Abstract
Whether the hepatitis delta virus (HDV) DNA vaccine can induce anti-HDV antibodies has been debatable. The role of the isoprenylated motif of hepatitis delta antigens (HDAg) in the generation of immune responses following DNA-based immunization has never been studied. Plasmids p2577L, encoding large HDAg (L-HDAg), p2577S, expressing small HDAg (S-HDAg), and p25L-211S, encoding a mutant form of L-HDAg with a cysteine-to-serine mutation at codon 211, were constructed in this study. Mice were intramuscularly injected with the plasmids. The anti-HDV antibody titers, T-cell proliferation responses, T-helper responses, and HDV-specific, gamma interferon (IFN-γ)-producing CD8+ T cells were analyzed. Animals immunized with p2577S showed a strong anti-HDV antibody response. Conversely, only a low titer of anti-HDV antibodies was detected in mice immunized with p2577L. Epitope mapping revealed that the anti-HDV antibodies generated by p2577L vaccination hardly reacted with epitope amino acids 174 to 194, located at the C terminus of S-HDAg. All of the HDAg-encoding plasmids could induce significant T-cell proliferation responses and generate Th1 responses and HDV-specific, IFN-γ-producing CD8+ T cells. In conclusion, HDAg-specific antibodies definitely exist following DNA vaccination. The magnitudes of the humoral immune responses generated by L-HDAg- and S-HDAg-encoding DNA vaccines are different. The isoprenylated motif can mask epitope amino acids 174 to 195 of HDAg but does not interfere with cellular immunity following DNA-based immunization. These findings are important for the choice of a candidate HDV DNA vaccine in the future.
Hepatitis delta virus (HDV) is a defective single-stranded RNA virus. The assembly and transmission of HDV require a supply of hepatitis B surface antigen (HBsAg) from hepatitis B virus (HBV) (21, 25). HDV superinfection can lead to fulminant hepatic failure and also has a high probability of progressing to chronic hepatitis or cirrhosis (8, 22, 24, 27, 28). Furthermore, HDV superinfection can increase the risk of hepatocellular carcinoma and mortality in patients with HBV-related cirrhosis (6). Although the present HBV vaccine is very effective, about 350 million individuals are already chronically infected by HBV worldwide (4). At present, interferon is the only licensed drug for treating chronic hepatitis D, but the relapse rate is high after discontinuation (5). The development of a prophylactic or therapeutic HDV vaccine has a potential use for HBV carriers who are at risk of HDV superinfection and for those who have been infected by HDV already. DNA vaccination is a promising method for preventing and treating persistent viral infections. Previous results suggested that DNA vaccines can produce Th1 immune responses against HDV (11).
HDV has two forms of viral proteins, large and small hepatitis D antigens (HDAg). The mRNA encoding large HDAg (L-HDAg) contains a UGG tryptophan codon at the site of the UAG amber termination codon of small HDAg (S-HDAg) because of an RNA editing event (1, 2, 3). Therefore, L-HDAg contains an additional 19 amino acids at the C terminus. L-HDAg can be isoprenylated at a unique cysteine located 4 amino acids from the C terminus (7). Mutation of this unique cysteine of L-HDAg to serine can block isoprenylation and HDV assembly (7). Evidence has shown that these additional 19 amino acids of L-HDAg can alter the overall conformation and hydrophobicity of HDAg (12, 13, 15, 18). S-HDAg also contains a unique conformation at the C terminus. This conformation is detectable with a monoclonal antibody (9E4) which is specific for S-HDAg and which does not react with L-HDAg. When isoprenylation is inhibited, this epitope become exposed in L-HDAg (12, 13). Based on this evidence, host immune responses may be different when immunization is carried out with endogenous L-HDAg versus S-HDAg.
A previous study demonstrated that an L-HDAg-encoding DNA vaccine could produce low titers of anti-HDV antibodies (11). However, in a subsequent study with the HDV DNA vaccine, no HDAg-specific antibody titers were detectable by a commercial enzyme-linked immunosorbent assay (ELISA) or by a Western blot assay (17). This discrepancy needs further study for clarification.
The immunogenic domain of HDV recognized by chronically HDV-infected patients includes amino acids 2 to 7, 63 to 74, 86 to 91, 94 to 100, 159 to 172, 174 to 195, and 197 to 207 (23). It also has been suggested that cytotoxic-T-cell epitopes of HDV may be located at the carboxyl end (amino acids 77 to 195) of S-HDAg (14). In a longitudinal analysis of the HDV genome at different time points during chronic HDV infection, the emergence of amino acid changes at the carboxyl end of S-HDAg (amino acids 170 to 195) usually occurred after a severe hepatitis attack (29). Thus, the C terminus of HDAg may contain important B- or T-cell epitopes. In this study, we confirmed that HDAg-specific antibodies certainly are inducible by HDV DNA vaccination. The isoprenylated motif can mask epitope amino acids 174 to 195 of HDAg but does not interfere with cellular immunity following DNA-based immunization.
