Abstract
In the title compound, C18H22N2O3S, the pyrimidine ring is not planar. It adopts a half-chair conformation The crystal structure is characterized by classical N—H⋯O and C—H⋯O inter- and intramolecular hydrogen bonds, respectively. The title compound exhibits a wide spectrum of biological activities.
Related literature
For related literature, see: Allen et al. (1987 ▶); Biginelli (1893 ▶); Cremer & Pople (1975 ▶); Gurskaya et al. (2003a
▶,b
▶); Kappe (1993 ▶); Kappe et al. (1997 ▶); Li (2006 ▶); Nardelli (1983 ▶); Nizam Mohideen et al. (2008 ▶); Overman et al. (1995 ▶); Snider et al. (1996 ▶).
Experimental
Crystal data
C18H22N2O3S
M r = 346.44
Monoclinic,

a = 28.325 (5) Å
b = 7.410 (2) Å
c = 20.202 (4) Å
β = 121.61 (3)°
V = 3610.9 (18) Å3
Z = 8
Mo Kα radiation
μ = 0.20 mm−1
T = 293 (2) K
0.4 × 0.2 × 0.1 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2004 ▶) T min = 0.954, T max = 0.983
16675 measured reflections
3183 independent reflections
2722 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.140
S = 1.04
3183 reflections
220 parameters
H-atom parameters constrained
Δρmax = 0.47 e Å−3
Δρmin = −0.33 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT and XPREP (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2003 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026664/pv2100sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026664/pv2100Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O2i | 0.86 | 2.16 | 2.990 (2) | 161 |
| C7—H7⋯O2 | 0.98 | 2.46 | 2.831 (3) | 102 |
Symmetry code: (i)  .
.
Acknowledgments
MNM, AR, and CAMAH thank the Management of The New College, Chennai, India, for providing the necessary facilities.
supplementary crystallographic information
Comment
The title compound, (I), belongs to the class of 5-substituted 1,2,3,4-tetrahydropyrimidin-2-ones, which are known as `Biginelli compounds' (Kappe, 1993). The Biginelli reaction is a classic multicomponent reaction (Biginelli, 1893). The biological activity of some isolated alkaloids has been attributed to the presence of the dihydropyrimidinone moiety in the molecules (Overman et al., 1995) and the conformation of the pyrimidine ring (Kappe et al., 1997; Gurskaya et al., 2003a,b). Most important among them are batzelladine alkaloids, which have been found to be potent HIVgp-120-CD4 inhibitors (Snider et al., 1996). The aim of the present work was to study classical and extended Biginelli reactions. As part of our ongoing investigation of pyrimidine derivatives, the title compound, (I), has been prepared and its crystal structure is presented here.
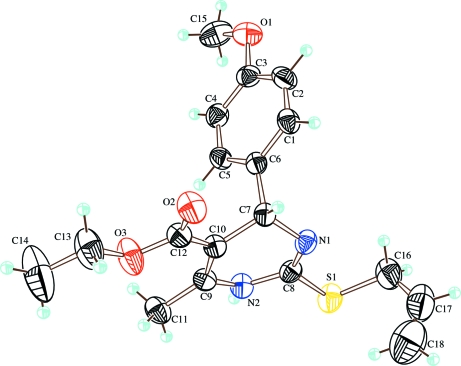
The bond lengths and angles in the title compound (Fig. 1) are comparable with ethyl 1,2,3,4-tetrahydro-6-methyl-2-oxo-4-phenylpyrimidine-5-carboxylate, a structure closely related to (I) (Nizam Mohideen et al., 2008). The torsion angles [C1—C6—C7—C10 = 153.1 (2), C5—C6—C7—C10 = -31.5 (2), C9—C10—C12—O2 = 171.3 (2), C7—C10—C12—O2 = -11.6 (3), C9—C10—C12—O3 = -10.1 (1) and C7—C10—C12—O3 = 167.1 (2) °] differs from the torsion angles [47.6 (2), -137.1 (2), 10.1 (2), -167.8 (2) -171.5 (2) and 10.5 (2) °] in the reported structure mentioned above.
