Abstract
The transposon Tn5090/Tn402 encodes a 559 amino acid transposase, TniA, with a DDE motif. Gel mobility shifting and cleavage protection analysis with DNase I and hydroxyl radical probes revealed that TniA binds to multiple repeat sequences on either terminus of Tn5090/Tn402. Four of these TniA-binding 19mers occurred on the left-hand (t) end and two on the right-hand (i) end. Hydroxyl radical cleavage protection demonstrated the presence of 3–6 bp contact sequences on one face of the DNA helix. The binding pattern and organisation of repeats suggested parallels between Tn5090/Tn402 and Mu, which controls its transpositional activity in the assembly step of a higher order transpososome complex. The complex terminal structure and genes of transposase and nucleotide-binding proteins in tandem are hallmarks of the handful of Mu-like elements that are known to date.
INTRODUCTION
Translocation of mobile DNA elements requires cleavage at each end of the element and subsequent joining of the exposed ends to a target site (1). The two reactions are mediated by transposase, which commonly contains a metal-connected DDE motif in its sequence (2–5). The specific contacts between the transposase and the ends of its mobile element are crucial for transposition (6).
Although most transposons carry one inverted repeat sequence on each end, the transposable phage Mu has been found to carry multiple sites with a complex organisation (7,8). These sites are arranged to allow assembly of the tetrameric, active form of the transposase in a higher order complex with substrate DNA, which provides checkpoints for efficient control of transposition (9–12). Through the course of transposition the nucleoprotein complex formed by binding to the repeats is gradually stabilised and converted into a conformation of tightly knit protomers sharing the active site (the DDE motif) (13,14). The complex catalysing the strand transfer step is very stable and disassembly requires a molecular chaperone and potentially other host factors (15,16).
Multiple transposase-binding repeat sequences organised in a Mu-like fashion have also been found on the termini of a small number of non-viral transposons such as Tn7 and Tn552 (17–20). Tn5090/Tn402 (3,21; Fig. 1) and the closely related Tn5053 (22) could be further examples because several repeats of 19 bp have been noticed on their ends. The transposase of Tn5090/Tn402 is 25% identical to those of Tn7 and Tn552, respectively, and <20% identical to the transposase of Mu (3). The principal aim of this study was to investigate if and how the transposase, TniA, binds to the arrayed sequences on the ends of Tn5090/Tn402. If repeated transposase-binding sites are shared among these DNA elements this could reflect a conserved strategy for the assembly and maturation of synaptic complexes.
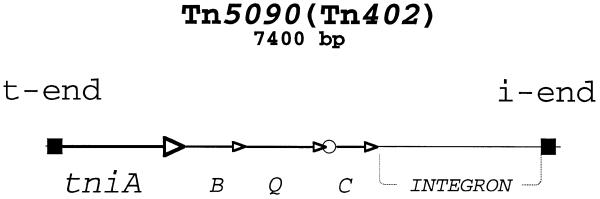
Figure 1.
Physical map of Tn5090/Tn402. Arrows indicate structural genes; an open circle denotes the resolvase (TniC) site, res. The integron carried by Tn5090/Tn402 is shown without details. Filled boxes represent transposon ends.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
Escherichia coli DH5α [supE44 ΔlacU169 (φ80lacZ′ΔM15) hsdR17 recA1 gyrA96 thi-1 relA] served as a host for subcloning of DNA fragments. The lysogenic host strain BL21(DE3) (23) was used to achieve transcription from the T7 promoter of pET vectors. DH5α was grown in 2× TY and BL21(DE3) in L broth.
The fragment cloned to express transposase was prepared by PCR amplification of the tniA gene. The forward primer 5′-CAAAGCGAGGTGCATATGGCGACGGACAC-3′, carrying an NdeI site that overlapped the translation start codon of tniA, was combined with the reverse primer 5′-ATGGATCCTACCACTCCTCAATCTGGTC-3′, carrying a linker site for restriction endonuclease BamHI. The amplified fragment was cloned in the NdeI and BamHI sites of pET11b (Novagen) to form pLKO420. The ends of Tn5090/Tn402 on plasmid R751 are identical to those of the derivative in R100.1 (Tn21) (3,24). The latter plasmid was used as template in PCR amplification of the transposon ends because the target duplication of 5 bp (TCCAT) is intact. The t-end of Tn5090/Tn402 was amplified using the forward primer 5′-ATTAAAGCTTGGGCATGGATGTCATTTTC-3′, containing a HindIII linker site, and the reverse primer 5′-TACTGCAGCCATGGTCACCTCGCTTTGGTGC-3′, containing a PstI site. The resulting amplicon, comprising 154 bp, was cloned in the HindIII and PstI sites of pUC19 (pLKO440) to represent the t-end in binding assays. A fragment containing the i-end of Tn5090/Tn402 was amplified using the forward primer 5′-ATGAATTCCACCTCCATTGTCGTTTTCAG-3′, containing a site for EcoRI, and the reverse primer 5′-TTAGGTACCCCTGCGCGAAGGCCATCGG-3′, including a KpnI recognition sequence. The amplified fragment, comprising 183 bp, was cloned in the EcoRI and KpnI sites of the pUC19 vector (pLKO441A). The inserted fragment was shortened to 82 bp in the subclone pLKO441B by digestion of pLKO441A with BamHI and religation. The fragment of plasmid pLKO441B included repeats i1 and i2 and represented the i-end in protein binding assays. The larger fragment of pLKO441A also included the atypical repeats i3 and i4 and was used in control experiments. All cloned fragments containing transposase-binding sites or the transposase gene, tniA, were checked by sequencing.
