Abstract
Cell entry by reoviruses requires a large, transcriptionally active subvirion particle to gain access to the cytoplasm. The features of this particle have been the subject of debate, but three primary candidates—the infectious subvirion particle (ISVP), ISVP*, and core particle forms—that differ in whether putative membrane penetration protein μ1 and adhesin σ1 remain particle bound have been identified. Experiments with antibody reagents in this study yielded new information about the steps in particle disassembly during cell entry. Monoclonal antibodies specific for the δ region of μ1 provided evidence for a conformational change in μ1 and for release of the δ proteolytic fragment from entering particles. Antiserum raised against cores provided evidence for entry-related changes in particle structure and identified entering particles that largely lack the δ fragment inside cells. Antibodies specific for σ1 showed that it is also largely shed from entering particles. Limited coimmunostaining with markers for late endosomes and lysosomes indicated the particles lacking δ and σ1 did not localize to those subcellular compartments, and other observations suggested that both the particles and free δ were released into the cytoplasm. Essentially equivalent findings were obtained with native ISVPs and highly infectious recoated particles containing wild-type proteins. Poorly infectious recoated particles containing a hyperstable mutant form of μ1, however, showed no evidence for the in vitro and intracellular changes in particle structure normally detected by antibodies, and these particles instead accumulated in late endosomes or lysosomes. Recoated particles with hyperstable μ1 were also ineffective at mediating erythrocyte lysis in vitro and promoting α-sarcin coentry and intoxication of cells in cultures. Based on these and other findings, we propose that ISVP* is a transient intermediate in cell entry which mediates membrane penetration and is then further uncoated in the cytoplasm to yield particles, resembling cores, that largely lack the δ fragment of μ1.
The entry of animal viruses into host cells is accompanied by proteolytic cleavage, protein conformational changes, and/or protein shedding events that result in partial or complete disassembly of entering particles. One key aspect of disassembly is the conversion of virions, which often have greater stability in the aqueous environments between cells, to particle forms with more hydrophobic surfaces that can interact with a cellular lipid bilayer and mediate the passage of viral components into the cytoplasm. Other key elements include the activation of viral particles or nucleocapsids for genome transcription, translation, and/or targeting to particular subcellular sites. We have been studying the disassembly cascade and cell entry mechanisms used by a group of large nonenveloped viruses, the mammalian orthoreoviruses (reoviruses).
Reoviruses belong to the family Reoviridae, an evolutionarily divergent group of double-stranded RNA viruses whose human-infecting members also include rota-, orbi-, colti-, and seadornaviruses (see references 52 and 68 for general reviews). Reovirus virions are 85 nm wide and comprise a genome enclosed by two concentric, icosahedral protein capsids (25, 48). The genome, inner-capsid proteins, and outer-capsid protein λ2 constitute the core particle (63), which is competent for genome-dependent synthesis, capping, and export of translatable viral mRNAs (2, 31, 62). The surface-exposed outer-capsid proteins λ2, μ1, σ3, and σ1 affect survival in the environment (24, 49) and mediate adherence to cells and entry into the cytoplasm (3, 13, 15, 16, 33, 35, 38, 43). The bulk of the outer capsid consists of 200 heterohexameric complexes of putative membrane penetration protein μ1 (76 kDa) and its protector protein, σ3 (41 kDa) (25, 40), arranged in a fenestrated T=13 (alevo) net (48) over the T=1 inner capsid (57). This lattice is substituted at the fivefold vertices by pentameric turrets of the mRNA-capping protein λ2 (144 kDa) (20, 28, 45, 57, 72). At the center of each turret is anchored a trimeric receptor-binding fiber of adhesin σ1 (∼50 kDa) (16, 18, 30, 64). The μ1 protein is apparently autolytic and cleaves itself into 4-kDa N-terminal fragment μ1N and 72-kDa C-terminal fragment μ1C (40, 53).
