Abstract
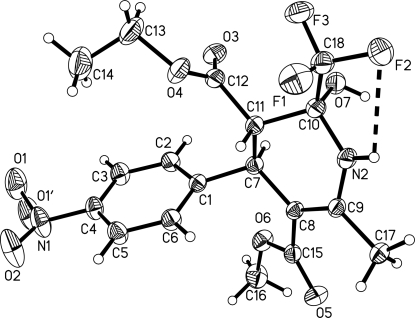
In the title compound, C18H19F3N2O7, the tetrahydropyridine ring adopts a half-chair conformation. The nitro group is disordered over two sites with occupancies of 0.780 (15) and 0.220 (15). An intramolecular N—H⋯F hydrogen bond is observed in the molecular structure. The molecules are linked into a two-dimensional network parallel to (100) by O—H⋯O, N—H⋯O and C—H⋯O hydrogen bonds.
Related literature
For related literature, see: Achiwa & Kato (1999 ▶); Dubur et al. (1989 ▶); Hermann et al. (2003 ▶); Ulrich (2004 ▶).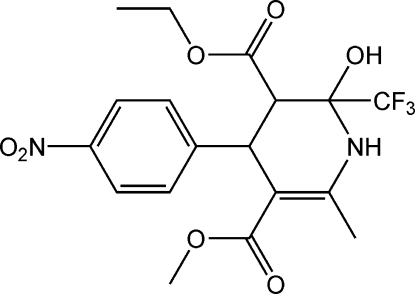
Experimental
Crystal data
C18H19F3N2O7
M r = 432.35
Monoclinic,
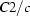
a = 28.678 (6) Å
b = 9.6678 (19) Å
c = 14.120 (3) Å
β = 95.72 (3)°
V = 3895.1 (13) Å3
Z = 8
Mo Kα radiation
μ = 0.13 mm−1
T = 293 (2) K
0.16 × 0.16 × 0.04 mm
Data collection
Rigaku Saturn diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2002 ▶) T min = 0.979, T max = 0.995
11673 measured reflections
3438 independent reflections
2572 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.150
S = 1.06
3438 reflections
284 parameters
14 restraints
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.20 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2002 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808024835/ci2642sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808024835/ci2642Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H7⋯O5i | 0.82 | 1.98 | 2.783 (3) | 166 |
| N2—H2⋯O3ii | 0.86 | 2.20 | 3.030 (3) | 163 |
| N2—H2⋯F2 | 0.86 | 2.42 | 2.735 (2) | 102 |
| C5—H5⋯O2iii | 0.93 | 2.51 | 3.432 (4) | 171 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii) 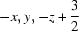 .
.
Acknowledgments
The authors thank the Natural Science Foundation of Xuzhou Normal University (grant No. 06XLB07) and the Natural Science Foundation of Xuzhou City (grant No. XJ07065) for financial support.
supplementary crystallographic information
Comment
1,4-Dihydropyridines are well known compounds as a consequence of their pharmacological profile as the most important calcium channel modulators (Achiwa & Kato, 1999). 4-Aryl-2,6-dimethyl-1,4-dihydropyridine -3,5-dicarboxylate derivatives are widely used for the treatment of cardiovascular diseases such as hypertension, angina pectoris and infarction (Dubur et al., 1989). In addition, compounds that contain fluorine have special bioactivity, for example, flumioxazin is a widely used herbicide (Hermann et al., 2003; Ulrich,2004). This led us to pay much attention to the synthesis and bioactivity of these fluoro-compounds. During the synthesis of trifluoromethylated 1,4-dihydropyridine derivatives, an intermediate, the title compound, was isolated. We report here the crystal structure of the title compound.
In the title molecule (Fig.1), the pyridine ring adopts a half-chair conformation, with atoms C10 and C11 deviating from the N2/C7/C8/C9 plane by 0.261 (4) Å and -0.544 (4)Å, respectively. The dihedral angle between the N2/C7/C8/C9 and C1-C6 planes is 72.30 (9)°. The N atom of the nitro group adopts a planar configuration in the major disorder component, while pyramidal configuration in the minor disorder component. An intramolecular N2—H2···F2 hydrogen bond is observed in the molecular structure.