Overexpression of soluble TniA
A single colony of BL21(DE3) transformed with pLKO420 was resuspended in 150 ml of L broth and incubated at 37°C with agitation. At mid-exponential growth phase the culture was transferred to a 16°C incubator. IPTG inducer was added to a concentration of 1 mM and cultivation continued for 20 h. The cells were harvested and resuspended in 5 ml of 50 mM Tris–HCl (pH 7.5), 20 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Cell lysis was achieved by four pulses of sonication for 30 s at an amplitude of 18 microns using a Soniprep 150 apparatus. The soluble and insoluble protein fractions were separated by centrifugation at 12 000 r.p.m. for 30 min. The soluble fraction was loaded onto a SP Sepharose® Fast Flow (Pharmacia) column which was washed with 30 ml of 50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 0.1 mM PMSF. TniA was eluted with 20 ml of 50 mM Tris–HCl (pH 7.5), 400 mM NaCl, 0.1 mM PMSF (elution buffer). TniA-containing fractions were identified by SDS–PAGE, pooled and concentrated using Centriprep YM-10 (Amicon). Next, the protein solution was loaded onto a Superdex® 200 HR chromatography FPLC column (Pharmacia) equilibrated with elution buffer (see above) and fractions containing TniA were collected. The purity and quantity of transposase were checked by SDS–PAGE and the method of Bradford (25).
End-labelling
Plasmids carrying transposon ends were transformed into DH5α and purified using Qiagen plasmid purification kits. DNA was labelled at the 3′-end using the Klenow fragment of DNA polymerase I and [α-32P]dATP as described by Sambrook et al. (26). The plasmid pLKO440 was digested with EcoRI and HindIII to label the top and bottom strands of the t-end, respectively. The plasmids pLKO441A and pLKO441B were cut by HindIII and EcoRI to label the top and bottom strand of the i-end, respectively. The labelled DNA fragments were cut with a second restriction enzyme and separated by electrophoresis in a 6% polyacrylamide gel as described by Dixon et al. (27).
Gel mobility shift assay
Freshly prepared TniA (5 µl), serially diluted in elution buffer, was mixed with 2 ng 32P-labelled transposon ends (70 c.p.s.) in 15 µl reactions containing final concentrations of 60 mM Tris–HCl (pH 7.5), 60 mM KCl, 50 µg/ml bovine serum albumin (BSA), 5 mM spermidine, 1 µg poly(dI·dC)·poly(dI·dC), 30 µM PMSF and 133 mM NaCl at 30°C for 10 min. After addition of 2 µl of dye solution (100 mM EDTA, 0.2% xylene cyanol, 0.2% bromophenol blue and 50% sucrose) the protein–DNA complexes were loaded onto a non-denaturing 6% polyacrylamide gel and run in TBE buffer at 8°C for 3 h at 10 V/cm. The gel was dried and the bands were visualised by autoradiography.
DNase I footprinting
The 3′-labelled DNA fragments (5 ng, 200 c.p.s.) were mixed with TniA (2.5 µg in 5 µl of elution buffer) in 100 µl reactions containing final concentrations of 47 mM Tris–HCl (pH 7.5), 60 mM KCl, 50 µg/ml BSA, 5 mM spermidine, 1 µg poly(dI·dC)·poly(dI·dC), 5 µM PMSF and 133 mM NaCl at 30°C for 10 min. The binding mixtures were supplemented with MgCl2 to 1 mM final concentration and treated with 5 µl of a 20 µg/ml solution of DNase I (final concentration 1 µg/ml) for 2 min at 20°C. The stock solution contained 1 mg/ml DNase I, 0.15 M NaCl, 50% glycerol, was routinely stored at –20°C and was 100-fold diluted in water prior to use. The cleavage reactions were stopped by adding 2 µl of 0.5 M EDTA, incubation at 37°C for 30 min with 1 mg/ml proteinase K and ethanol precipitation. The cleaved DNA samples were dissolved in 4 µl of formamide dye solution (48% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol FF), boiled for 2 min and loaded onto a denaturing 6% polyacrylamide gel. The acrylamide:bisacrylamide ratio in the gel was 19:1 and the urea concentration was 8 M. The bands were detected by autoradiography.
Hydroxyl radical footprinting
TniA (2.5 µg in 5 µl of elution buffer) was incubated at 30°C for 10 min with 10 ng 3′-labelled transposon ends (400 c.p.s.) in 10 µl reactions containing 69 mM Tris–HCl (pH 7.5), 60 mM KCl, 50 µg/ml BSA, 5 mM spermidine, 1 µg poly(dI·dC)·poly(dI·dC), 5 µM PMSF, 200 mM NaCl (final concentrations). Hydroxyl radical cleavage was carried out by adding 10 µl of a solution containing 70 µM ferrous ammonium sulphate, 0.33 mM EDTA, 0.1% H2O2, 10 mM ascorbic acid to the binding reaction mixture. After 10 min the reaction was quenched by adding glycerol to 5% final concentration. For analysis of the i-end the reaction mixtures were treated with proteinase K as described above and the DNA was recovered by ethanol precipitation, dissolved in 4 µl of formamide dye (see above), boiled for 2 min and analysed on a denaturing 6% polyacrylamide gel (further specified above). Hydroxyl radical-treated t-end substrates were quenched as described above and loaded on a non-denaturing 6% polyacrylamide gel. Free DNA and protein–DNA complexes were eluted from the gel as described by Dixon et al. (27). The recovered DNA was then dissolved in 4 µl of formamide dye, boiled for 2 min and loaded on a denaturing 6% polyacrylamide gel. The bands were visualised by autoradiography.
RESULTS
Expression of transposase
The Tn5090/Tn402 encoded transposase, TniA, was overexpressed using the pET11b expression vector. Aggregation of the protein was reduced by decreasing the induction temperature (see Materials and Methods). A significant fraction of the induced TniA was soluble under these conditions and was purified to near homogeneity using ion exchange chromatography and gel filtration (Fig. 2). At a concentration of 1 mg/ml TniA co-eluted in the gel filtration step with BSA (67 kDa; the calculated size of TniA is 62.95 kDa). This observation suggests that TniA is a monomer in solution.
Figure 2.