MATERIALS AND METHODS
Construction of expression vectors.
The L-HDAg gene was amplified by PCR with pairs of primers (δx25 [5′-GGCTCTAGAGTAAGAGTACTGAGG-3′] and δEcoRV [5′-ATGATATCCCGACCCGAAGAG-3′]) from plasmid TW2577-1L (GenBankaccession no.AF540888), which contained the HDV coding region (genotype I) in vector PCRII, and then cloned into the XbaI/EcoRV sites in plasmid pcDNA3.1(−) (Invitrogen, San Diego, Calif.) to produce plasmid p2577L. The S-HDAg gene was amplified by PCR with pairs of primers (δx25 and δHindIII [5′-ATAAGCTTCCGACCCGAAGAG-3′]) from plasmid TW2577-4S (GenBank accession no. AF530090) and then cloned into the XbaI/HindIII sites in plasmid pcDNA3.1(−) to produce plasmid p2577S. Both plasmid TWD2577-1L and plasmid TW2577-4S were cloned from the same chronically HDV-infected patient. Plasmid p25L-211S, encoding L-HDAg sequences with a cysteine-to-serine mutation at codon 211, was amplified by PCR and cloned into pCMV-EBNA as previously described (7). Plasmid pcDNA3.1(−) was used as a negative control. Plasmid DNA was purified from transformed Escherichia coli DH5α (Gibco BRL, Life Technologies, Gaithersburg, Md.) by using a Qiagen Giga plasmid purification kit (Taigen Bioscience Corporation).
In vitro studies.
Huh-7 cells were transfected by the calcium phosphate-DNA coprecipitation method as reported previously (10, 19, 26). To detect HDAgs by Western blotting, Huh-7 cell lysates were harvested at 48 h after transfection. Proteins obtained by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) separation and blotting onto nitrocellulose membranes were stained for HDAgs with anti-HDV antibody-positive human serum. The antigen-bound antibodies on the membranes were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies. Finally, the color was developed with a Western blotting chemiluminescence reagent (MEN Life Science, Boston, Mass.).
Purification of L-HDAg and S-HDAg.
Recombinant HDAg fusion proteins were purified as previously described (9, 11, 16). Recombinant L-HDAg and S-HDAg fusion proteins were both made with E. coli maltose-binding protein (MBP).
Generation of an HDAg-expressing cell line.
The P815 mastocytoma cell line congenic for BALB/c mice (H-2d) was used to generate an HDAg-expressing cell line. P815 transfectants were established after transfection of P815 cells with plasmid p2577L by electroporation (BTX 830 apparatus; 350 V, 99 μs, pulse number 5). A stably expressed HDAg clone (P815/2577L) was selected by adding G418 and screened by Western blotting as described above.
Immunization of mice.
Female BALB/c mice were obtained from the National Laboratory Animal Breeding and Research Center, Taipei, Taiwan. Mice were housed at the Laboratory Animal Facility, Taipei Veterans General Hospital. Mice were immunized at 6 to 8 weeks of age. Cardiotoxin (Sigma) was given to each mouse 1 week before immunization (11). Groups of mice were anesthetized and given intramuscular (bilateral quadriceps) injections of a total dose of 100 μg of plasmid DNA dissolved in 100 μl of sterilized normal saline. Mice were immunized as follows: group 1, 100 μg of p2577L; group 2, 100 μg of p2577S; group 3, 100 μg of p25L-211S; and group 4, 100 μg of pcDNA3.1(−). Each mouse was given booster doses at 3 and 6 weeks after the first immunization. All of the experiments in this study were repeated at least twice for validation.
ELISA of antibodies.
Serum samples from groups of 10 mice were analyzed for the presence of HDAg-specific antibodies as previously described (11). The absorbance at 490 nm was measured with an ELISA reader. The results were considered significant when the optical density (OD) of the tested sera was higher than the mean OD and 3 standard deviations of the control sera.
Confirmation of anti-HDV antibodies by Western blotting.
We used Western blotting to identify whether the antibodies detected by the ELISA were HDAg specific. Huh-7 cells were transfected with p2577L and p2577S by the calcium phosphate-DNA coprecipitation method as reported previously (10, 19, 26). Huh-7 cell lysates were harvested at 48 h after transfection. Proteins obtained by SDS-PAGE separation and blotting onto nitrocellulose membranes were stained for HDAgs with mouse serum samples. The antigen-bound antibodies on the membranes were detected with HRP-conjugated goat anti-mouse immunoglobulin G (Sigma). Finally, the color was developed with the Western blotting chemiluminescence reagent described above.
Synthesis of HDV peptides and anti-HDV antibody epitope mapping.