The crystal packing is stabilized by O—H···O, N—H···O and C—H···O intermolecular hydrogen bonds (Table 1) which link the molecules into a two-dimensional network parallel to the (1 0 0) (Fig. 2).
Experimental
The title compound was synthesized by the reaction of 4-nitrobenzaldehyde (1 mmol), methyl 3-amino-but-2-enoate (1 mmol) and ethyl 4,4,4-trifluoro-3-oxobutanoate (1 mmol) catalyzed by triethylbenzylaminium chloride (0.02 g) in water (10 ml) at 363 K. After cooling, the reaction mixture was washed with water and recrystallized from ethanol, to obtain single crystals suitable for X-ray diffraction.
Refinement
All H atoms were placed in calculated positions (C-H = 0.93–0.98 Å, O-H = 0.82 Å and N-H = 0.86 Å) and included in the final cycles of refinement using a riding model, with Uiso(H) = 1.2Ueq(parent atom). Atom O1 is disordered over two positions with site-occupancy factors of 0.220 (15) and 0.780 (15), respectively. The Uij components of disordered atoms were restrained to an approximate isotropic behaviour. The N1—O1 and N1—O1' bond lengths were restrained to 1.22 (2) Å.
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% probability displacement ellipsoids and the atom-numbering scheme. Both disorder components are shown. The dashed line indicates a hydrogen bond.
Fig. 2.
The packing diagram of the title compound. Intermolecular hydrogen bonds are shown as dashed lines and H atoms not involved in hydrogen bonding have been omitted for clarity.
Crystal data
| C18H19F3N2O7 | F000 = 1792 |
| Mr = 432.35 | Dx = 1.475 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 3562 reflections |
| a = 28.678 (6) Å | θ = 2.2–25.0º |
| b = 9.6678 (19) Å | µ = 0.13 mm−1 |
| c = 14.120 (3) Å | T = 293 (2) K |
| β = 95.72 (3)º | Prism, colourless |
| V = 3895.1 (13) Å3 | 0.16 × 0.16 × 0.04 mm |
| Z = 8 |
Data collection
| Rigaku Saturn diffractometer | 3438 independent reflections |
| Radiation source: rotating anode | 2572 reflections with I > 2σ(I) |
| Monochromator: confocal | Rint = 0.043 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 25.0º |
| T = 293(2) K | θmin = 2.2º |
| ω scans | h = −33→34 |
| Absorption correction: multi-scan(CrystalClear; Rigaku/MSC, 2002) | k = −11→11 |
| Tmin = 0.979, Tmax = 0.995 | l = −16→12 |
| 11673 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.052 | H-atom parameters constrained |