Stepwise remodeling and/or disassembly of the outer capsid is a feature of cell entry by reoviruses and can be at least partly recapitulated in vitro. Under some conditions, protease treatment of virions yields infectious subvirion particles (ISVPs), which have lost σ3 to degradation and contain a cleaved form of μ1 (59-kDa N-terminal fragment δ and 13-kDa C-terminal fragment φ generated from μ1C) as the major surface protein (7, 22, 25, 36, 51, 57, 59). Under other conditions, protease treatment of virions yields cores, which have additionally lost the μ1 fragments and σ1 (25, 36, 42, 51, 53, 57, 63). ISVP-like particles are a required intermediate for cell entry (1, 46, 65) and are generated from virions either by extracellular proteolysis in the intestinal lumen (4, 6) or by endosomal or lysosomal proteolysis following endocytic uptake from the cell surface (1, 26, 65). The capacity of ISVPs, but not virions or cores, to induce membrane permeabilization both in vitro and in cell cultures (8, 13-15, 35, 43, 67) has been interpreted as evidence that ISVP-like particles initiate membrane penetration, resulting in cytoplasmic delivery of “transcriptase particles” activated to synthesize viral mRNAs for translation and packaging. However, ISVPs have hydrophilic surfaces (7, 25, 30, 36), suggesting they must undergo further changes before insertion into a lipid bilayer. Furthermore, because the viral transcriptases are inhibited in virions and ISVPs but activated in cores (7, 9, 23, 27, 36, 73), entry must be accompanied or followed by structural changes that result in transcriptase activation. Transcriptase particles derived from infecting virions or ISVPs have been isolated from cells by several groups but have been variously reported to resemble ISVPs or cores in protein composition (e.g., to contain or lack, respectively, large amounts of the μ1 fragments) (8, 9, 60, 61).
In an attempt to understand the membrane penetration step in cell entry by reoviruses, Chandran et al. recently examined the mechanism by which ISVPs induce the permeabilization of erythrocyte membranes (hemolysis) in vitro (13). They found that hemolysis is preceded by a suite of structural changes in the ISVP, including a conformational change in μ1 that exposes hydrophobic regions and increases the sensitivity of the δ fragment to further proteolysis. The structural changes also include the shedding of σ1 and an inferred conformational change in λ2 that promotes σ1 release. In sum, these changes convert the ISVP to a distinct particle form, the ISVP*, which is both necessary and sufficient to mediate hemolysis and is activated for viral mRNA synthesis. These results led to the proposal that the ISVP→ISVP* transition must precede membrane penetration and transcriptase activation in cells and that ISVP*s, rather than ISVPs or cores, may most closely resemble the particles that penetrate the cytoplasm and initiate mRNA synthesis (13).
In the present study, we used a combination of approaches to test whether the ISVP→ISVP* transition must indeed accompany productive cell entry by reoviruses. We found that a conformational change involving the δ region of μ1 precedes viral protein synthesis in ISVP-infected cells and that σ1 is shed from the particles during entry, as predicted by the previous in vitro work. We also found that particles resembling cores are efficiently generated inside cells by the additional shedding of the δ proteolytic fragment and that both these particles and free δ localize to the cytoplasm early after infection. Recoated particles containing a mutant, hyperstable form of μ1 were found to be defective in both the ISVP→ISVP* change in vitro and infection of cells in cultures. Results obtained with recoated particles furthermore established a strong link between productive cell entry and the μ1 conformational change and disassembly events discovered in this and previous work. We conclude that the ISVP* is a necessary but transient intermediate in the disassembly cascade that accompanies cell entry by reoviruses.
MATERIALS AND METHODS
Cells.
Spinner-adapted L929 cells were grown in Joklik's modified minimal essential medium (Irvine Scientific, Irvine, Calif.) supplemented to contain 2% fetal and 2% calf bovine sera (HyClone, Logan, Utah) as well as 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Irvine Scientific). Mv1Lu and CV-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, Calif.) supplemented to contain 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Sf21 and Tn High Five cells (Invitrogen) were grown in TC-100 medium (Invitrogen) supplemented to contain 10% heat-inactivated fetal bovine serum.
Virions, ISVPs, ISVP*s, and cores.