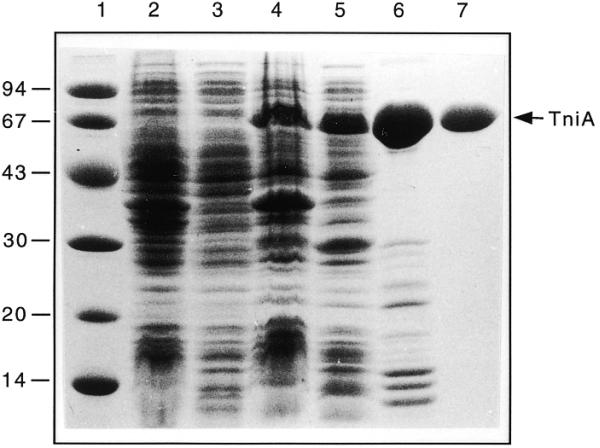
SDS–PAGE (12.5%) demonstrating solubility and purity of TniA expressed in E.coli BL21(DE3). The protein bands were stained with Coomassie Brilliant Blue R. Lane 1, molecular mass standard (molecular weights given in kDa); lane 2, insoluble proteins in a crude extract of BL21(DE3); lane 3, soluble proteins in a crude extract of BL21(DE3); lane 4, insoluble proteins in an extract from BL21(DE3) expressing TniA from pLKO420; lane 5, soluble proteins in an extract from BL21(DE3) expressing TniA from pLKO420; lane 6, pooled and concentrated TniA peak fractions purified by ion exchange chromatography; lane 7, TniA further purified by gel filtration.
Gel mobility retardation
TniA of increasing concentration was incubated with either transposon end and several electrophoretic mobility shifts were observed. This suggested the presence of multiple binding sites on both ends (Fig. 3). The complexes formed between TniA and the fragments containing the t-end resulted in six retarded bands (Fig. 3A). Three of these complexes (I, II and III) were formed with 16-fold diluted TniA and two further complexes, IV and V, with 4-fold diluted protein. The binding mixtures with undiluted TniA contained the slowest migrating complexes, V and VI.
Figure 3.
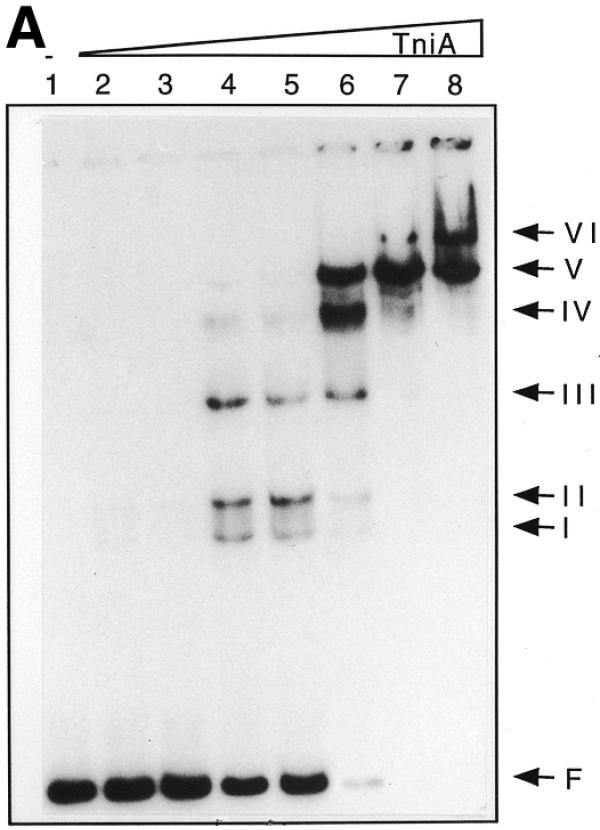
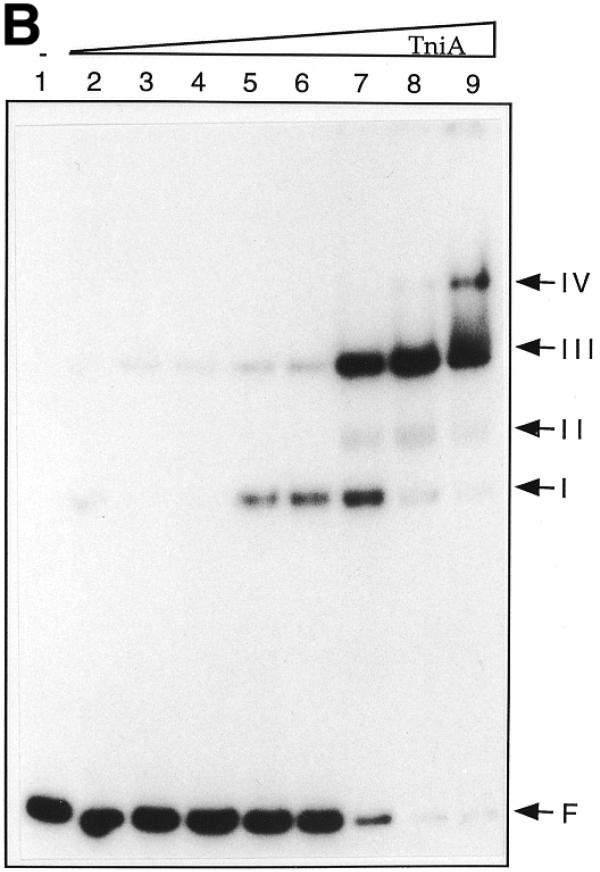
Electrophoretic mobility shifts of the ends of Tn5090/Tn402 after incubation with TniA at increasing concentration. 32P-labelled transposon ends were incubated with different amounts of TniA and analysed on non-denaturing 6% polyacrylamide gels. (A) The 154 bp fragment including the four 19 bp repeats of the t-end: lane 1, a control with no TniA; lanes 2–8, 64-, 32-, 16-, 8-, 4- and 2-fold diluted and undiluted TniA (0.5 µg/µl). (B) The 82 bp fragment containing repeats i1 and i2 of the i-end: lane 1, no TniA; lanes 2–9, 128-, 64-, 32-, 16-, 8-, 4- and 2-fold diluted and undiluted TniA (0.5 µg/µl).
Gel retardation of the mixtures containing the i-end fragment of 82 bp from pLKO441B gave a different band pattern than those containing the t-end fragments (Fig. 3B). Band I and, particularly, band III were intense and appeared at a lower concentration of transposase than band II, which was faint but observed reproducibly. Repeats i1 and i2 resemble the repeats on the t-end whereas repeats i3 and i4 differ significantly from the consensus sequence (3). In order to assess whether these atypical repeats influence the binding of transposase gel retardation of the i-end fragments from pLKO441B (excluding i3 and i4) and pLKO441A (including repeats i3 and i4) were compared. The electrophoretic retardation patterns (not shown) were very similar, indicating that the transposase, TniA, binds to repeats i1 and i2 but does not bind to i3 and i4.