| wR(F2) = 0.150 | w = 1/[σ2(Fo2) + (0.0873P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 3438 reflections | Δρmax = 0.22 e Å−3 |
| 284 parameters | Δρmin = −0.20 e Å−3 |
| 14 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| F1 | 0.08689 (6) | 0.58977 (17) | 0.93658 (13) | 0.0775 (5) | |
| F2 | 0.15032 (7) | 0.70065 (15) | 0.97856 (12) | 0.0800 (6) | |
| F3 | 0.12464 (6) | 0.68534 (15) | 0.83137 (11) | 0.0725 (5) | |
| O1 | 0.0452 (7) | −0.100 (2) | 0.5112 (8) | 0.086 (6) | 0.220 (15) |
| O1' | 0.06713 (19) | −0.1837 (7) | 0.5322 (3) | 0.090 (2) | 0.780 (15) |
| O2 | 0.01478 (8) | −0.1793 (3) | 0.63009 (19) | 0.0972 (9) | |
| O3 | 0.13847 (6) | 0.44788 (18) | 0.67834 (12) | 0.0545 (5) | |
| O4 | 0.06795 (6) | 0.44145 (19) | 0.73469 (12) | 0.0594 (5) | |
| O5 | 0.21673 (6) | 0.00503 (18) | 1.01751 (13) | 0.0546 (5) | |
| O6 | 0.21490 (6) | 0.01870 (17) | 0.85947 (12) | 0.0512 (5) | |
| O7 | 0.19972 (6) | 0.52220 (18) | 0.86560 (12) | 0.0512 (5) | |
| H7 | 0.2218 | 0.5189 | 0.9071 | 0.077* | |
| N1 | 0.05038 (9) | −0.1349 (3) | 0.6024 (2) | 0.0738 (8) | |
| N2 | 0.16656 (7) | 0.42268 (18) | 0.99703 (13) | 0.0432 (5) | |
| H2 | 0.1630 | 0.4725 | 1.0463 | 0.052* | |
| C1 | 0.13495 (7) | 0.1409 (2) | 0.77500 (16) | 0.0353 (5) | |
| C2 | 0.14347 (8) | 0.1173 (2) | 0.68116 (16) | 0.0424 (6) | |
| H2A | 0.1683 | 0.1621 | 0.6565 | 0.051* | |
| C3 | 0.11549 (9) | 0.0281 (2) | 0.62409 (18) | 0.0485 (6) | |
| H3 | 0.1210 | 0.0132 | 0.5611 | 0.058* | |
| C4 | 0.07957 (8) | −0.0378 (2) | 0.66212 (18) | 0.0469 (6) | |
| C5 | 0.07026 (9) | −0.0190 (3) | 0.75489 (19) | 0.0502 (6) | |
| H5 | 0.0458 | −0.0659 | 0.7793 | 0.060* | |
| C6 | 0.09829 (8) | 0.0715 (2) | 0.81067 (17) | 0.0446 (6) | |
| H6 | 0.0924 | 0.0860 | 0.8735 | 0.053* | |
| C7 | 0.16340 (7) | 0.2485 (2) | 0.83367 (15) | 0.0365 (5) | |
| H7A | 0.1902 | 0.2747 | 0.7996 | 0.044* | |
| C8 | 0.18168 (7) | 0.2018 (2) | 0.93315 (15) | 0.0354 (5) | |
| C9 | 0.18004 (7) | 0.2873 (2) | 1.00940 (15) | 0.0368 (5) | |
| C10 | 0.15830 (8) | 0.4834 (2) | 0.90448 (17) | 0.0427 (6) | |
| C11 | 0.13175 (8) | 0.3763 (2) | 0.83985 (16) | 0.0373 (5) | |
| H11 | 0.1044 | 0.3473 | 0.8711 | 0.045* | |
| C12 | 0.11414 (9) | 0.4275 (2) | 0.74130 (17) | 0.0418 (6) | |
| C13 | 0.04231 (12) | 0.4735 (4) | 0.6429 (2) | 0.0755 (9) | |
| H13A | 0.0637 | 0.5090 | 0.5997 | 0.091* | |
| H13B | 0.0189 | 0.5439 | 0.6509 | 0.091* | |
| C14 | 0.01957 (13) | 0.3484 (4) | 0.6032 (2) | 0.0918 (11) | |