HDV peptides were synthesized commercially by Sigma-Genosys (Woodlands, Tex.). The 11 HDV peptides represented amino acids 2 to 18, 15 to 42, 39 to 61, 57 to 81, 77 to 100, 96 to 122, 118 to 144, 140 to 159, 155 to 182, 174 to 195, and 196 to 214 of genotype I HDAg and had the same sequences as the plasmids used in this study. Serum samples with anti-HDV antibody titers equal to or greater than 400:1 at week 9 were further analyzed for epitope mapping. Microtiter plates were coated with 100 μl (10 μg/ml) of 17- to 27-mer overlapping peptides. After blocking, 100-μl samples of 1:100 dilutions of tested sera in triplicate were added to wells. Bound proteins were detected with HRP-conjugated goat anti-mouse immunoglobulin G. Color was generated by adding 0.1 M citric acid (pH 5) and phenylenediamine (Sigma). The absorbance at 490 nm was measured with an ELISA reader.
Lymphocyte proliferation assay.
To determine the HDAg-specific lymphoproliferative response, groups of three mice were immunized with the same doses and schedules as those mentioned above. On day 7 after the last immunization, immune splenocytes were collected for a proliferation assay. T-cell-enriched splenocytes were prepared by collecting cells from a nylon wool column. For the lymphocyte proliferation assay, 100 μl of 2 × 106 splenocytes per ml was added to each well of a 96-well U-bottom plate. Stimulated wells received purified recombinant L-HDAg (MBP-DL2577), S-HDAg (MBP-DS2577), and MBP at 10 μg/ml; transferrin (120 μg/ml; Sigma) served as a negative control antigen, and concanavalin A (5 μg/ml; Pierce, Rockford, Ill.) served as a positive mitogenic control. Control wells received cells only. After 3 days in culture, the cells were pulsed with [3H]thymidine (1 μCi/well) for 16 h and harvested with Filter-Mate (Packard); incorporated radioactivity was determined by using Top-Count (Packard). The stimulation index (SI) was calculated as the mean counts per minute in the stimulated wells divided by the mean counts per minute in the control wells. An SI of greater than 2 was defined as significant (11).
ELISPOT assay for IFN-γ.
DNA vaccines can produce Th1 immune responses against HDV (11). To determine the number of HDAg-specific, gamma interferon (IFN-γ)-producing T-helper cells, a mouse IFN-γ ELISPOT assay kit (R&D Systems, Minneapolis, Minn.) was used in accordance with the manufacturer's protocol. In summary, splenocytes were stimulated with L-HDAg, S-HDAg, and control protein for 3 days. A total of 2 × 105 splenocytes in 100 μl of medium were pipetted into wells and incubated at 37°C for 6 h. Biotinylated polyclonal antibody specific for mouse IFN-γ was added, followed by alkaline phosphatase conjugated to streptavidin. 5-Bromo-4-chloro-3-indolylphosphate p-toluidine salt-nitroblue tetrazolium chloride was used as a substrate. The images of spots were captured with a dissection microscope, and then counts were determined with ImageMaster TotalLab version 1.10 software (Amersham Pharmacia Biotech). The number of specific spot-forming cells (SFC) was determined as the mean number of spots in the presence of antigen minus the mean number of spots in wells containing medium only.
Intracellular IFN-γ staining and fluorescein-activated cell sorting analysis.
To determine the number of HDAg-specific, IFN-γ-producing CD8+ T cells, direct intracellular IFN-γ and cellular surface marker staining of immunized splenocytes was carried out. In summary, at 14 days after the last immunization, immune splenocytes from groups of three mice were incubated in 24-well culture plates (5 × 106 splenocytes per well) in the presence of irradiated P815/2577L cells (10,000 rads, 105 cells/well) at 37°C for 16 h. GolgiStop (Pharmingen, San Diego, Calif.) was added to the culture medium, and the mixture was incubated for a further 4 h at 37°C. The cells were harvested and incubated with rat anti-mouse CD16/CD32 monoclonal antibody (clone 2.4G2; Pharmingen) for 15 min on ice to block nonspecific binding to the Fc receptor. The cells were surface stained with fluorescein isothiocyanate-conjugated rat anti-mouse CD8a monoclonal antibody (clone 53-6.7; Pharmingen) for 30 min on ice. After being washed to remove unbound antibodies, the cells were fixed with Cytofix/Cytoperm solution (Pharmingen) for 20 min at 4°C. Finally, the cells were stained with R-phycoerythrin-conjugated rat anti-mouse IFN-γ monoclonal antibody (clone XMG1.2; Pharmingen). Samples were acquired on a FACScan flow cytometer, and the data were analyzed with CELLQuest software (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
Statistical analysis.
Fisher's exact test was used when appropriate in this study. To compare the results among the three groups, a Kruskal-Wallis one-way anaylsis of variance was used. When the P value was <0.05, a multiple-comparison test with the Dunnett method was used to compare the two groups (11). A P value of <0.05 was considered significant for all tests.