In (I), the heterocyclic ring (atoms N1, N2, C7, C8, C9, C10) of the dihydropyrimidine group is not planar, as indicated by the displacement of atom C7 from the least-squares plane [0.212 (1) Å] and by the C8—N1—C7—C10 torsion angle [31.1 (1) °]. Atom C11 deviating by -0.204 (1) Å from the least squares plane of the pyrimidine ring. The pyrimidine ring adopts half chair conformation; the puckering parameters are q2 = 0.312 (1) Å, φ = 236.3 (2)°, and θ = 104.2 (1)° (Cremer & Pople, 1975), and the lowest displacement asymmetry parameters ΔS(C7) is 2.3 (1)°, Δ2(C10) is 22.4 (1)° (Nardelli, 1983).
The benzene ring is planar, the larget displacement observed being -0.008 (1) Å for atom C6. The dihedral angle between the pyrimidine and benzene rings is 89.5 (1)°, close to the value of 86.5 (1)° found in ethyl 1,2,3,4-tetrahydro-6-methyl-2-oxo-4-phenylpyrimidine-5-carboxylate.
The crystal packing is characterized by classical N—H···O and C—H···O inter and intramolecular hydrogen bonds (Table 1).
Experimental
To a suspension of NaH (0.100 g, 2 mmol, 50% dispersion in mineral oil washed with hexane) in dry THF (25 ml) was added a solution of dihydropyrimidone, (0.594 g, 2 mmol) in dry THF (10 ml) and stirred in an atmosphere of N2 for one hour. Then a solution of allyl bromide (0.2 ml, 2.5 mmol) in dry THF (5 ml) was added drop wise and stirred for futher four hours. (TLC control, silica, ethyl acetate: hexane 1:9 as eluent). Evaporation of solvent under reduced pressure, followed by purification of the residue by column chromatography gave a yellow solid. Single crystals of the title compound suitable for X-ray diffraction were obtained by slow evaporation of a solution in ethanol (mp 368–369 K).
Refinement
All H atoms were positioned geometrically and allowed to ride on their parent C atoms, with C—H distances fixed in the range 0.93–0.98 Å and N—H distance of 0.86 Å, with Uiso(H) = 1.5Ueq(C) for methyl H atoms and Uiso(H) = 1.2Ueq(C, N) for other H atoms.
Figures
Fig. 1.
The molecular configuration and atom-numbering scheme for (I). Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C18H22N2O3S | F000 = 1472 |
| Mr = 346.44 | Dx = 1.275 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 7889 reflections |
| a = 28.325 (5) Å | θ = 2.6–25º |
| b = 7.410 (2) Å | µ = 0.20 mm−1 |
| c = 20.202 (4) Å | T = 293 (2) K |
| β = 121.61 (3)º | Needle, yellow |
| V = 3610.9 (18) Å3 | 0.4 × 0.2 × 0.1 mm |
| Z = 8 |
Data collection
| Bruker Kappa APEXII CCD diffractometer' | 3183 independent reflections |
| Radiation source: fine-focus sealed tube | 2722 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.026 |
| T = 293(2) K | θmax = 25.0º |
| ω and φ scan | θmin = 1.7º |
| Absorption correction: Multi-scan(SADABS; Bruker, 2004) | h = −33→33 |