TniA at the highest concentration did not influence the electrophoretic mobility of a negative control fragment from the tniC area of Tn5090/Tn402 (3).
Enzymatic cleavage protection
DNase I footprinting revealed that TniA recognises four 19 bp repeats on the t-end and two repeats at the i-end (Fig. 4). The fourth repeat at the t-end (shown as t4 in the figures) escaped notice in the original sequence report of Tn5090/Tn402 (3) because the sequence of its first part differs from the consensus sequence. Consistent with the gel retardation data (see above), TniA binds to repeats i1 and i2 but did not, even at the highest concentration, protect repeats further in the element, i3 and i4, from nuclease digestion. Equivalents of these two unverified repeats are, furthermore, missing in the closely related and functional transposon Tn5053 (22).
Figure 4.
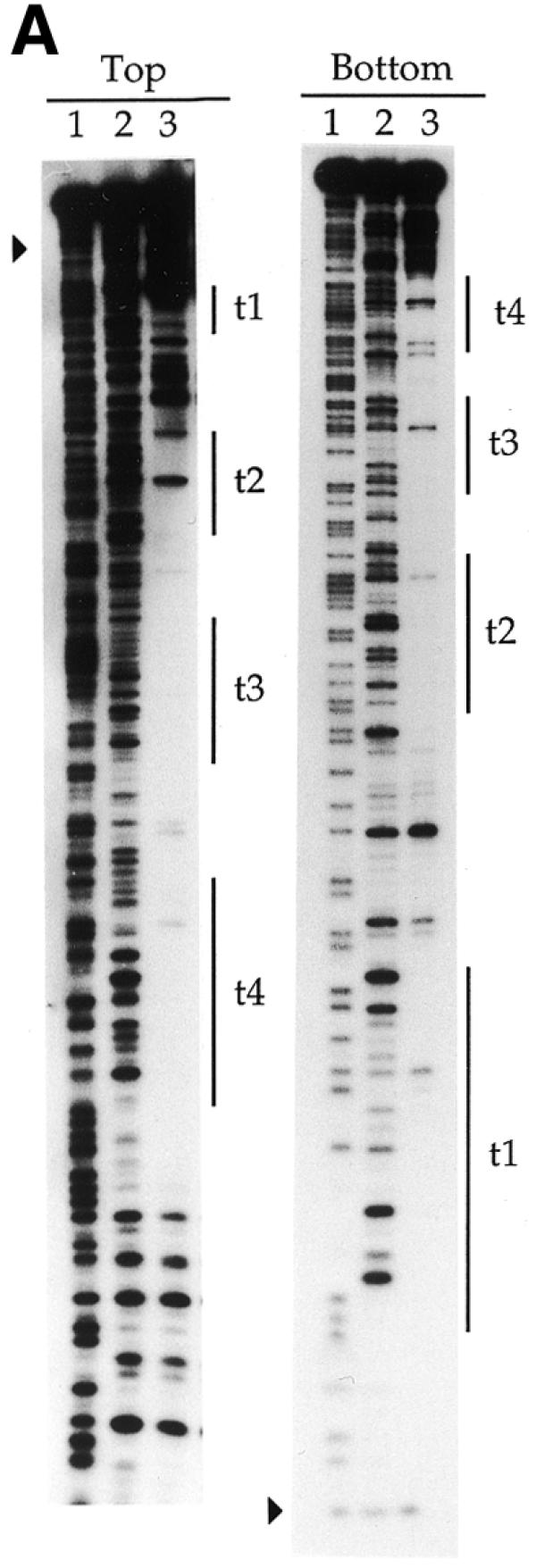
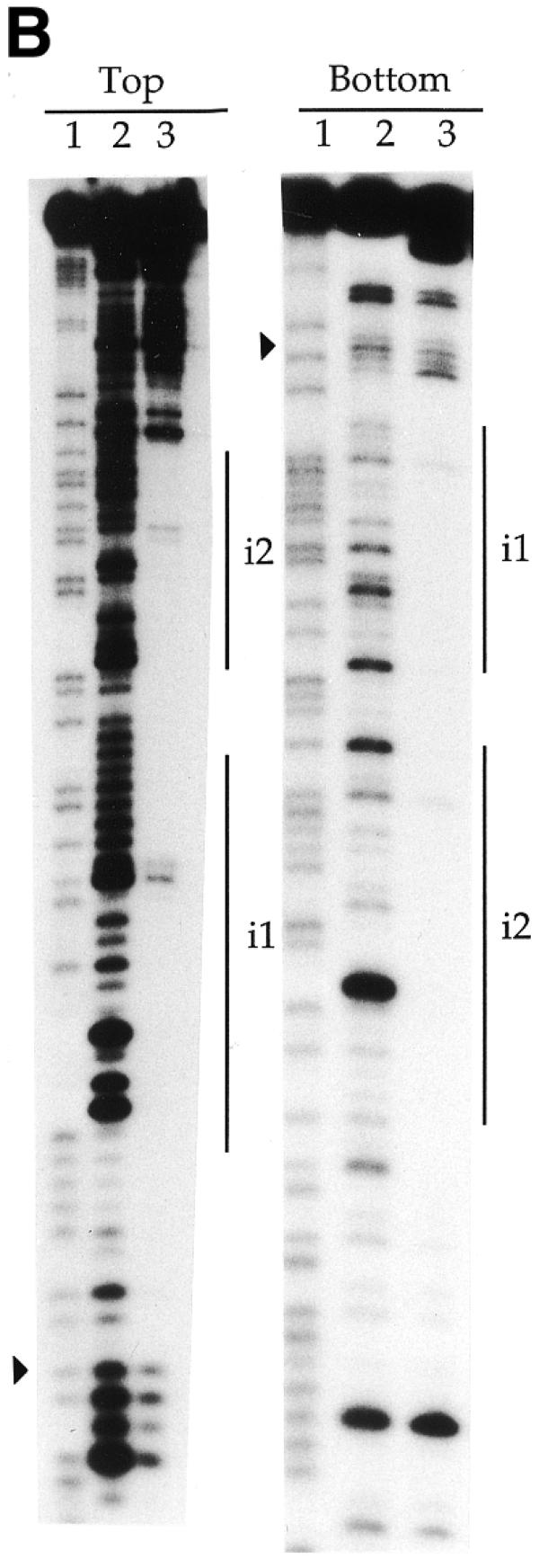
Denaturing polyacrylamide gels showing DNase I cleavage close to the transposon ends under protection by TniA. (A) The footprints on the 3′-labelled top and bottom strands, respectively, of a fragment containing 154 bp of the t-end. (B) Protections on the 3′-labelled top and bottom strands of an 82 bp fragment containing the i-end. Vertical lines indicate the 19 bp repeats and filled triangles indicate terminal nucleotides of the transposons. Lane 1, chemical cleavage standard A + G; lane 2, DNase I cleavage in the absence of TniA; lane 3, DNase I cleavage under protection by TniA.