RESULTS
Viral protein expression in vitro.
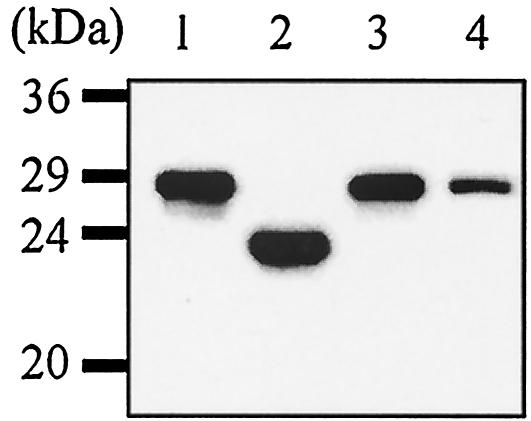
Huh-7 cells were transfected with equal amounts of p2577L, p2577S, and p25L-211S. The HDAgs expressed from cell lysates were detected by Western blotting. All of the plasmids could express their encoded viral proteins (Fig. 1). SDS-PAGE and Western blotting could detect HDAg in lysates of P815/2577L cells (Fig. 1). This finding confirmed the generation of a stable HDAg-expressing cell line (P815/2577L).
FIG. 1.
Viral protein expression. Huh-7 cells were transfected with p2577L, p2577S, and p25L-211S. Cell lysates were harvested 48 h after transfection. Equal volumes of samples were loaded for SDS-PAGE (lanes 1 to 3). HDAg was also detected in lysates of P815/2577L cells (lane 4).
Anti-HDV antibody responses.
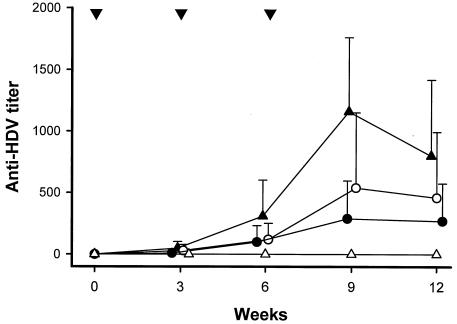
As shown in Fig. 2, animals immunized with 100 μg of p2577S showed the strongest anti-HDV antibody response. The antibody titers gradually increased, and a maximal titer was achieved at week 9. The antibody titers (mean and standard deviation) generated at week 9 by p2577L, p2577S, and p25L-211S were 290 ± 307.1, 1,160 ± 602.2, and 540 ± 611.4, respectively. Plasmid p2577S induced significantly higher anti-HDV antibody titers than plasmid p2577L did (P value, <0.05, as determined by Kruskal-Wallis and Dunnett tests). All mice immunized with p2577S could generate anti-HDV antibodies, and the seroconversion rate was 70% for mice immunized with p2577L or p25L-211S. Interestingly, 6 of 10 mice immunized with plasmid p2577S produced high titers of anti-HDV antibodies (>1,000:1), but none of the mice immunized with plasmid p2577L produced high anti-HDV antibody titers (P value, 0.0054, as determined by Fisher's exact test). Two of 10 mice in the p25L-211S-immunized group generated high anti-HDV antibody titers. We also used Student's t test to analyze the antibody responses mounted by each of the groups. The titers of anti-HDV antibodies generated by p2577S were significantly higher than those produced by p2577L not only at week 9 but also up to week 12. The P values were 0.058 at week 3, 0.06 at week 6, 0.001 at week 9, and 0.03 at week 12. No anti-HDV antibody response was detectable in animals immunized with pcDNA3.1(−).
FIG. 2.
Kinetics of anti-HDV antibodies in mice immunized with plasmid p2577L (•), p2577S (▴), p25L-211S (○), or pcDNA3.1(−) (▵). Titers of anti-HDV antibodies were assayed by an ELISA and determined by serial dilution of sera. Titers below 50:1 were considered representative of nonresponders.Data are presented as the mean and standard deviation for all immunized animals per time point.
Western blotting with anti-mouse serum samples.
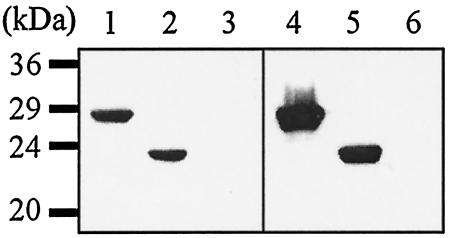
As the report from Mauch et al. indicated that no HDAg-specific antibody titers were detectable after L-HDAg or S-HDAg DNA vaccination (17), we used Western blotting to confirm whether the anti-HDV antibodies detected by the ELISA were HDAg specific. L-HDAg and S-HDAg obtained from lysates of p2577L- and p2577S-transfected Huh-7 cells were loaded at the same volumes for SDS-PAGE. After blotting onto nitrocellulose membranes, the HDAgs were stained with mouse serum samples at a 200:1 dilution. As shown in Fig. 3, serum samples from mice immunized with L-HDAg- and S-HDAg-encoding plasmids could specifically bind to HDAgs. We confirmed again that an HDV DNA vaccine did generate anti-HDV antibodies at a higher titer with S-HDAg immunization.