| Tmin = 0.954, Tmax = 0.983 | k = −8→8 |
| 16675 measured reflections | l = −24→24 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.045 | H-atom parameters constrained |
| wR(F2) = 0.140 | w = 1/[σ2(Fo2) + (0.0785P)2 + 3.5811P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.002 |
| 3183 reflections | Δρmax = 0.48 e Å−3 |
| 220 parameters | Δρmin = −0.33 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.15041 (3) | 0.63300 (8) | 0.44134 (3) | 0.0551 (2) | |
| O1 | 0.33272 (7) | 1.1779 (2) | 0.34204 (10) | 0.0580 (4) | |
| O2 | 0.06407 (7) | 1.4020 (2) | 0.24877 (9) | 0.0522 (4) | |
| O3 | 0.01426 (7) | 1.2100 (2) | 0.15153 (9) | 0.0617 (5) | |
| N1 | 0.14728 (7) | 0.9848 (2) | 0.40787 (9) | 0.0385 (4) | |
| N2 | 0.09759 (7) | 0.7816 (2) | 0.30377 (10) | 0.0387 (4) | |
| H2 | 0.0954 | 0.6715 | 0.2889 | 0.046* | |
| C1 | 0.23063 (8) | 1.2512 (3) | 0.39532 (11) | 0.0399 (5) | |
| H1 | 0.2280 | 1.3171 | 0.4325 | 0.048* | |
| C2 | 0.27875 (9) | 1.2591 (3) | 0.39437 (13) | 0.0456 (5) | |
| H2A | 0.3082 | 1.3292 | 0.4308 | 0.055* | |
| C3 | 0.28341 (8) | 1.1622 (3) | 0.33898 (12) | 0.0406 (5) | |
| C4 | 0.23922 (9) | 1.0596 (3) | 0.28488 (12) | 0.0424 (5) | |
| H4 | 0.2418 | 0.9951 | 0.2474 | 0.051* | |
| C5 | 0.19079 (8) | 1.0533 (3) | 0.28672 (11) | 0.0387 (5) | |
| H5 | 0.1611 | 0.9844 | 0.2499 | 0.046* | |
| C6 | 0.18558 (8) | 1.1467 (2) | 0.34195 (11) | 0.0331 (4) | |
| C7 | 0.13489 (8) | 1.1274 (2) | 0.34918 (11) | 0.0334 (4) | |
| H7 | 0.1298 | 1.2415 | 0.3692 | 0.040* | |
| C8 | 0.13171 (8) | 0.8265 (3) | 0.38169 (11) | 0.0359 (4) | |
| C9 | 0.06696 (8) | 0.9154 (3) | 0.25004 (11) | 0.0355 (4) | |
| C10 | 0.08227 (8) | 1.0897 (3) | 0.27177 (11) | 0.0341 (4) | |
| C11 | 0.02055 (9) | 0.8432 (3) | 0.17415 (13) | 0.0484 (5) | |
| H11A | −0.0117 | 0.9173 | 0.1562 | 0.073* | |
| H11B | 0.0123 | 0.7217 | 0.1813 | 0.073* | |
| H11C | 0.0315 | 0.8444 | 0.1365 | 0.073* | |
| C12 | 0.05336 (8) | 1.2485 (3) | 0.22475 (12) | 0.0382 (5) | |
| C13 | −0.01773 (13) | 1.3590 (4) | 0.10104 (16) | 0.0718 (8) | |
| H13A | 0.0067 | 1.4473 | 0.0989 | 0.086* | |
| H13B | −0.0387 | 1.4178 | 0.1204 | 0.086* | |
| C14 | −0.05525 (19) | 1.2832 (6) | 0.0238 (2) | 0.1255 (18) | |
| H14A | −0.0341 | 1.2191 | 0.0066 | 0.188* | |
| H14B | −0.0755 | 1.3789 | −0.0120 | 0.188* | |
| H14C | −0.0807 | 1.2017 | 0.0261 | 0.188* | |
| C15 | 0.33997 (12) | 1.0703 (4) | 0.28976 (19) | 0.0700 (8) | |
| H15A | 0.3327 | 0.9462 | 0.2949 | 0.105* | |
| H15B | 0.3774 | 1.0823 | 0.3016 | 0.105* | |
| H15C | 0.3147 | 1.1098 | 0.2374 | 0.105* | |
| C16 | 0.19334 (12) | 0.7289 (4) | 0.53535 (15) | 0.0662 (7) | |
| H16A | 0.2215 | 0.6410 | 0.5676 | 0.079* | |
| H16B | 0.2122 | 0.8324 | 0.5304 | 0.079* | |