The phosphodiester linkages in the DNA outside both ends of the transposon were fully exposed to cleavage by DNase I. Protection by transposase started very close to either end and progressed inward through repeats i1 and t1, respectively. The protected area started more precisely at the phosphodiester linkage inside the dinucleotides TG/AC on either terminus of the transposon except for the bottom strand of the i-end. For that strand the protection by TniA was recessed by another nucleotide to leave two internal phosphodiester bonds exposed to cleavage (Figs 4 and 6).
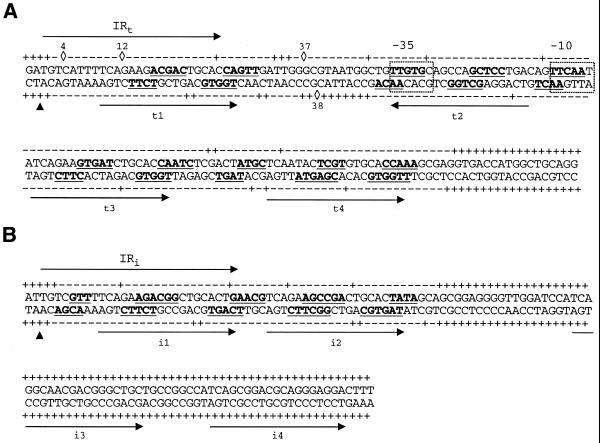
Figure 6.
Summary and details of the chemical and enzymatic probe protection analyses of the TniA contacts with the t-end (A) and the i-end (B). The arrows above the sequences indicate the terminal 25 bp inverted repeats while the arrows under the sequences indicate the 19 bp repeats. Transposase nicking sites are indicated with filled triangles. Sequences protected from hydroxyl radical cleavage are in bold and underlined. The linkages marked – were protected from DNase I cleavage. Phosphodiester linkages cleaved by DNase I are indicated with +, while DNase I hypersensitive phosphates are indicated with diamonds and the number of the nucleotide starting from the transposon termini. The data for DNase I footprinting of i3 and i4, illustrated here, are not shown in Figure 4. Promoter hexamers are boxed.
The binding of TniA induced DNase I hypersensitivity of a few phosphodiester bonds on the t-end but such hypersensitive sites were not observed on the i-end. At the t-end we observed four phosphodiester linkages with increased DNase I sensitivity. These occurred on the 5′-sides of C4, G12 and G37 in the upper strand and G38 in the bottom strand (Figs 4 and 6).
The 19 bp repeats on the transposon ends were protected from nuclease by TniA in a concentration-dependent manner. TniA protected the t1 and t3 repeats on the t-end at the same concentration, followed by the t2 repeat, which was protected at a higher concentration (data not shown). A still higher concentration of transposase was needed for protection of t4. The two repeats on the i-end (i1 and i2) were protected at the same dilution of TniA (data not shown).
Hydroxyl radical footprinting
To further define the nucleotides in contact with the transposase, hydroxyl radical probing (27) was performed on the transposase–transposon ends complexes. This revealed two transposase contact regions in each 19 bp repeat (Fig. 5). Each protected region comprised less than 6 bp. One of them comprised the central section of each 19 bp unit, whereas the other overlapped one repeat end (Fig. 6). Highly conserved triplets (TCA and CTG; 3) were unprotected parts of the 19 bp repeats. Footprinting of both strands showed DNA protection with an offset of a few nucleotides in the complementary strand, which indicates that TniA binds to one side of the DNA that might be wrapped around the protein. Two additional sites located outside the 19 bp repeats were weakly protected, one site between t2 and t3 and the other located between t3 and t4. A short protected region between the transposon end and repeat i1 was also observed (Figs 5 and 6).
Figure 5.