FIG. 3.
Western blotting with sera from immunized mice. L-HDAg (lanes 1 and 4) and S-HDAg (lanes 2 and 5) obtained from lysates of p2577L- and p2577S-transfected Huh-7 cells were loaded at the same volumes for SDS-PAGE. Lanes 3 and 6 represented a nontransfected cell lysate used as a negative control. After blotting onto nitrocellulose membranes, the antigens were stained with p2577L (lanes 1 to 3)- or p2577S (lanes 4 to 6)-immunized mouse sera (200:1 dilution).
Epitope mapping for anti-HDV antibodies.
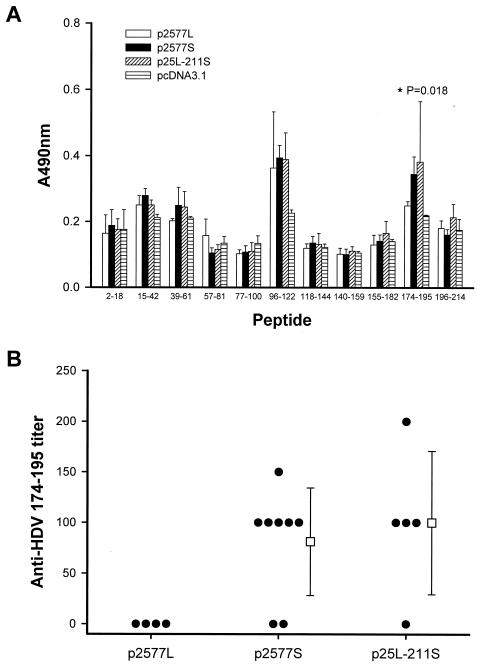
The anti-HDV antibody-positive sera with titers equal to or greater than 400:1 at week 9 were further analyzed for epitope mapping. Four samples in the p2577L group, eight in the p2577S-immunized group, and five in the p25L-211S-immunized group fulfilled the criteria for epitope mapping. As shown in Fig. 4A, antibodies induced by vaccination with plasmids p2577L, p2577S, and p25L-211S could bind to amino acids 96 to 122. In addition, one weak epitope at amino acids 15 to 42 was noted. Of note, antibodies induced by immunization with plasmids p2577S and p25L-211S could recognize the C terminus of S-HDAg (amino acids 174 to 195), but antibodies generated by plasmid p2577L immunization reacted only weakly with this epitope (the P value for a comparison of OD values among the three groups was 0.018, as determined by the Kruskal-Wallis test). We further analyzed the antibody titers against this epitope after serial dilution of serum samples from each group. As shown in Fig. 4B, only mice immunized with p2577S or p25L-211S mounted antibodies against the epitope at amino acids 174 to 195. This finding suggests that the isoprenylated motif of L-HDAg can interfere with antibody recognition of the unique epitope at the C terminus of S-HDAg.
FIG. 4.
Epitope mapping. (A) Serum samples with anti-HDV antibody titers equal to or greater than 400:1 at week 9 were analyzed for epitope mapping. Two major epitopes, at amino acids 96 to 122 and 174 to 195, were identified. The antibodies generated after immunization with p2577L could not bind to the epitope at amino acids 174 to 195. (B) Anti-HDV (amino acids 174 to 195) antibody titers at week 9 determined by serial dilution of sera. The results were considered significant when the OD of the tested sera was higher than the mean OD and 3 standard deviations of the control sera.
T-cell proliferation responses.
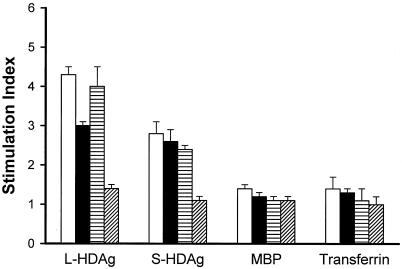
Groups of three mice were given injections of 100 μg of different plasmids. Splenic lymphocytes derived from p2577L-, p2577S-, and p25L-211S-inoculated animals had positive proliferation responses to L-HDAg, with peak SIs of 4.3, 3.0, and 4.0, respectively (Fig. 5); the corresponding peak SIs for S-HDAg were 2.8, 2.6, and 2.4. All mice failed to respond to MBP and transferrin included as control antigens. Mice vaccinated with pcDNA3.1(−) did not respond to L-HDAg or S-HDAg.
FIG. 5.
