0.0242 (7) | −0.0081 (8) |
| O2 | 0.0462 (7) | 0.0755 (10) | 0.0514 (8) | 0.0223 (7) | −0.0049 (6) | −0.0018 (7) |
| N1 | 0.0413 (8) | 0.0515 (9) | 0.0350 (7) | −0.0009 (7) | −0.0056 (6) | 0.0016 (7) |
| C1 | 0.0475 (10) | 0.0575 (12) | 0.0381 (9) | 0.0151 (9) | 0.0005 (8) | 0.0109 (8) |
| C2 | 0.0635 (13) | 0.0623 (14) | 0.0514 (11) | 0.0022 (11) | 0.0064 (10) | 0.0072 (10) |
| C3 | 0.0754 (17) | 0.0786 (17) | 0.0776 (17) | −0.0049 (14) | 0.0120 (14) | 0.0157 (14) |
| C4 | 0.0658 (16) | 0.095 (2) | 0.0812 (18) | 0.0141 (15) | 0.0191 (14) | 0.0270 (16) |
| C5 | 0.0709 (16) | 0.100 (2) | 0.0647 (15) | 0.0353 (16) | 0.0233 (13) | 0.0080 (14) |
| C6 | 0.0665 (14) | 0.0663 (14) | 0.0553 (12) | 0.0216 (11) | 0.0047 (11) | 0.0011 (11) |
| C7 | 0.0436 (9) | 0.0517 (11) | 0.0410 (9) | −0.0013 (8) | −0.0029 (8) | 0.0032 (8) |
| C8 | 0.0616 (13) | 0.0608 (13) | 0.0518 (11) | 0.0015 (11) | −0.0108 (10) | −0.0044 (10) |
| C9 | 0.0864 (17) | 0.0596 (14) | 0.0690 (15) | 0.0060 (13) | −0.0117 (13) | −0.0124 (12) |
| C10 | 0.0851 (18) | 0.0611 (15) | 0.0773 (17) | 0.0192 (13) | −0.0111 (14) | −0.0043 (13) |
| C11 | 0.0618 (13) | 0.0677 (15) | 0.0635 (14) | 0.0129 (11) | −0.0145 (11) | 0.0051 (12) |
| C12 | 0.0450 (10) | 0.0561 (12) | 0.0446 (10) | 0.0007 (9) | −0.0052 (8) | 0.0056 (9) |
| C13 | 0.0387 (9) | 0.0575 (11) | 0.0376 (9) | −0.0068 (8) | −0.0033 (7) | 0.0085 (8) |
| C14 | 0.0437 (10) | 0.0622 (12) | 0.0433 (10) | −0.0063 (9) | −0.0102 (8) | 0.0104 (9) |
| C15 | 0.0440 (10) | 0.0675 (13) | 0.0359 (9) | −0.0200 (9) | −0.0030 (8) | 0.0083 (9) |
| C16 | 0.0556 (12) | 0.0824 (16) | 0.0422 (10) | −0.0268 (11) | −0.0098 (9) | 0.0062 (11) |
| C17 | 0.0692 (14) | 0.0837 (17) | 0.0425 (11) | −0.0398 (13) | −0.0033 (10) | −0.0006 (11) |
| C18 | 0.0757 (15) | 0.0689 (15) | 0.0499 (11) | −0.0344 (12) | 0.0056 (11) | −0.0080 (11) |
| C19 | 0.0628 (12) | 0.0549 (12) | 0.0462 (10) | −0.0237 (10) | 0.0041 (9) | 0.0006 (9) |
| C20 | 0.0479 (10) | 0.0532 (11) | 0.0366 (9) | −0.0171 (8) | 0.0035 (8) | 0.0041 (8) |
| C21 | 0.0446 (10) | 0.0500 (10) | 0.0358 (9) | −0.0065 (8) | −0.0017 (7) | 0.0058 (8) |
| C22 | 0.0381 (9) | 0.0522 (11) | 0.0329 (8) | −0.0072 (8) | −0.0026 (7) | 0.0051 (7) |
| C23 | 0.099 (2) | 0.117 (2) | 0.0532 (13) | −0.0450 (19) | −0.0173 (13) | −0.0105 (15) |
| C24 | 0.0975 (19) | 0.0495 (13) | 0.0662 (15) | −0.0065 (13) | 0.0034 (13) | −0.0067 (11) |
Geometric parameters (Å, °)
| S1—O1 | 1.4170 (15) | C11—H11 | 0.9300 |
| S1—O2 | 1.4190 (15) | C12—C13 | 1.442 (3) |
| S1—N1 | 1.6636 (15) | C13—C14 | 1.377 (3) |