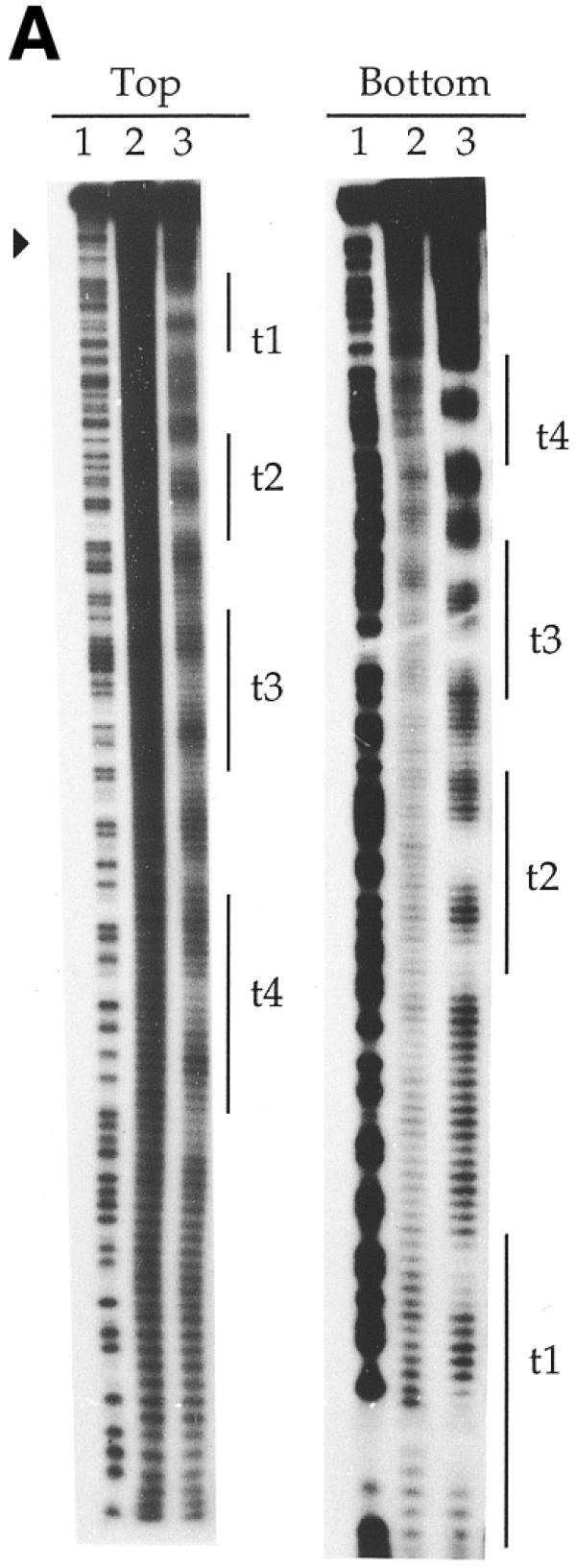
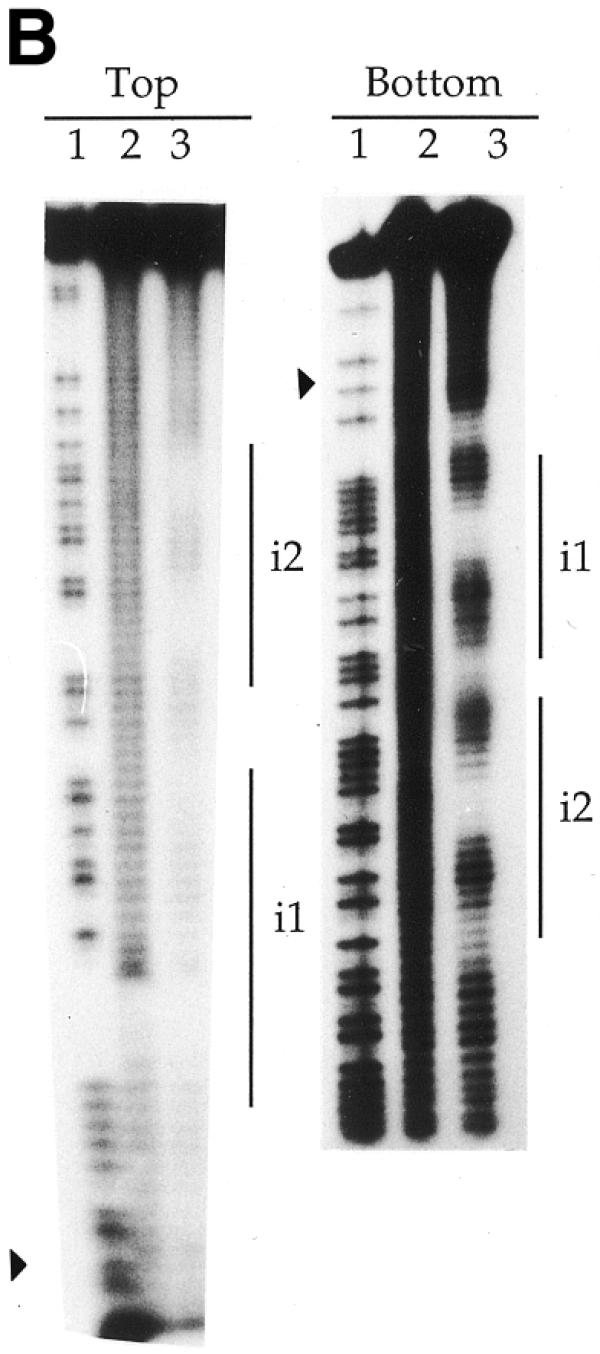
Hydroxyl radical footprinting gel depicting the contacts between transposase and transposon ends. 3′-Labelled top and bottom strands of fragments of the t-end (A) and the i-end (B) as in Figure 4. Vertical lines indicate the 19 bp repeats and filled triangles indicate terminal nucleotides of the transposons. Lane 1, chemical cleavage standard A + G; lane 2, cleavage in the absence of TniA; lane 3, hydroxyl radical cleavage at a protective concentration of TniA.
DISCUSSION
In this work we have assessed a pattern of several transposase-binding sequences on the ends of transposon Tn5090/Tn402. The small group of transposons which carry multiple repeats on the ends similar to those of Mu (7,8,17–20) obviously includes several non-viral members. Further study of transposons of this class, for instance Tn5090/Tn402, could bridge the mechanistic observations made with the Mu model system to other transposons.
DNase I footprinting analysis revealed four 19 bp repeats on the t-end of Tn5090/Tn402 and two on the i-end. Each repeat may be bound by one monomer of TniA, but it is not possible at this stage to determine to what extent the interactions occur in a cooperative manner. Repeats t1 and t3 as well as the two repeats on the i-end were protected at the same concentration of TniA, which suggests, but does not prove, that these repeats are bound cooperatively. Awaiting more direct experimental support, the mentioned four repeats are perhaps the best candidates for coordinating transposase in the expected tetramerical configuration. Figure 8 shows diagramatically how TniA might multimerise upon interaction with the repeats on the paired transposon ends (not fully supported by the data presented in this work). A region immediately inside repeat t1 showed a run of strand-specific cleavages with approximate helical periodicity, followed by repeat t2, which is inverted with respect to the others. This could comprise a loop region or other distortion that protrudes from an otherwise regular shape of the complex (Figs 6 and 8). The pattern of protection obtained in higher resolution with the hydroxyl radical probe indicates that TniA binds to 10 short sequences at the t-end and five at the i-end (Fig. 6). These contact sequences occur on one face of the DNA helix, perhaps in a conformation in which it is wrapped around oligomers of the transposase. Notably, the potentially distorted region between t1 and t2 lacked one of the hydroxyl radical footprints that otherwise appeared with an approximately helical periodicity.