Virions of reovirus T1L were grown and purified by the standard protocol (30) and stored in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris [pH 7.5]). To generate virions containing [35S]methionine-cysteine-labeled proteins, Tran35S label (7 mCi) (ICN, Costa Mesa, Calif.) was added at the onset of infection. Nonpurified ISVPs were used in all experiments and were obtained by digesting virions in virion buffer at concentrations of 5 × 1012 to 1 × 1013 particles/ml with Nα-p-tosyl-l-lysine chloromethyl ketone-treated α-chymotrypsin (200 μg/ml) (Sigma-Aldrich, St. Louis. Mo.) for 10 to 20 min at 32 or 37°C. Digestion was stopped by the addition of ethanolic phenylmethylsulfonyl fluoride (2 to 5 mM) (Sigma-Aldrich) at 4°C. Parallel ISVP and ISVP* samples (see Fig. 1 and 2) were obtained by treatment of nonpurified ISVPs at a concentration of 4 × 1012 particles/ml with NaCl and CsCl (300 mM), respectively, for 10 to 20 min at 32 or 37°C, followed by removal to 4°C. Purified cores were obtained from virions as described previously (15, 54). Particle concentrations were estimated by measuring the A260 (22). Infection of L929 cells and plaque assays to determine viral titers were carried out as described previously (30, 49).
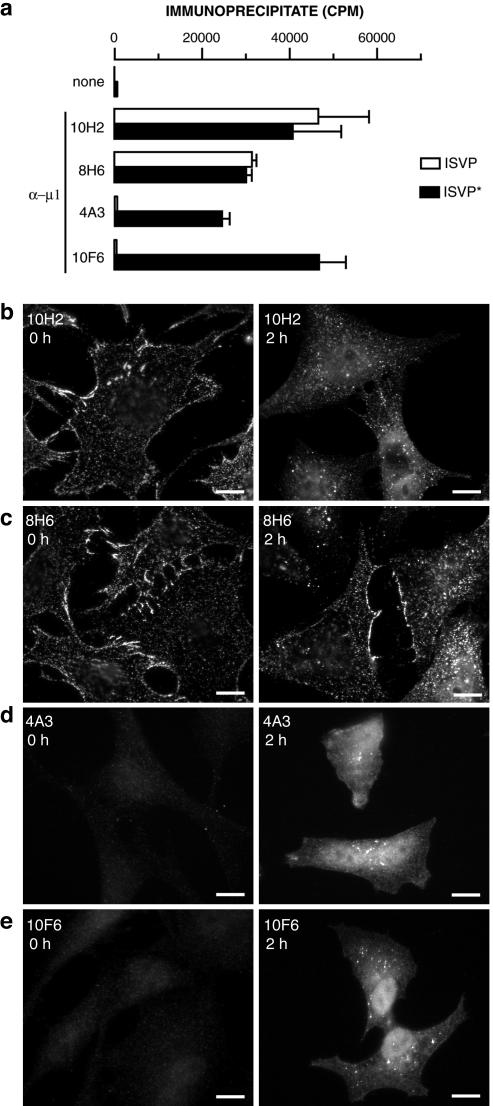
FIG. 1.
Protein μ1-specific MAbs detect μ1 conformational change in vitro and within infected cells. (a) 35S-labeled ISVPs and ISVP*s were immunoprecipitated with protein A-conjugated magnetic beads alone (none) or beads bound to μ1-specific MAb 10H2, 8H6, 4A3, or 10F6. Viral particles bound to antibody were quantitated by liquid scintillation counting. Error bars indicate standard deviations. (b to e) ISVPs were allowed to bind to CHX-pretreated Mv1Lu cells at 4°C, and cells were fixed at 0 or 2 h p.i. at 37°C. Cells were permeabilized and immunostained with MAb 10H2 (b), 8H6 (c), 4A3 (d), or 10F6 (e), followed by goat anti-mouse IgG conjugated to Alexa 488. Bars, 10 μm.
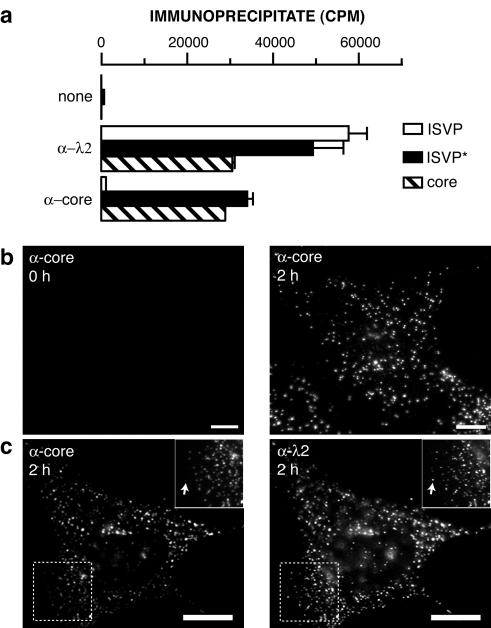
FIG. 2.