| S1—C1 | 1.760 (2) | C13—C22 | 1.414 (3) |
| N1—C7 | 1.430 (2) | C14—C15 | 1.404 (3) |
| N1—C22 | 1.434 (2) | C14—H14 | 0.9300 |
| C1—C2 | 1.377 (3) | C15—C16 | 1.424 (3) |
| C1—C6 | 1.382 (3) | C15—C20 | 1.426 (3) |
| C2—C3 | 1.379 (3) | C16—C17 | 1.360 (4) |
| C2—H2 | 0.9300 | C16—H16 | 0.9300 |
| C3—C4 | 1.355 (4) | C17—C18 | 1.412 (4) |
| C3—H3 | 0.9300 | C17—C23 | 1.513 (3) |
| C4—C5 | 1.363 (4) | C18—C19 | 1.371 (3) |
| C4—H4 | 0.9300 | C18—H18 | 0.9300 |
| C5—C6 | 1.395 (4) | C19—C20 | 1.428 (3) |
| C5—H5 | 0.9300 | C19—C24 | 1.506 (3) |
| C6—H6 | 0.9300 | C20—C21 | 1.412 (3) |
| C7—C8 | 1.382 (3) | C21—C22 | 1.362 (3) |
| C7—C12 | 1.402 (3) | C21—H21 | 0.9300 |
| C8—C9 | 1.378 (3) | C23—H23A | 0.9600 |
| C8—H8 | 0.9300 | C23—H23B | 0.9600 |
| C9—C10 | 1.381 (4) | C23—H23C | 0.9600 |
| C9—H9 | 0.9300 | C24—H24A | 0.9600 |
| C10—C11 | 1.368 (3) | C24—H24B | 0.9600 |
| C10—H10 | 0.9300 | C24—H24C | 0.9600 |
| C11—C12 | 1.388 (3) | ||
| O1—S1—O2 | 120.10 (9) | C7—C12—C13 | 107.97 (17) |
| O1—S1—N1 | 106.26 (9) | C14—C13—C22 | 119.30 (19) |
| O2—S1—N1 | 106.79 (8) | C14—C13—C12 | 132.68 (19) |
| O1—S1—C1 | 109.07 (10) | C22—C13—C12 | 107.91 (16) |
| O2—S1—C1 | 108.47 (10) | C13—C14—C15 | 119.73 (18) |
| N1—S1—C1 | 105.14 (8) | C13—C14—H14 | 120.1 |
| C7—N1—C22 | 107.48 (14) | C15—C14—H14 | 120.1 |
| C7—N1—S1 | 122.17 (12) | C14—C15—C16 | 121.2 (2) |
| C22—N1—S1 | 121.34 (13) | C14—C15—C20 | 120.20 (17) |
| C2—C1—C6 | 121.3 (2) | C16—C15—C20 | 118.6 (2) |
| C2—C1—S1 | 119.61 (16) | C17—C16—C15 | 121.7 (2) |
| C6—C1—S1 | 119.11 (19) | C17—C16—H16 | 119.2 |
| C1—C2—C3 | 119.2 (2) | C15—C16—H16 | 119.2 |
| C1—C2—H2 | 120.4 | C16—C17—C18 | 118.7 (2) |
| C3—C2—H2 | 120.4 | C16—C17—C23 | 121.7 (3) |
| C4—C3—C2 | 120.3 (3) | C18—C17—C23 | 119.5 (3) |
| C4—C3—H3 | 119.8 | C19—C18—C17 | 122.9 (2) |
| C2—C3—H3 | 119.8 | C19—C18—H18 | 118.6 |
| C3—C4—C5 | 120.8 (3) | C17—C18—H18 | 118.6 |
| C3—C4—H4 | 119.6 | C18—C19—C20 | 118.6 (2) |
| C5—C4—H4 | 119.6 | C18—C19—C24 | 120.8 (2) |
| C4—C5—C6 | 120.6 (2) | C20—C19—C24 | 120.51 (19) |
| C4—C5—H5 | 119.7 | C21—C20—C15 | 119.36 (18) |
| C6—C5—H5 | 119.7 | C21—C20—C19 | 121.15 (19) |
| C1—C6—C5 | 117.9 (3) | C15—C20—C19 | 119.46 (18) |
| C1—C6—H6 | 121.1 | C22—C21—C20 | 118.67 (18) |
| C5—C6—H6 | 121.1 | C22—C21—H21 | 120.7 |
| C8—C7—C12 | 121.70 (19) | C20—C21—H21 | 120.7 |
| C8—C7—N1 | 129.54 (17) | C21—C22—C13 | 122.74 (16) |
| C12—C7—N1 | 108.65 (17) | C21—C22—N1 | 129.15 (17) |
| C9—C8—C7 | 117.5 (2) | C13—C22—N1 | 107.99 (16) |
| C9—C8—H8 | 121.3 | C17—C23—H23A | 109.5 |