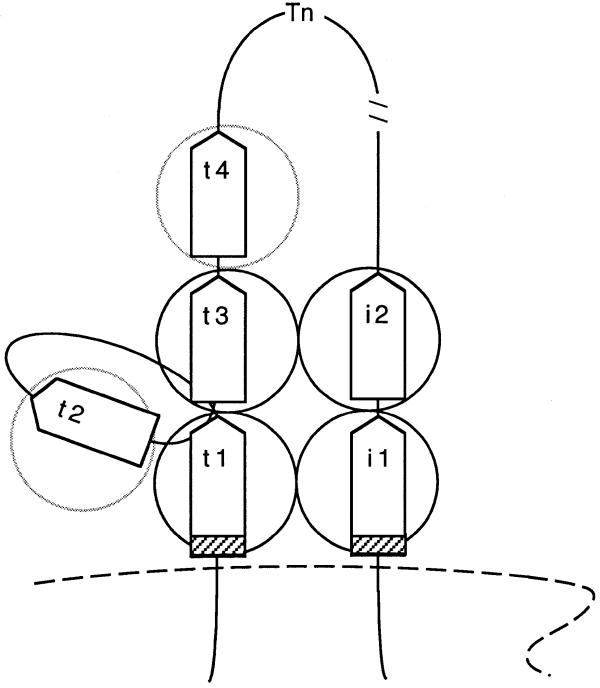
Figure 8.
A diagram suggesting possible interactions between TniA and its cognate transposon termini in a tentative, early synaptic complex. Boxes represent the repeats on the i-end or t-end and circles illustrate transposase protomers. The 8 bp spacer between the terminal nucleotides (bold line) and the first 19 bp repeats are shown as hatched boxes. The early synaptic complex is speculated to be a dynamic structure in which sites (arbitrarily selected) on the t-end might bind TniA protomers (shadowed) transiently. A broken line symbolises the target DNA.
The complex structure of several repeats might have evolved on the ends of Tn5090/Tn402 due to a need for efficient regulation. One aspect of this control is at the interactive level. The catalytic capacity of the transposase could depend on regulative contacts between protomers and between protomers and DNA. Another way to achieve control is by regulating the transcriptional activity of the genes involved in transposition. A very likely promoter of the tniABQ operon overlaps one of the repeats, t2. Furthermore, two of the contact sequences defined by hydroxyl radical footprinting overlap the –35 and –10 hexamers of the postulated promoter (Fig. 6). Repeat t4 shows least similarity with the consensus among the six TniA-binding repeats and binds only when the transposase is present in high concentration. Therefore, t4 might have a distinct function from the others.
Tn5090/Tn402 was recently reported to have a unique targeting mechanism with a strong preference for inserting into or close to binding sites of site-specific resolvases (24,28). It has been suggested that a sequence overlapping the cryptic repeats i3 and i4 plays a role in rescuing the interrupted res site by substituting for the lost portion (29).
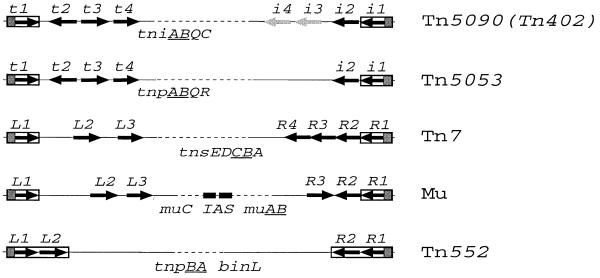
Despite some structural and functional similarities among members of the Mu-like transposon family, the sequences of the repeats are only conserved with respect to each element and the organisation of repeat clusters is largely different (Fig. 7). The most conserved feature is the length of the sequence (6–8 bp) from the ends, with a constant 5′-TG, to the beginning of the first repeat. Tn552 shows least complexity of its ends, with only two 23 bp tandem repeats on either end (20). Each end of Mu contains three repeats (7,8). The right end of Tn7 contains four repeats, whereas there are three repeats on the left end (17,19). Tn5090/Tn402 carries four repeats on each end but two repeats on the i-end need not be considered because they do not bind transposase (this report). In both Mu and Tn5090/Tn402 the repeats are not the same distances apart and include one repeat in the opposite orientation from the others (Fig. 7).
Figure 7.
The organisation of significant repeats and genes among the Mu-like transposons. Terminal inverted repeats are boxed and the 6–8 bp spacers between the termini (including the conserved 5′-terminal TG) and the first 19–23 bp repeats are marked with shadowed boxes. Each arrow represents one repeat and its orientation. The arrows showing repeats i3 and i4 that were negative for binding TniA are shadowed. The internal activator sequence of Mu (IAS) is indicated with two filled boxes. The transposition genes encoded by each element are mentioned by their designations in physical order. The gene designations of the tandem genes encoding a transposase and a nucleotide-binding protein, common for the Mu-related elements, are underlined.
The transposases of Mu, Tn5 and other DNA transposons form a structural family with the retroviral IN proteins (integrases) (30,31). Despite a very low degree of amino acid identity these proteins have a common fold and often carry the catalytic DDE motif (2–5). The transposases encoded by Tn7, Tn552, Mu and Tn5090/Tn402 are indeed more closely related to each other than to other transposases. However, the amino acid sequence identity among these four proteins is <25% (3).