Anticore serum detects changes in particle structure in vitro and within infected cells. (a) 35S-labeled ISVPs, ISVP*s, and cores were immunoprecipitated with protein A-conjugated magnetic beads alone (none) or beads bound to λ2-specific MAb 7F4 (α-λ2) or anticore serum (α-core). Viral particles bound to antibody were quantitated by liquid scintillation counting. Error bars indicate standard deviations. (b) ISVPs were allowed to adsorb to CHX-pretreated Mv1Lu cells at 4°C, and cells were fixed at 0 or 2 h p.i. at 37°C. Cells were permeabilized and immunostained with anticore serum, followed by goat anti-rabbit IgG conjugated to Alexa 594. (c) Cells were infected as described for panel b, fixed at 2 h p.i. at 37°C, permeabilized, and coimmunostained with anticore serum and 7F4, followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. Arrows in panel c insets indicate punctate spots detected by 7F4 but not anticore serum. Bars, 10 μm.
Antibodies.
Monoclonal antibodies (MAbs) specific for reovirus outer-capsid proteins μ1 (10H2, 8H6, 4A3, and 10F6), σ1 (5C6), and λ2 (7F4) were grown and purified as described previously (70). Human LAMP-2-specific MAb H4B4 (44) was purchased from the Developmental Studies Hybridoma Bank (Department of Biological Sciences, University of Iowa, Iowa City). Human LAMP-1-specific rabbit polyclonal antiserum (11) was a generous gift from M. Fukuda (The Burnham Institute, La Jolla, Calif.). The production of μNS-specific rabbit polyclonal antiserum has been described elsewhere (10). Core- and σ1-specific rabbit polyclonal antisera were produced by the Polyclonal Antibody Service (Animal Care Unit, University of Wisconsin Medical School, Madison) from purified T1L cores that had been heat inactivated by incubation for 10 min at 70°C (15) and from purified glutathione S-transferase- T1L σ1 fusion protein (13), respectively. In some cases, primary antibodies were directly conjugated to Alexa 488 (Molecular Probes, Eugene, Oreg.) in accordance with the manufacturer's instructions. All primary antibodies were titrated to maximize the signal-to-noise ratio. Goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG conjugated to Alexa 488 or Alexa 594 were obtained from Molecular Probes and used at a dilution of 1:500.
Plasmid constructs encoding protein μ1-HS.
Plasmid pBKS-M2L encoding T1L μ1 with the P344L and L359F mutations (15) was the template for site-directed mutagenesis by the QuikChange protocol (Stratagene, La Jolla, Calif.). Complementary mutagenic primers with the sequences 5′-GCCATTCCACCTAAGCCAGAAGACTATAATGTGCGTAC-3′ and 5′-GTACGCACATTATAGTCTTCTGGCTTAGGTGGAATGGC-3′ (nucleotide changes underlined) were used to change alanine 319 to glutamic acid (A319E) and to create a BbsI site for screening mutant clones. A Bsu36I-MluI restriction fragment encoding the desired amino acid changes (A319E, P344L, and L359F) was sequenced and subcloned into shuttle plasmid pFastbacDUAL-M2L-S4L (15) for recombinant baculovirus production (see below). The construct expressing μ1 with the A319E, P344L, and L359F mutations is henceforth referred to as μ1-HS.
Recombinant μ1, σ3, and σ1.
A recombinant baculovirus coexpressing T1L μ1-HS and σ3 was generated in Sf21 cells by using the Bac-to-Bac system (Invitrogen) as previously described for T1L with wild-type μ1 (μ1-WT) and σ3 (15). A recombinant baculovirus separately expressing T1L σ1 has been described elsewhere (17). Tn High Five cells were infected with third- or fourth-passage baculovirus stocks (5 to 10 PFU/cell) and harvested at 65 h postinfection (p.i.). Cytoplasmic lysates of baculovirus-infected cells expressing only μ1 and σ3 or expressing μ1, σ3, and σ1 were prepared by lysis with Triton X-100 or probe sonication, respectively, as described previously (15, 16).
Recoated cores.