| C17 | 0.16579 (19) | 0.7873 (5) | 0.57661 (19) | 0.0867 (10) | |
| H17 | 0.1891 | 0.8174 | 0.6287 | 0.104* | |
| C18 | 0.1139 (2) | 0.8017 (6) | 0.5498 (3) | 0.1070 (13) | |
| H18A | 0.0884 | 0.7736 | 0.4981 | 0.128* | |
| H18B | 0.1015 | 0.8404 | 0.5820 | 0.128* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0785 (5) | 0.0335 (3) | 0.0492 (4) | −0.0010 (3) | 0.0305 (3) | 0.0058 (2) |
| O1 | 0.0468 (9) | 0.0614 (11) | 0.0707 (11) | −0.0059 (8) | 0.0342 (8) | −0.0012 (9) |
| O2 | 0.0615 (10) | 0.0249 (8) | 0.0522 (9) | 0.0007 (7) | 0.0174 (8) | −0.0004 (7) |
| O3 | 0.0692 (11) | 0.0348 (9) | 0.0446 (9) | 0.0068 (8) | 0.0045 (8) | 0.0012 (7) |
| N1 | 0.0498 (10) | 0.0318 (9) | 0.0347 (9) | −0.0012 (7) | 0.0226 (8) | 0.0015 (7) |
| N2 | 0.0510 (10) | 0.0219 (8) | 0.0409 (9) | 0.0009 (7) | 0.0224 (8) | −0.0030 (7) |
| C1 | 0.0456 (11) | 0.0325 (11) | 0.0356 (10) | −0.0020 (8) | 0.0171 (9) | −0.0050 (8) |
| C2 | 0.0414 (11) | 0.0391 (12) | 0.0444 (11) | −0.0100 (9) | 0.0142 (9) | −0.0054 (9) |
| C3 | 0.0393 (11) | 0.0361 (11) | 0.0454 (11) | 0.0010 (8) | 0.0215 (9) | 0.0083 (9) |
| C4 | 0.0521 (12) | 0.0385 (11) | 0.0400 (11) | −0.0019 (9) | 0.0264 (10) | −0.0029 (9) |
| C5 | 0.0419 (11) | 0.0337 (11) | 0.0358 (10) | −0.0082 (8) | 0.0171 (9) | −0.0065 (8) |
| C6 | 0.0395 (10) | 0.0237 (9) | 0.0313 (9) | 0.0004 (7) | 0.0151 (8) | 0.0028 (7) |
| C7 | 0.0415 (10) | 0.0240 (9) | 0.0331 (9) | −0.0002 (8) | 0.0184 (8) | −0.0013 (7) |
| C8 | 0.0436 (11) | 0.0294 (10) | 0.0383 (10) | 0.0022 (8) | 0.0239 (9) | 0.0033 (8) |
| C9 | 0.0372 (10) | 0.0307 (10) | 0.0387 (10) | −0.0005 (8) | 0.0200 (9) | −0.0024 (8) |
| C10 | 0.0376 (10) | 0.0270 (10) | 0.0359 (10) | 0.0006 (8) | 0.0180 (8) | −0.0002 (8) |
| C11 | 0.0488 (13) | 0.0345 (12) | 0.0484 (12) | −0.0045 (9) | 0.0161 (10) | −0.0061 (9) |
| C12 | 0.0380 (10) | 0.0333 (11) | 0.0405 (11) | 0.0007 (8) | 0.0187 (9) | 0.0004 (8) |
| C13 | 0.0752 (18) | 0.0447 (15) | 0.0550 (15) | 0.0156 (13) | 0.0063 (13) | 0.0100 (12) |
| C14 | 0.133 (3) | 0.085 (3) | 0.065 (2) | 0.022 (2) | −0.013 (2) | 0.0010 (19) |
| C15 | 0.0728 (17) | 0.0696 (18) | 0.094 (2) | 0.0004 (14) | 0.0614 (17) | 0.0047 (16) |
| C16 | 0.0784 (18) | 0.0486 (15) | 0.0485 (14) | −0.0047 (13) | 0.0173 (13) | 0.0134 (11) |
| C17 | 0.129 (3) | 0.067 (2) | 0.0587 (17) | −0.014 (2) | 0.045 (2) | −0.0015 (15) |
| C18 | 0.141 (4) | 0.098 (3) | 0.103 (3) | 0.017 (3) | 0.078 (3) | 0.004 (2) |
Geometric parameters (Å, °)
| S1—C8 | 1.766 (2) | C7—H7 | 0.9800 |
| S1—C16 | 1.780 (3) | C9—C10 | 1.359 (3) |
| O1—C3 | 1.370 (3) | C9—C11 | 1.501 (3) |
| O1—C15 | 1.421 (3) | C10—C12 | 1.464 (3) |
| O2—C12 | 1.211 (2) | C11—H11A | 0.9600 |
| O3—C12 | 1.334 (3) | C11—H11B | 0.9600 |
| O3—C13 | 1.453 (3) | C11—H11C | 0.9600 |