| H14A | 0.0430 | 0.2830 | 0.5884 | 0.138* | |
| H14B | −0.0001 | 0.3712 | 0.5462 | 0.138* | |
| H14C | 0.0008 | 0.3086 | 0.6488 | 0.138* | |
| C15 | 0.20522 (8) | 0.0677 (2) | 0.94376 (17) | 0.0396 (5) | |
| C16 | 0.23732 (10) | −0.1148 (3) | 0.8569 (2) | 0.0651 (8) | |
| H16A | 0.2607 | −0.1231 | 0.9101 | 0.098* | |
| H16B | 0.2518 | −0.1237 | 0.7987 | 0.098* | |
| H16C | 0.2143 | −0.1864 | 0.8601 | 0.098* | |
| C17 | 0.19319 (8) | 0.2520 (2) | 1.11186 (15) | 0.0460 (6) | |
| H17A | 0.1907 | 0.3331 | 1.1502 | 0.069* | |
| H17B | 0.2249 | 0.2184 | 1.1198 | 0.069* | |
| H17C | 0.1725 | 0.1818 | 1.1313 | 0.069* | |
| C18 | 0.12955 (10) | 0.6156 (2) | 0.91287 (19) | 0.0543 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0761 (12) | 0.0770 (11) | 0.0819 (13) | 0.0222 (9) | 0.0199 (10) | −0.0030 (9) |
| F2 | 0.1242 (15) | 0.0509 (9) | 0.0612 (11) | 0.0076 (9) | −0.0094 (10) | −0.0176 (8) |
| F3 | 0.1081 (13) | 0.0524 (9) | 0.0544 (10) | 0.0161 (8) | −0.0046 (9) | 0.0094 (7) |
| O1 | 0.085 (8) | 0.118 (10) | 0.055 (7) | −0.032 (7) | 0.008 (5) | −0.026 (6) |
| O1' | 0.093 (3) | 0.111 (4) | 0.069 (2) | −0.038 (3) | 0.023 (2) | −0.052 (2) |
| O2 | 0.0728 (14) | 0.1166 (18) | 0.1066 (19) | −0.0437 (14) | 0.0303 (14) | −0.0568 (16) |
| O3 | 0.0674 (11) | 0.0622 (11) | 0.0344 (10) | −0.0019 (8) | 0.0074 (9) | 0.0101 (8) |
| O4 | 0.0528 (11) | 0.0799 (12) | 0.0425 (11) | 0.0022 (9) | −0.0094 (9) | 0.0098 (9) |
| O5 | 0.0621 (11) | 0.0555 (10) | 0.0458 (11) | 0.0078 (8) | 0.0028 (9) | 0.0114 (9) |
| O6 | 0.0595 (11) | 0.0521 (10) | 0.0420 (10) | 0.0109 (8) | 0.0042 (9) | −0.0055 (8) |
| O7 | 0.0536 (10) | 0.0592 (10) | 0.0403 (10) | −0.0162 (9) | 0.0018 (8) | 0.0045 (8) |
| N1 | 0.0681 (16) | 0.0885 (18) | 0.0668 (18) | −0.0311 (14) | 0.0167 (14) | −0.0349 (15) |
| N2 | 0.0620 (13) | 0.0418 (10) | 0.0254 (10) | 0.0000 (9) | 0.0027 (9) | −0.0041 (8) |
| C1 | 0.0372 (12) | 0.0379 (11) | 0.0307 (12) | 0.0012 (9) | 0.0025 (10) | −0.0011 (9) |
| C2 | 0.0468 (13) | 0.0470 (13) | 0.0348 (13) | −0.0064 (11) | 0.0109 (11) | −0.0042 (11) |
| C3 | 0.0543 (15) | 0.0555 (14) | 0.0368 (14) | −0.0044 (12) | 0.0099 (12) | −0.0133 (11) |
| C4 | 0.0476 (14) | 0.0504 (13) | 0.0425 (14) | −0.0070 (11) | 0.0032 (12) | −0.0148 (11) |
| C5 | 0.0478 (14) | 0.0553 (14) | 0.0487 (15) | −0.0135 (12) | 0.0103 (12) | −0.0045 (12) |
| C6 | 0.0506 (14) | 0.0524 (13) | 0.0318 (13) | −0.0075 (11) | 0.0097 (11) | −0.0045 (11) |