T-cell proliferation responses. BALB/c mice were given an intramuscular injection of p2577L (□), p2577S (▪), p25L-211S (▤), or pcDNA3.1(−) (▨). At 7 days after the last immunization, splenocytes from three immunized mice were used in proliferation assays. Stimulated wells received purified L-HDAg (10 μg/ml), purified S-HDAg (10 μg/ml), purified MBP (10 μg/ml), and transferrin (120 μg/ml). Data are presented as the mean and standard deviation SI. An SI of greater than 2 was defined as significant.
ELISPOT assay.
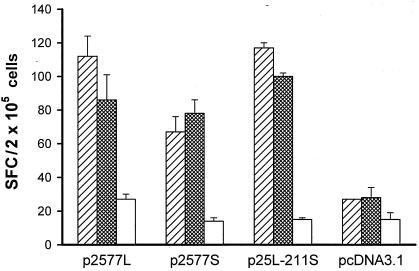
To analyze the T-cell responses against HDAg, an ELISPOT assay for IFN-γ was also performed. Splenocytes from groups of three mice were stimulated with recombinant L-HDAg and S-HDAg or MBP. The number of HDV-specific SFC was calculated by subtracting the number of spots in the absence of antigen from that in the presence of antigen. As shown in Fig. 6, all HDAg-encoding plasmids could produce significantly higher numbers of IFN-γ-positive SFC against L-HDAg and S-HDAg than against MBP or against the control.
FIG. 6.
IFN-γ ELISPOT assay for quantification of T-helper responses in mice immunized with various plasmid constructs. Symbols: ▨, L-HDAg; ▩, S-HDAg; □, MBP. Data are presented as the mean and standard deviation.
Intracellular IFN-γ staining.
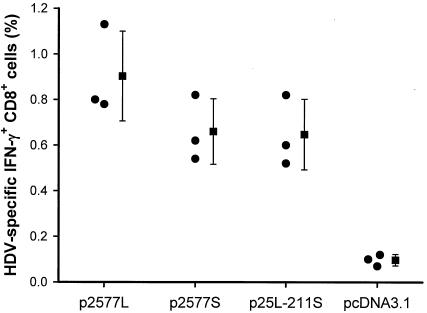
We tried to quantify the cytotoxic-T-lymphocyte responses among mice immunized with various HDAg-encoding DNA vaccines. After stimulation with irradiated P815/2577L cells, HDV-specific, IFN-γ-producing CD8+ cells represented 0.9% ± 0.2% (mean and standard deviation), 0.66% ± 0.14%, 0.65% ± 0.15%, and 0.1% ± 0.03% of the total CD8 cells after vaccination with p2577L, p2577S, p25L-211S, and pcDNA3.1(−), respectively (Fig. 7). HDV-specific, IFN-γ-producing CD8+ T cells represented approximately 0.7 to 0.9% of the CD8 cells in mice after immunization with plasmids encoding various forms of HDAg.
FIG. 7.
CD8+ cytotoxic-T-lymphocyte responses determined by intracellular IFN-γ staining. At 2 weeks after the last immunization, immune splenocytes were cultured in the presence of irradiated P815/2577L cells. The cells were surface stained with fluorescein isothiocyanate-conjugated rat anti-mouse CD8a monoclonal antibody. Finally, the cells were stained with R-phycoerythrin-conjugated rat anti-mouse IFN-γ monoclonal antibody. Samples were acquired for flow cytometry analysis. Data are presented as the mean and standard deviation.
DISCUSSION
L-HDAg of HDV is 19 amino acids longer at the C terminus than S-HDAg and contains unique properties for isoprenylation and HDV assembly (1, 7, 25). The 15 amino acids upstream from the isoprenylation site are also critical for HDV packaging. Even though L-HDAg contains the complete sequence of S-HDAg, L-HDAg lacks trans-activating activity for RNA replication. An earlier study suggested that S-HDAg has a unique conformation at the C terminus (12). Isoprenylation can mask a conformation epitope of HDAg. The B-cell epitope is usually conformation dependent, and an antibody can recognize the specific structure domain of an antigen. In this study, we found that DNA-based immunization with endogenous S-HDAg could generate a more significant humoral immune response against HDV than could L-HDAg. After detailed epitope mapping, the antibodies produced by DNA vaccination with HDAg-encoded plasmids could recognize a major epitope, at residues 96 to 122, containing the arginine-rich motif (amino acids 97 to 107) of HDAg. Another epitope, at residues 174 to 195, could bind to antibodies induced by p2577S and p25L-211S but had only a weak affinity for antibodies induced by p2577L. These results imply that the isoprenylated motif of HDAg can mask the presentation of the epitope at the C terminus of S-HDAg. The masking effect of isoprenylation may partially explain why the plasmid encoding S-HDAg could induce high anti-HDV antibody titers and seroconversion rates. However, the serine-211 mutant form of the L-HDAg-encoding plasmid did not induce antibodies as strongly as did the S-HDAg-encoding plasmid. We speculate that not only the isoprenylated motif but also the additional 19 amino acids of L-HDAg might interfere with antigenic region presentation following DNA-based immunization.