| N1—C8 | 1.267 (3) | C12—O2 | 1.211 (2) |
| N1—C7 | 1.486 (2) | C13—C14 | 1.464 (4) |
| N2—C8 | 1.388 (3) | C13—H13A | 0.9700 |
| N2—C9 | 1.389 (3) | C13—H13B | 0.9700 |
| N2—H2 | 0.8600 | C14—H14A | 0.9600 |
| C1—C2 | 1.374 (3) | C14—H14B | 0.9600 |
| C1—C6 | 1.395 (3) | C14—H14C | 0.9600 |
| C1—H1 | 0.9300 | C15—H15A | 0.9600 |
| C2—C3 | 1.393 (3) | C15—H15B | 0.9600 |
| C2—H2A | 0.9300 | C15—H15C | 0.9600 |
| C3—C4 | 1.380 (3) | C16—C17 | 1.474 (5) |
| C4—C5 | 1.392 (3) | C16—H16A | 0.9700 |
| C4—H4 | 0.9300 | C16—H16B | 0.9700 |
| C5—C6 | 1.385 (3) | C17—C18 | 1.277 (5) |
| C5—H5 | 0.9300 | C17—H17 | 0.9300 |
| C6—C7 | 1.524 (3) | C18—H18A | 0.9300 |
| C7—C10 | 1.517 (3) | C18—H18B | 0.9300 |
| C8—S1—C16 | 101.35 (11) | C9—C11—H11B | 109.5 |
| C3—O1—C15 | 117.37 (19) | H11A—C11—H11B | 109.5 |
| C12—O3—C13 | 117.77 (18) | C9—C11—H11C | 109.5 |
| C8—N1—C7 | 116.11 (16) | H11A—C11—H11C | 109.5 |
| C8—N2—C9 | 119.55 (16) | H11B—C11—H11C | 109.5 |
| C8—N2—H2 | 120.2 | O2—C12—O3 | 122.22 (18) |
| C9—N2—H2 | 120.2 | O2—C12—O3 | 122.22 (18) |
| C2—C1—C6 | 121.64 (19) | O2—C12—C10 | 123.91 (19) |
| C2—C1—H1 | 119.2 | O2—C12—C10 | 123.91 (19) |
| C6—C1—H1 | 119.2 | O3—C12—C10 | 113.86 (17) |
| C1—C2—C3 | 120.11 (19) | O3—C13—C14 | 107.1 (2) |
| C1—C2—H2A | 119.9 | O3—C13—H13A | 110.3 |
| C3—C2—H2A | 119.9 | C14—C13—H13A | 110.3 |
| O1—C3—C4 | 124.2 (2) | O3—C13—H13B | 110.3 |
| O1—C3—C2 | 116.34 (19) | C14—C13—H13B | 110.3 |
| C4—C3—C2 | 119.44 (19) | H13A—C13—H13B | 108.5 |
| C3—C4—C5 | 119.65 (19) | C13—C14—H14A | 109.5 |
| C3—C4—H4 | 120.2 | C13—C14—H14B | 109.5 |
| C5—C4—H4 | 120.2 | H14A—C14—H14B | 109.5 |
| C6—C5—C4 | 121.81 (18) | C13—C14—H14C | 109.5 |
| C6—C5—H5 | 119.1 | H14A—C14—H14C | 109.5 |
| C4—C5—H5 | 119.1 | H14B—C14—H14C | 109.5 |
| C5—C6—C1 | 117.34 (18) | O1—C15—H15A | 109.5 |
| C5—C6—C7 | 122.25 (17) | O1—C15—H15B | 109.5 |
| C1—C6—C7 | 120.26 (17) | H15A—C15—H15B | 109.5 |
| N1—C7—C10 | 112.63 (15) | O1—C15—H15C | 109.5 |
| N1—C7—C6 | 107.63 (15) | H15A—C15—H15C | 109.5 |
| C10—C7—C6 | 112.61 (15) | H15B—C15—H15C | 109.5 |
| N1—C7—H7 | 107.9 | C17—C16—S1 | 116.9 (2) |
| C10—C7—H7 | 107.9 | C17—C16—H16A | 108.1 |
| C6—C7—H7 | 107.9 | S1—C16—H16A | 108.1 |
| N1—C8—N2 | 125.34 (17) | C17—C16—H16B | 108.1 |
| N1—C8—S1 | 123.53 (15) | S1—C16—H16B | 108.1 |
| N2—C8—S1 | 111.13 (14) | H16A—C16—H16B | 107.3 |
| C10—C9—N2 | 117.60 (18) | C18—C17—C16 | 128.1 (3) |
| C10—C9—C11 | 128.98 (19) | C18—C17—H17 | 115.9 |
| N2—C9—C11 | 113.43 (17) | C16—C17—H17 | 115.9 |
| C9—C10—C12 | 125.37 (18) | C17—C18—H18A | 120.0 |
| C9—C10—C7 | 118.86 (17) | C17—C18—H18B | 120.0 |
| C12—C10—C7 | 115.70 (16) | H18A—C18—H18B | 120.0 |
| C9—C11—H11A | 109.5 | ||
| C6—C1—C2—C3 | 0.3 (3) | C16—S1—C8—N2 | 179.62 (16) |