Tn5090/Tn402 is the primary carrier element of class 1 integrons, recognised as containing different antibiotic resistance genes acquired by site-specific recombination (3,32). The transposon encodes, in addition to the transposase TniA, two auxiliary proteins TniB and TniQ (3) and is, furthermore, dependent on two serine recombinase systems. One of these is a part of the transposon and is used to resolve co-integrates formed by replicative transposition (22; M.Kamali-Moghaddam et al., unpublished results). The other serine recombinase system belongs to the target DNA and specifies the site where the transposon is inserted (24,28). The protein TniB is potentially a nucleotide-binding switch protein putatively equivalent to the B protein of phage Mu (33) and TnsC of Tn7 (34). TniQ, in contrast, is unique and has no known analogue or homologue except that of the closely related transposon Tn5053 (22). Future experiments will be carried out to investigate whether the target resolvase and the proteins TniB and TniQ interact with each other and with the transposase assembled on the clustered repeats on the ends to form a functional nucleoprotein complex with the target.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant (12638) to L.S. from the Swedish Medical Research Council. We thank David Morgan and Hiroshi Nakai for critically reading the manuscript and Erik Larsson, Maria Larsson, John Flensburg, Hans Widlund, Mikael Kubista and Sofia Edlund for valuable help.
References
- 1.Mizuuchi K. (1992) Tranpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- 2.Baker T.A. and Luo,L. (1994) Identification of residues in the Mu transposase essential for catalysis. Proc. Natl Acad. Sci. USA, 91, 6654–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rådström P., Sköld,O., Swedberg,G., Flensburg,J., Roy,P.H. and Sundström,L. (1994) Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu and the retroelements. J. Bacteriol ., 176, 3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fayet O., Ramond,P., Polard,P., Prere,M.F. and Chandler,M. (1990) Functional similarities beteween retroviruses and the IS3 family of bacterial insertion sequences. Mol. Microbiol., 4, 1771–1777. [DOI] [PubMed] [Google Scholar]
- 5.Rowland S.-J. and Dyke,K.G.H. (1990) Tn552, a novel transposible element from Staphylococcus aureus. Mol. Microbiol., 4, 961–965. [DOI] [PubMed] [Google Scholar]
- 6.Derbyshire K.M., Hwang,L. and Grindley,N.D. (1987) Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc. Natl Acad. Sci. USA, 84, 8049–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou A.H., Leung,P.C. and Harshey,R.M. (1991) Transposase contacts with mu DNA ends. J. Biol. Chem., 266, 20476–20482. [PubMed] [Google Scholar]
- 8.Craigie R., Mizuuchi,M. and Mizuuchi,K. (1984) Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell, 39, 387–394. [DOI] [PubMed] [Google Scholar]
- 9.Surette M.G., Buch,S.J. and Chaconas,G. (1987) Transpososomes: stable protein–DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell, 49, 253–262. [DOI] [PubMed] [Google Scholar]
- 10.Baker T.A. and Mizuuchi,K. (1992) DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev., 6, 2221–2232. [DOI] [PubMed] [Google Scholar]
- 11.Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1991) DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc. Natl Acad. Sci. USA, 15, 9031–9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavoie BD, Chan,B.S., Allison,R.G. and Chaconas,G. (1991) Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J., 10, 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams T.L., Jackson,E.L., Carritte,A. and Baker,T.A. (1999) Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev., 13, 2725–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J.Y., Jayaram,M. and Harshey,R.M. (1996) Positional information within the Mu transposase tetramer: catalytic contributions of individual monomers. Cell, 85, 447–455. [DOI] [PubMed] [Google Scholar]
- 15.Jones J.M., Welty,D.J. and Nakai,H. (1998) Versatile action of Escherichia coli ClpXP as protease or molecular chaperone for bacteriophage Mu transposition. J. Biol. Chem., 273, 459–465. [DOI] [PubMed] [Google Scholar]
- 16.Mhammedi-Alaoui A., Pato,M., Gama,M.J. and Toussaint,A. (1994) A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol. Microbiol., 11, 1109–1116. [DOI] [PubMed] [Google Scholar]
- 17.Lichtenstein C. and Brenner,S. (1982) Unique insertion site of Tn7 in the E. coli chromosome. Nature, 297, 601–603. [DOI] [PubMed] [Google Scholar]
- 18.Arciszewska L.K. and Craig,N.L. (1991) Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res., 25, 5021–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y., Lichtenstein,C. and Cotterill,S. (1991) Purification and characterisation of the TnsB protein of Tn7. Nucleic Acids Res., 19, 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowland S.-J., Sherratt,D.J., Stark,W.M. and Boocock,M. (1995) Tn552 transposase purification and in vitro activities. EMBO J., 14, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro J.A. and Sporn,P. (1977) Tn402: a new transposible element determining trimethoprim resistance that inserts in bacteriophage lambda. J. Bacteriol ., 129, 1632–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kholodii G.Y., Mindlin,S.Z., Bass,I.A., Yurieva,O.V., Minakhina,S.V. and Nikiforov,V.G. (1995) Four genes, two ends and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol., 17, 1189–1200. [DOI] [PubMed] [Google Scholar]
- 23.Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 24.Kamali-Moghaddam M. and Sundström,L. (2000) Transposon targeting determined by resolvase. FEMS Microbiol. Lett., 186, 55–59. [DOI] [PubMed] [Google Scholar]
- 25.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Dixon W.J., Hayes,J.J., Levin,J.R., Weidner,M.F., Dombroski,B.A. and Tullius,T.D. (1991) Hydroxyl radical footprintig. Methods Enzymol., 208, 380–413. [DOI] [PubMed] [Google Scholar]
- 28.Minakhina S., Kholodii,G., Mindlin,S., Yurieva,O. and Nikiforov,V. (1999) Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol., 33, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 29.Tosini F., Venanzi,S., Boschi,A. and Battaglia,P.A. (1998) The uvp1 gene on the R46 plasmid encodes a resolvase that catalyzes site-specific resolution involving the 5′-conserved segment of the adjacent integron In1. Mol. Gen. Genet., 258, 404–411. [DOI] [PubMed] [Google Scholar]
- 30.Rice P. and Mizuuchi,K. (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- 31.Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- 32.Sundström L., Rådström,P., Swedberg,G. and Sköld,O. (1988) Site-specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet., 213, 191–201. [DOI] [PubMed] [Google Scholar]
- 33.Teplow D.B., Nakayama,C., Leung,P.C. and Harshey,R.M. (1988) Structure–function relationships in the transposition protein B of bacteriophage Mu. J. Biol. Chem., 263, 10851–10857. [PubMed] [Google Scholar]
- 34.Gamas P. and Craig,N.L. (1992) Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res., 20, 2525–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]