Recently, Mauch et al. reported that no anti-HDV antibody response could be detected after HDV DNA immunization (17). In that study, both L-HDAg- and S-HDAg-encoding plasmids were used in different strains of mice. The lack of a humoral immune response was discrepant from the results of Polo et al., who demonstrated significant humoral immunity to HDAg induced by intramuscular injection of DNA (20). In the present study, we confirmed the existence of anti-HDV antibodies not only by an ELISA but also by a Western blot assay. All of the experiments in this study were performed at least twice. There is no doubt that the HDV DNA vaccine can induce HDAg-specific antibodies, and a high titer of anti-HDV antibodies was generated by S-HDAg DNA immunization. It should be noted that the immunization schedules and dosages of plasmids were different. From sequence analysis, 25 amino acid differences were found between the plasmids used (GenBank accession no. M21012 and AF540888). The sequence variation also might have resulted in the discrepancy.
DNA-based immunization can produce a Th1 immune response to HDV (11). In this study, all of the plasmids encoding HDAg sequences could produce a Th1 immune response and HDV-specific, IFN-γ-secreting CD8+ T cells. These findings imply that the T-cell epitopes for major histocompatibility complex class I of mice might not be located within the additional 19 amino acids of L-HDAg.
In general, both humoral immunity and cellular immunity are necessary for a protective vaccine. In chronically HDV-infected patients, amino acids 94 to 100 and 174 to 195 of HDAg are immunodominant regions (23). In the present study, HDV DNA vaccination could induce antibodies that directly reacted with these two major epitopes in mice. Although the neutralizing ability of the anti-HDV antibodies was unclear, HDV variants with amino acid changes near or within amino acids 170 to 195 usually emerged after severe hepatitis attacks in chronically HDV-infected patients, suggesting that anti-HDV antibodies might have immune selection effects (29). So far, the titers of anti-HDV antibodies needed for a candidate HDV prophylactic or therapeutic DNA vaccine have not been determined. In the present study, we demonstrated that the magnitudes of humoral immune responses generated by L-HDAg- and S-HDAg-encoding DNA vaccines are different. This finding provides information relevant for the choice of L-HDAg or S-HDAg as a candidate DNA vaccine in the future.
In conclusion, HDAg-specific antibodies definitely exist following DNA vaccination. The immune responses generated by L-HDAg- and S-HDAg-encoding DNA vaccines are different. The isoprenylated motif can mask the epitope at residues 174 to 195 of HDAg but does not interfere with cellular immunity following DNA-based immunization.
Acknowledgments
This study was supported by grants from the National Science Council (NSC91-2315-B-075-005), Department of Health (DOH90-TD-1042), and Taipei Veterans General Hospital, Taipei, Taiwan, Republic of China.
We thank Cheng-Po Hu (Department of Medical Education and Research, Taipei Veterans General Hospital) and Mi-Hua Tao (Division of Cancer Research, Institute of Biomedical Science, Academia Sinica) for critical review and Pui-Ching Lee (Department of Medicine, Taipei Veterans General Hospital) for preparation of the figures.
REFERENCES
- 1.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casey, J. L., K. F. Bergmann, T. L. Brown, and J. L. Gerin. 1992. Structural requirement for RNA editing in hepatitis D virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. USA 89:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davey, S. 1996. State of the world's vaccines and immunization, p. 76-82. World Health Organization, Geneva, Switzerland.