| C7 | 0.0392 (12) | 0.0414 (11) | 0.0291 (11) | −0.0038 (9) | 0.0047 (10) | 0.0004 (10) |
| C8 | 0.0356 (11) | 0.0421 (11) | 0.0286 (12) | −0.0036 (9) | 0.0028 (9) | 0.0022 (9) |
| C9 | 0.0360 (11) | 0.0447 (12) | 0.0294 (12) | −0.0058 (10) | 0.0008 (9) | 0.0023 (10) |
| C10 | 0.0536 (14) | 0.0420 (12) | 0.0320 (13) | −0.0026 (10) | 0.0022 (11) | 0.0012 (10) |
| C11 | 0.0420 (12) | 0.0408 (11) | 0.0291 (12) | −0.0032 (10) | 0.0030 (10) | 0.0001 (9) |
| C12 | 0.0521 (14) | 0.0390 (12) | 0.0338 (13) | −0.0027 (10) | 0.0016 (12) | 0.0007 (10) |
| C13 | 0.076 (2) | 0.095 (2) | 0.0499 (18) | 0.0053 (18) | −0.0195 (16) | 0.0128 (17) |
| C14 | 0.091 (2) | 0.114 (3) | 0.065 (2) | −0.019 (2) | −0.0149 (19) | −0.008 (2) |
| C15 | 0.0373 (12) | 0.0449 (12) | 0.0363 (13) | −0.0063 (10) | 0.0015 (10) | −0.0001 (11) |
| C16 | 0.0681 (18) | 0.0536 (15) | 0.072 (2) | 0.0125 (13) | 0.0009 (16) | −0.0161 (14) |
| C17 | 0.0544 (14) | 0.0520 (13) | 0.0309 (13) | −0.0051 (11) | 0.0000 (11) | 0.0020 (11) |
| C18 | 0.0761 (19) | 0.0455 (14) | 0.0403 (15) | 0.0047 (13) | 0.0008 (14) | −0.0029 (12) |
Geometric parameters (Å, °)
| F1—C18 | 1.324 (3) | C5—C6 | 1.380 (3) |
| F2—C18 | 1.335 (3) | C5—H5 | 0.93 |
| F3—C18 | 1.329 (3) | C6—H6 | 0.93 |
| O1—N1 | 1.325 (13) | C7—C8 | 1.518 (3) |
| O1'—N1 | 1.237 (4) | C7—C11 | 1.541 (3) |
| O2—N1 | 1.208 (3) | C7—H7A | 0.98 |
| O3—C12 | 1.200 (3) | C8—C9 | 1.362 (3) |
| O4—C12 | 1.326 (3) | C8—C15 | 1.462 (3) |
| O4—C13 | 1.458 (3) | C9—C17 | 1.498 (3) |
| O5—C15 | 1.221 (3) | C10—C18 | 1.532 (3) |
| O6—C15 | 1.336 (3) | C10—C11 | 1.532 (3) |
| O6—C16 | 1.444 (3) | C11—C12 | 1.515 (3) |
| O7—C10 | 1.408 (3) | C11—H11 | 0.98 |
| O7—H7 | 0.82 | C13—C14 | 1.459 (5) |
| N1—C4 | 1.467 (3) | C13—H13A | 0.97 |
| N2—C9 | 1.371 (3) | C13—H13B | 0.97 |
| N2—C10 | 1.431 (3) | C14—H14A | 0.96 |
| N2—H2 | 0.86 | C14—H14B | 0.96 |
| C1—C6 | 1.384 (3) | C14—H14C | 0.96 |
| C1—C2 | 1.390 (3) | C16—H16A | 0.96 |
| C1—C7 | 1.516 (3) | C16—H16B | 0.96 |
| C2—C3 | 1.381 (3) | C16—H16C | 0.96 |
| C2—H2A | 0.93 | C17—H17A | 0.96 |
| C3—C4 | 1.366 (3) | C17—H17B | 0.96 |
| C3—H3 | 0.93 | C17—H17C | 0.96 |
| C4—C5 | 1.375 (4) | ||
| C12—O4—C13 | 119.7 (2) | N2—C10—C11 | 107.09 (18) |
| C15—O6—C16 | 118.2 (2) | C18—C10—C11 | 111.8 (2) |
| C10—O7—H7 | 109.5 | C12—C11—C10 | 115.27 (18) |
| O2—N1—O1' | 122.1 (3) | C12—C11—C7 | 110.74 (18) |
| O1'—N1—C4 | 117.4 (2) | C10—C11—C7 | 108.39 (18) |