| C7—C8—H8 | 121.3 | C17—C23—H23B | 109.5 |
| C8—C9—C10 | 121.6 (2) | H23A—C23—H23B | 109.5 |
| C8—C9—H9 | 119.2 | C17—C23—H23C | 109.5 |
| C10—C9—H9 | 119.2 | H23A—C23—H23C | 109.5 |
| C11—C10—C9 | 120.7 (2) | H23B—C23—H23C | 109.5 |
| C11—C10—H10 | 119.6 | C19—C24—H24A | 109.5 |
| C9—C10—H10 | 119.6 | C19—C24—H24B | 109.5 |
| C10—C11—C12 | 119.3 (2) | H24A—C24—H24B | 109.5 |
| C10—C11—H11 | 120.3 | C19—C24—H24C | 109.5 |
| C12—C11—H11 | 120.3 | H24A—C24—H24C | 109.5 |
| C11—C12—C7 | 119.10 (19) | H24B—C24—H24C | 109.5 |
| C11—C12—C13 | 132.82 (18) | ||
| O1—S1—N1—C7 | −43.11 (16) | C7—C12—C13—C14 | −176.6 (2) |
| O2—S1—N1—C7 | −172.43 (14) | C11—C12—C13—C22 | 175.6 (2) |
| C1—S1—N1—C7 | 72.46 (16) | C7—C12—C13—C22 | −0.6 (2) |
| O1—S1—N1—C22 | 173.90 (14) | C22—C13—C14—C15 | −0.3 (3) |
| O2—S1—N1—C22 | 44.58 (16) | C12—C13—C14—C15 | 175.41 (19) |
| C1—S1—N1—C22 | −70.53 (16) | C13—C14—C15—C16 | −179.16 (17) |
| O1—S1—C1—C2 | −158.20 (16) | C13—C14—C15—C20 | −0.4 (3) |
| O2—S1—C1—C2 | −25.75 (18) | C14—C15—C16—C17 | 177.57 (19) |
| N1—S1—C1—C2 | 88.19 (17) | C20—C15—C16—C17 | −1.2 (3) |
| O1—S1—C1—C6 | 22.46 (19) | C15—C16—C17—C18 | 0.4 (3) |
| O2—S1—C1—C6 | 154.91 (16) | C15—C16—C17—C23 | −179.2 (2) |
| N1—S1—C1—C6 | −91.15 (17) | C16—C17—C18—C19 | 0.6 (3) |
| C6—C1—C2—C3 | −0.1 (3) | C23—C17—C18—C19 | −179.9 (2) |
| S1—C1—C2—C3 | −179.44 (18) | C17—C18—C19—C20 | −0.6 (3) |
| C1—C2—C3—C4 | −0.4 (4) | C17—C18—C19—C24 | −179.3 (2) |
| C2—C3—C4—C5 | 0.4 (4) | C14—C15—C20—C21 | 0.7 (3) |
| C3—C4—C5—C6 | 0.0 (4) | C16—C15—C20—C21 | 179.44 (17) |
| C2—C1—C6—C5 | 0.5 (3) | C14—C15—C20—C19 | −177.66 (17) |
| S1—C1—C6—C5 | 179.85 (17) | C16—C15—C20—C19 | 1.1 (3) |
| C4—C5—C6—C1 | −0.5 (4) | C18—C19—C20—C21 | −178.54 (18) |
| C22—N1—C7—C8 | −176.3 (2) | C24—C19—C20—C21 | 0.2 (3) |
| S1—N1—C7—C8 | 36.3 (3) | C18—C19—C20—C15 | −0.2 (3) |
| C22—N1—C7—C12 | −0.2 (2) | C24—C19—C20—C15 | 178.47 (18) |
| S1—N1—C7—C12 | −147.55 (14) | C15—C20—C21—C22 | −0.2 (3) |
| C12—C7—C8—C9 | −0.1 (3) | C19—C20—C21—C22 | 178.10 (16) |
| N1—C7—C8—C9 | 175.6 (2) | C20—C21—C22—C13 | −0.5 (3) |
| C7—C8—C9—C10 | 0.0 (4) | C20—C21—C22—N1 | −176.05 (17) |
| C8—C9—C10—C11 | 0.0 (4) | C14—C13—C22—C21 | 0.8 (3) |
| C9—C10—C11—C12 | 0.0 (4) | C12—C13—C22—C21 | −175.89 (17) |
| C10—C11—C12—C7 | −0.1 (3) | C14—C13—C22—N1 | 177.13 (16) |
| C10—C11—C12—C13 | −175.9 (2) | C12—C13—C22—N1 | 0.5 (2) |
| C8—C7—C12—C11 | 0.2 (3) | C7—N1—C22—C21 | 175.86 (18) |
| N1—C7—C12—C11 | −176.30 (18) | S1—N1—C22—C21 | −36.4 (2) |