- 5.Farci, P., A. Mandas, A. Colana, M. E. Lai, V. Desmet, P. Van Eyken, Y. Gibo, L. Caruso, S. Scaccabarozzi, D. Criscuolo, J. C. Ryff, and A. Balestrieri. 1994. Treatment of chronic hepatitis D with interferon alfa-2a. N. Engl. J. Med. 330:88-94. [DOI] [PubMed] [Google Scholar]
- 6.Fattovich, G., G. Giustina, E. Christensen, M. Pantalena, I. Zagni, G. Realdi, and S. W. Schalm. 2000. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. Gut 46:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 256:1331-1333. [DOI] [PubMed] [Google Scholar]
- 8.Govindarajan, S., K. M. De Cock, and A. G. Redeker.1986. . Natural course of delta superinfection in chronic hepatitis B virus-infected patients: histologic study with multiple liver biopsies. Gastroenterology 6:640-644. [DOI] [PubMed] [Google Scholar]
- 9.Hsu, S. C., B. S. Yan, J. M. Pan, and W. J. Syu. 1997. A monoclonal antibody reacts with maltose-binding protein of Escherichia coli and related enteric bacteria. J. Immunol. Methods 204:169-174. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, S. C., W. J. Syu, L. T. Ting, and J. C. Wu. 2000. Immunohistochemical differentiation of hepatitis D virus genotypes.Hepatology 32:1111-1116. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y. H., J. C. Wu, M. H. Tao, W. J. Syu, H. C. Hsu, W. K. Chi, F. Y. Chang, and S. D. Lee. 2000. DNA-based immunization produces Th1 immune response to hepatitis delta virus in a mouse model. Hepatology 32:104-110. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, S. B., and M. M. C. Lai.1993. . A unique conformation at the carboxyl terminus of the small hepatitis delta antigen revealed by a specific monoclonal antibody. Virology 193:924-931. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, S. B., and M. M. C. Lai.1994. . Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J. Virol. 68:2958-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karayiannis, P., J. Saldanha, A. M. Jackson, S. Luther, R. Goldin, J. Monjardino, and H. C. Thomas. 1993. Partial control of hepatitis delta virus superinfection by immunisation of woodchucks (Marmota monax) with hepatitis delta antigen expressed by a recombinant vaccinia or baculovirus. J. Med. Virol. 41:210-214. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. Z., P. J. Chen, M. M. C. Lai, and D. S. Chen. 1994. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology 199:169-175. [DOI] [PubMed] [Google Scholar]
- 16.Lin, H. P., S. C. Hsu, J. C. Wu, I. J. Sheen, B. S. Yan, and W. J. Syu.1999. . Localization of isoprenylated antigen of hepatitis delta virus by anti-farnesyl antibodies. J. Gen. Virol. 80:91-96. [DOI] [PubMed] [Google Scholar]
- 17.Mauch, C., C. Grimm, S. Meckel, J. R. Wands, H. E. Blum, M. Roggendorf, and M. Geissler. 2001. Induction of cytotoxic T lymphocyte responses against hepatitis delta virus antigens which protect against tumor formation in mice. Vaccine 20:170-180. [DOI] [PubMed] [Google Scholar]
- 18.Moraleda, G., S. Seeholzer, V. Bichko, R. Dunbrack, J. Otto, and J. Taylor.1999. . Unique properties of the large antigen of hepatitis delta virus. J. Virol. 73:7147-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato.1982. . Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 20.Polo, J. M., B. Lim, S. Govindarajan, and M. M. C. Lai. 1995. Replication of hepatitis delta virus RNA in mice after intramuscular injection of plasmid DNA.J. Virol. 69:5203-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzetto, M., M. G. Canese, J. L. Gerin, W. T. London, D. L. Sly, and R. H. Purcell.1980. . Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J. Infect. Dis. 141:590-602. [DOI] [PubMed] [Google Scholar]
- 22.Rizzetto, M., G. Verme, S. Recchia, F. Bonino, P. Farci, S. Arico, R. Calzia, A. Picciotto, M. Colombo, and H. Popper. 1983. Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen. An active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern. Med. 98:437-441. [DOI] [PubMed] [Google Scholar]
- 23.Wang, J. G., R. W. Jansen, E. A. Brown, and S. M. Lemon. 1990. Immunogenic domains of hepatitis delta virus antigens: peptide mapping of epitopes recognized by human and woodchuck antibodies. J. Virol. 64:1108-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, J. C., S. D. Lee, S. Govindarajan, T. W. Kung, Y. T. Tsai, K. J. Lo, and L. P. Ting. 1990. Correlation of serum delta RNA with clinical course of acute delta virus superinfection in Taiwan: a longitudinal study. J. Infect. Dis. 161:1116-1120. [DOI] [PubMed] [Google Scholar]
- 25.Wu, J. C., P. J. Chen, M. Y. P. Kuo, S. D. Lee, D. S. Chen, and L. P. Ting. 1991. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line.J. Virol. 65:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, J. C., C. L. Chen, S. D. Lee, I. J. Sheen, and L. P. Ting. 1992. Expression and localization of the small and large delta antigens during the replication cycle of hepatitis D virus.Hepatology 16:1120-1127. [PubMed] [Google Scholar]
- 27.Wu, J. C., C. H. Chen, M. C. Hou, T. Z. Chen, S. D. Lee, and K. J. Lo.1994. . Multiple viral infections as the most common cause of fulminant and subfulminant viral hepatitis in an endemic area for hepatitis B: application and limitations of polymerase chain reaction.Hepatology 19:836-840. [PubMed] [Google Scholar]
- 28.Wu, J. C., T. Z. Chen, Y. S. Huang, F. S. Yen, L. T. Ting, W. Y. Sheng, S. H. Tsay, and S. D. Lee. 1995. Natural history of hepatitis D viral superinfection—significance of viremia detected by polymerase chain reaction. Gastroenterology 108:796-802. [DOI] [PubMed] [Google Scholar]
- 29.Wu, J. C., T. Y. Chiang, W. K. Shiue, S. Y. Wang, I. J. Sheen, Y. H. Huang, and W. J. Syu. 1999. Recombination of hepatitis D virus RNA sequences and its implications. Mol. Biol. Evol. 16:1622-1632. [DOI] [PubMed] [Google Scholar]