| O2—N1—C4 | 119.6 (2) | C12—C11—H11 | 107.4 |
| O2—N1—O1 | 112.9 (7) | C10—C11—H11 | 107.4 |
| O1—N1—C4 | 113.5 (6) | C7—C11—H11 | 107.4 |
| C9—N2—C10 | 121.81 (19) | O3—C12—O4 | 125.6 (2) |
| C9—N2—H2 | 119.1 | O3—C12—C11 | 124.7 (2) |
| C10—N2—H2 | 119.1 | O4—C12—C11 | 109.7 (2) |
| C6—C1—C2 | 118.6 (2) | O4—C13—C14 | 109.4 (3) |
| C6—C1—C7 | 121.39 (19) | O4—C13—H13A | 109.8 |
| C2—C1—C7 | 119.82 (19) | C14—C13—H13A | 109.8 |
| C3—C2—C1 | 120.8 (2) | O4—C13—H13B | 109.8 |
| C3—C2—H2A | 119.6 | C14—C13—H13B | 109.8 |
| C1—C2—H2A | 119.6 | H13A—C13—H13B | 108.2 |
| C4—C3—C2 | 118.6 (2) | C13—C14—H14A | 109.5 |
| C4—C3—H3 | 120.7 | C13—C14—H14B | 109.5 |
| C2—C3—H3 | 120.7 | H14A—C14—H14B | 109.5 |
| C3—C4—C5 | 122.6 (2) | C13—C14—H14C | 109.5 |
| C3—C4—N1 | 118.8 (2) | H14A—C14—H14C | 109.5 |
| C5—C4—N1 | 118.6 (2) | H14B—C14—H14C | 109.5 |
| C4—C5—C6 | 118.0 (2) | O5—C15—O6 | 121.4 (2) |
| C4—C5—H5 | 121.0 | O5—C15—C8 | 127.6 (2) |
| C6—C5—H5 | 121.0 | O6—C15—C8 | 111.0 (2) |
| C5—C6—C1 | 121.3 (2) | O6—C16—H16A | 109.5 |
| C5—C6—H6 | 119.3 | O6—C16—H16B | 109.5 |
| C1—C6—H6 | 119.3 | H16A—C16—H16B | 109.5 |
| C1—C7—C8 | 114.77 (17) | O6—C16—H16C | 109.5 |
| C1—C7—C11 | 107.06 (17) | H16A—C16—H16C | 109.5 |
| C8—C7—C11 | 109.70 (17) | H16B—C16—H16C | 109.5 |
| C1—C7—H7A | 108.4 | C9—C17—H17A | 109.5 |
| C8—C7—H7A | 108.4 | C9—C17—H17B | 109.5 |
| C11—C7—H7A | 108.4 | H17A—C17—H17B | 109.5 |
| C9—C8—C15 | 120.6 (2) | C9—C17—H17C | 109.5 |
| C9—C8—C7 | 121.00 (19) | H17A—C17—H17C | 109.5 |
| C15—C8—C7 | 118.15 (19) | H17B—C17—H17C | 109.5 |
| C8—C9—N2 | 120.7 (2) | F1—C18—F3 | 107.1 (2) |
| C8—C9—C17 | 126.9 (2) | F1—C18—F2 | 107.5 (2) |
| N2—C9—C17 | 112.41 (19) | F3—C18—F2 | 106.9 (2) |
| O7—C10—N2 | 113.3 (2) | F1—C18—C10 | 112.3 (2) |
| O7—C10—C18 | 106.83 (19) | F3—C18—C10 | 111.6 (2) |
| N2—C10—C18 | 108.25 (19) | F2—C18—C10 | 111.2 (2) |
| O7—C10—C11 | 109.64 (18) | ||
| C6—C1—C2—C3 | 1.0 (3) | O7—C10—C11—C12 | −64.1 (3) |
| C7—C1—C2—C3 | −174.7 (2) | N2—C10—C11—C12 | 172.51 (19) |
| C1—C2—C3—C4 | −0.7 (4) | C18—C10—C11—C12 | 54.1 (3) |
| C2—C3—C4—C5 | −0.2 (4) | O7—C10—C11—C7 | 60.6 (2) |
| C2—C3—C4—N1 | −178.7 (2) | N2—C10—C11—C7 | −62.8 (2) |
| O2—N1—C4—C3 | −169.9 (3) | C18—C10—C11—C7 | 178.84 (19) |
| O1'—N1—C4—C3 | 20.9 (6) | C1—C7—C11—C12 | −56.5 (2) |
| O1—N1—C4—C3 | −32.6 (13) | C8—C7—C11—C12 | 178.39 (18) |