| C15—O1—C3—C4 | −5.1 (3) | C8—N2—C9—C10 | 17.0 (3) |
| C15—O1—C3—C2 | 175.6 (2) | C8—N2—C9—C11 | −163.18 (18) |
| C1—C2—C3—O1 | 179.86 (19) | N2—C9—C10—C12 | −176.65 (17) |
| C1—C2—C3—C4 | 0.5 (3) | C11—C9—C10—C12 | 3.6 (3) |
| O1—C3—C4—C5 | −179.79 (19) | N2—C9—C10—C7 | 6.3 (3) |
| C2—C3—C4—C5 | −0.5 (3) | C11—C9—C10—C7 | −173.39 (19) |
| C3—C4—C5—C6 | −0.4 (3) | N1—C7—C10—C9 | −29.8 (2) |
| C4—C5—C6—C1 | 1.2 (3) | C6—C7—C10—C9 | 92.1 (2) |
| C4—C5—C6—C7 | −174.30 (18) | N1—C7—C10—C12 | 152.88 (16) |
| C2—C1—C6—C5 | −1.2 (3) | C6—C7—C10—C12 | −85.2 (2) |
| C2—C1—C6—C7 | 174.42 (18) | C13—O3—C12—O2 | −2.9 (3) |
| C8—N1—C7—C10 | 31.2 (2) | C13—O3—C12—O2 | −2.9 (3) |
| C8—N1—C7—C6 | −93.6 (2) | C13—O3—C12—C10 | 178.3 (2) |
| C5—C6—C7—N1 | 93.2 (2) | C9—C10—C12—O2 | 171.3 (2) |
| C1—C6—C7—N1 | −82.1 (2) | C7—C10—C12—O2 | −11.6 (3) |
| C5—C6—C7—C10 | −31.5 (2) | C9—C10—C12—O2 | 171.3 (2) |
| C1—C6—C7—C10 | 153.14 (17) | C7—C10—C12—O2 | −11.6 (3) |
| C7—N1—C8—N2 | −10.0 (3) | C9—C10—C12—O3 | −10.0 (3) |
| C7—N1—C8—S1 | 170.98 (14) | C7—C10—C12—O3 | 167.10 (17) |
| C9—N2—C8—N1 | −16.0 (3) | C12—O3—C13—C14 | 177.0 (3) |
| C9—N2—C8—S1 | 163.17 (14) | C8—S1—C16—C17 | 90.0 (2) |
| C16—S1—C8—N1 | −1.2 (2) | S1—C16—C17—C18 | −10.8 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O2i | 0.86 | 2.16 | 2.990 (2) | 161 |
| C7—H7···O2 | 0.98 | 2.46 | 2.831 (3) | 102 |
Symmetry codes: (i) x, y−1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2100).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Biginelli, P. (1893). Gazz. Chim. Ital.23, 360–413.
- Bruker (2004). APEX2, SAINT, XPREP and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1358.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Gurskaya, G. V., Zavodnik, V. E. & Shutalev, A. D. (2003a). Crystallogr. Rep.48, 92–97.
- Gurskaya, G. V., Zavodnik, V. E. & Shutalev, A. D. (2003b). Crystallogr. Rep.48, 416–421.
- Kappe, C. O. (1993). Tetrahedron, 49, 6937–6963.
- Kappe, C. O., Fabian, W. M. F. & Semones, M. A. (1997). Tetrahedron, 53, 2803–2816.
- Nardelli, M. (1983). Acta Cryst. C39, 1141–1142.
- Nizam Mohideen, M., Rasheeth, A., Huq, C. A. M. A. & Nizar, S. S. (2008). Acta Cryst. E64, o1752. [DOI] [PMC free article] [PubMed]
- Overman, L. E., Michael, H., Rabinowitz, M. H. & Renhowe, P. A. (1995). J. Am. Chem. Soc.117, 2657–2658.
- Li, R. (2006). Acta Cryst. E62, o5480–o5481.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Snider, B. B., Chen, J., Patil, A. D. & Freyer, A. J. (1996). Tetrahedron Lett.37, 6977–6980.
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808026664/pv2100sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808026664/pv2100Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report