Recoated cores containing recombinant μ1 and σ3 (r-cores) or μ1, σ3, and σ1 (r-cores+σ1) were prepared by incubation of insect cell lysates containing these proteins (see above) with purified T1L cores, followed by repurification of particles on CsCl gradients as described previously (15, 16). Nonpurified ISVP-like and ISVP*-like particles derived from r-cores and r-cores+σ1 (pr-cores and pr-cores+σ1, respectively) were obtained as described above for virions.
Immunoprecipitation.
Protein A-conjugated magnetic beads (10 μl) (Dynal, Lake Success, N.Y.) were incubated with antibody (15 μg of MAb or 35 μl of antiserum) for 2 h at room temperature and washed three times with immunoprecipitation buffer (IP buffer) (10 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40). Viral particles (5 × 1010 per 10 μl) were added to the antibody-bound beads with ice-cold IP buffer (180 μl). Samples were incubated with rotary mixing for 1 h at 4°C, washed six times with ice-cold IP buffer, and boiled in gel sample buffer (30) for 3 to 5 min. Proteins released from the beads were detected by liquid scintillation counting or sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie brilliant blue R-250 (Sigma-Aldrich) or phosphorimaging with a Storm system (Molecular Dynamics, Sunnyvale, Calif.). ISVP-infected Mv1Lu cells were lysed by incubation with ice-cold IP buffer for 30 min at 4°C in the presence of protease inhibitors (Roche, Indianapolis, Ind.) and centrifuged at 8,000 × g for 10 min to remove nuclei and cell debris (see Fig. 4). Viral particles and proteins within the resulting cytoplasmic lysates were immunoprecipitated essentially as described above, except that 20 μl of antibody-bound beads was used per reaction with a cytoplasmic lysate generated from ∼2 × 106 cells.
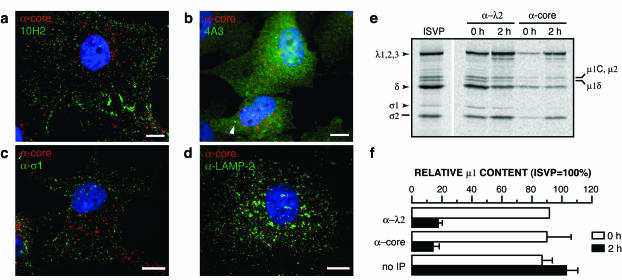
FIG. 4.
Particles resembling cores are generated in infected cells. (a to c) ISVPs were allowed to adsorb to CHX-pretreated Mv1Lu cells at 4°C, and cells were fixed at 2 h p.i. at 37°C. Cells were permeabilized and coimmunostained with anticore serum and μ1-specific MAb 10H2 (a), 4A3 (b), or σ1-specific MAb 5C6 (c), followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. The arrowhead in panel b indicates a punctate spot detected by both anticore serum and 4A3. (d) Intracellular localization of particles. CV-1 cells were infected as described above and fixed at 2 h p.i. Cells were permeabilized and coimmunostained with anticore serum and anti-LAMP-2 MAb H4B4 (α-LAMP-2), followed by goat anti-rabbit IgG conjugated to Alexa 488 and goat anti-mouse IgG conjugated to Alexa 594. Bars, 10 μm. (e and f) Biochemical evidence for particles resembling cores in infected cells. (e) 35S-labeled ISVPs (∼5 × 105 particles/cell) were allowed to adsorb to CHX-pretreated Mv1Lu cells (∼2 × 106) in 60-mm dishes at 4°C, and cells were harvested and lysed in IP buffer at 0 or 2 h p.i. at 37°C. Cytoplasmic lysates were immunoprecipitated with λ2-specific MAb 7F4 (α-λ2) or anticore serum (α-core). Proteins within the immunoprecipitates were resolved by SDS-PAGE, and viral capsid proteins were detected by phosphorimaging. A reference lane for ISVPs is also shown. Positions of viral proteins are indicated to the left and right. (f) Bands in each lane in panel e corresponding to the μ1 proteolytic fragment δ and core proteins λ1, λ2, and λ3 were quantitated by densitometry as described previously (14). Protein μ1 content within each lane was computed as a ratio of band volumes [δ/(λ1 + λ2 + λ3)] and is shown as a percentage of that obtained for the ISVP reference lane. A similar quantitative analysis was also carried out with cytoplasmic lysates prior to immunoprecipitation (no IP). Averages and standard deviations for two (α-λ2, 0 h) or three lanes from two independent experiments are shown.