| O2—N1—C4—C5 | 11.5 (4) | C1—C7—C11—C10 | 176.15 (17) |
| O1'—N1—C4—C5 | −157.7 (5) | C8—C7—C11—C10 | 51.0 (2) |
| O1—N1—C4—C5 | 148.8 (12) | C13—O4—C12—O3 | 5.3 (4) |
| C3—C4—C5—C6 | 0.7 (4) | C13—O4—C12—C11 | −172.6 (2) |
| N1—C4—C5—C6 | 179.3 (2) | C10—C11—C12—O3 | 74.1 (3) |
| C4—C5—C6—C1 | −0.4 (4) | C7—C11—C12—O3 | −49.4 (3) |
| C2—C1—C6—C5 | −0.4 (3) | C10—C11—C12—O4 | −108.0 (2) |
| C7—C1—C6—C5 | 175.2 (2) | C7—C11—C12—O4 | 128.5 (2) |
| C6—C1—C7—C8 | 50.4 (3) | C12—O4—C13—C14 | 102.9 (3) |
| C2—C1—C7—C8 | −134.0 (2) | C16—O6—C15—O5 | 3.2 (3) |
| C6—C1—C7—C11 | −71.6 (3) | C16—O6—C15—C8 | −178.7 (2) |
| C2—C1—C7—C11 | 104.0 (2) | C9—C8—C15—O5 | 16.0 (3) |
| C1—C7—C8—C9 | −137.4 (2) | C7—C8—C15—O5 | −169.7 (2) |
| C11—C7—C8—C9 | −16.9 (3) | C9—C8—C15—O6 | −161.96 (19) |
| C1—C7—C8—C15 | 48.4 (2) | C7—C8—C15—O6 | 12.3 (3) |
| C11—C7—C8—C15 | 168.92 (18) | O7—C10—C18—F1 | 171.4 (2) |
| C15—C8—C9—N2 | 166.91 (19) | N2—C10—C18—F1 | −66.2 (3) |
| C7—C8—C9—N2 | −7.1 (3) | C11—C10—C18—F1 | 51.5 (3) |
| C15—C8—C9—C17 | −11.1 (3) | O7—C10—C18—F3 | 51.2 (3) |
| C7—C8—C9—C17 | 174.88 (19) | N2—C10—C18—F3 | 173.5 (2) |
| C10—N2—C9—C8 | −6.4 (3) | C11—C10—C18—F3 | −68.8 (3) |
| C10—N2—C9—C17 | 171.9 (2) | O7—C10—C18—F2 | −68.1 (3) |
| C9—N2—C10—O7 | −79.4 (3) | N2—C10—C18—F2 | 54.3 (3) |
| C9—N2—C10—C18 | 162.2 (2) | C11—C10—C18—F2 | 172.0 (2) |
| C9—N2—C10—C11 | 41.6 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7···O5i | 0.82 | 1.98 | 2.783 (3) | 166 |
| N2—H2···O3ii | 0.86 | 2.20 | 3.030 (3) | 163 |
| N2—H2···F2 | 0.86 | 2.42 | 2.735 (2) | 102 |
| C5—H5···O2iii | 0.93 | 2.51 | 3.432 (4) | 171 |
Symmetry codes: (i) −x+1/2, −y+1/2, −z+2; (ii) x, −y+1, z+1/2; (iii) −x, y, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI2642).
References
- Achiwa, K. & Kato, T. (1999). Curr. Org. Chem.3, 77–106.
- Dubur, G. J., Veveris, M. M., Weinheimer, G., Bisenieks, E. A., Makarova, N. R., Kimenis, A. A., Uldrikis, J. R., Lukevics, E. J., Dooley, D. & Osswald, H. (1989). Arzneim. Forsch.39, 1185–1189. [PubMed]
- Hermann, B., Erwin, H. & Hansjorg, K. (2003). US patent 2 003 176 284.
- Rigaku/MSC (2002). CrystalClear Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Ulrich, H. (2004). US patent 2 004 033 897.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808024835/ci2642sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808024835/ci2642Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report