| C8—C7—C12—C13 | 176.93 (19) | C7—N1—C22—C13 | −0.19 (19) |
| N1—C7—C12—C13 | 0.5 (2) | S1—N1—C22—C13 | 147.52 (13) |
| C11—C12—C13—C14 | −0.5 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C8—H8···O1 | 0.93 | 2.37 | 2.945 (3) | 120 |
| C21—H21···O2 | 0.93 | 2.34 | 2.935 (2) | 122 |
| C8—H8···Cg1i | 0.93 | 3.00 | 3.859 | 155 |
| C11—H11···Cg2ii | 0.93 | 2.90 | 3.693 | 144 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x−1/2, y−1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT2759).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2004). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Chakkaravarthi, G., Dhayalan, V., Mohanakrishnan, A. K. & Manivannan, V. (2007). Acta Cryst. E63, o3673.
- Chakkaravarthi, G., Dhayalan, V., Mohanakrishnan, A. K. & Manivannan, V. (2008). Acta Cryst. E64, o542. [DOI] [PMC free article] [PubMed]
- Diaz, J. L., Villacampa, B., Lopez-Calahorra, F. & Velasco, D. (2002). Chem. Mater.14, 2240–2251.
- Etter, M. C., MacDonald, J. C. & Bernstein, J. (1990). Acta Cryst. B46, 256–262. [DOI] [PubMed]
- Friend, R. H., Gymer, R. W., Holmes, A. B., Burroughes, J. H., Mark, R. N., Taliani, C., Bradley, D. D. C., Dos Santos, D. A., Bredas, J. L., Logdlund, M. & Salaneck, W. R. (1999). Nature (London), 397, 121–127.
- Govindasamy, L., Velmurugan, D., Ravikumar, K. & Mohanakrishnan, A. K. (1998). Acta Cryst. C54, 277–279.
- Hökelek, T., Gündüz, H., Patir, S. & Uludaug, N. (1998). Acta Cryst. C54, 1297–1299.
- Hosomi, H., Ohba, S. & Ito, Y. (2000). Acta Cryst. C56, e144–e146. [DOI] [PubMed]
- Itoigawa, M., Kashiwada, Y., Ito, C., Furukawa, H., Tachibana, Y., Bastow, K. F. & Lee, K. H. (2000). J. Nat. Prod.63, 893–897. [DOI] [PubMed]
- Ramsewak, R. S., Nair, M. G., Strasburg, G. M., DeWitt, D. L. & Nitiss, J. L. (1999). J. Agric. Food Chem.47, 444–447. [DOI] [PubMed]
- Rodriguez, J. G., del Valle, C., Esteban-Calderon, C. & Martinez-Repoll, M. (1995). J. Chem. Crystallogr.25, 249–257.
- Sankaranarayanan, R., Velmurugan, D., Shanmuga Sundara Raj, S., Fun, H.-K., Babu, G. & Perumal, P. T. (2000). Acta Cryst. C56, 475–476. [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Tachibana, Y., Kikuzaki, H., Lajis, N. H. & Nakatani, N. (2001). J. Agric. Food Chem.49, 5589–5594. [DOI] [PubMed]
- Zhang, Q., Chen, J., Cheng, Y., Wang, L., Ma, D., Jing, X. & Wang, F. (2004). J. Mater. Chem.14, 895–900.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808024380/bt2759sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808024380/bt2759Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