Immunostaining and immunofluorescence (IF) microscopy.
Mv1Lu or CV-1 cells (2 × 105) were seeded on the day before infection in six-well plates containing 18-mm round glass coverslips. In most experiments, cells were preincubated with cycloheximide (CHX) (100 μg/ml) (Sigma-Aldrich) in DMEM for 1 h at 37°C to block protein synthesis. Nonpurified T1L ISVPs suspended in phosphate-buffered saline (PBS) supplemented with 2 mM MgCl2 (PBS-MC) (∼2.5 × 104 particles/cell, unless otherwise noted) were allowed to bind to cells on coverslips for 1 h at 4°C. Coverslips were then rinsed in PBS-MC and fixed immediately or placed in DMEM containing CHX and returned to 37°C. Infected cells were fixed by incubation with 2% paraformaldehyde for 10 to 15 min at room temperature and washed three times with PBS. Fixed cells were permeabilized by incubation with saponin (0.5%) in PBS supplemented with 0.5% bovine serum albumin (PBSA) for immunostaining of LAMP-1 or LAMP-2 or with Triton X-100 (0.1%) in PBSA for immunostaining of viral proteins. Permeabilized cells were incubated with primary antibodies for 30 min in PBSA, washed three times with PBS, and incubated with secondary antibodies for 30 min in PBSA. After three more washes with PBS, coverslips were mounted on glass slides with Prolong antifade reagent (Molecular Probes) and examined by using a TE-300 inverted microscope with fluorescence optics (Nikon, Melville, N.Y.). Wide-field epifluorescence images were collected as described previously (55) and prepared for presentation by using Adobe Photoshop and Illustrator (Adobe Systems, San Jose, Calif.). Confocal images were acquired with an Axiovert 200M inverted microscope (Carl Zeiss, Inc., Thornwood, N.Y.) under the control of SlideBook (Intelligent Imaging Innovations, Denver, Colo.). The microscope was equipped with a motorized filter turret and lens holder, a ×63 lens (Pan Apochromat, 1.4 NA; Carl Zeiss), and a spinning-disk confocal head (Perkin-Elmer, Wellesley, Mass.). Three-dimensional image stacks were recorded by acquisition of sequential sections recorded along the z axis by varying the position of the objective lens holder (step size, 0.3 μm). Images were processed with SlideBook (two-dimensional maximum-intensity projections). Prior to quantitation, images were deconvolved with the Near-Neighbors algorithm (SlideBook) by using calculated point spread functions (39).
Flow cytometry.
Untreated or CHX-pretreated Mv1Lu cells were suspended in PBS-MC at 2 × 107 cells/ml and incubated with ISVPs (∼5 × 104 particles/cell, unless otherwise noted) for 1 h at 4°C. After being washed with PBS to remove unbound virus, cells were suspended at a concentration of 106/ml in DMEM containing CHX and returned to 37°C. Samples were agitated every 10 to 15 min to keep cells in suspension and were removed to ice at various times. Infected cells were pelleted at 500 × g to remove growth medium and fixed, permeabilized, and immunostained in suspension as described above for cell monolayers. Immunostained cells were suspended in PBS, and data were collected with a FACScalibur flow cytometer and analyzed with CellQuest software (Beckton Dickinson, San Jose, Calif.). Isotype or preimmune serum controls were used to determine background fluorescence with each antibody.
α-Sarcin coentry assay.
The methods of Liprandi and colleagues (41) were used to measure virus-induced intoxication of cells by α-sarcin,but with several changes. Confluent L929 cell monolayers in 48-well microplates were preincubated in methionine-free DMEM (Invitrogen) for 2 h at 37°C and washed with ice-cold PBS-MC. Viral particles (ISVPs, ∼5.0 × 104 particles/cell; pr-cores+σ1, ∼5.0 × 105 particles/cell) were allowed to bind to cells for 1 h at 4°C. After two washes with ice-cold PBS-MC to remove unbound virus, prewarmed methionine-free DMEM supplemented with [35S]methionine-cysteine (22 μCi/ml) (Dupont NEN, Wilmington, Del.) and α-sarcin (50 μg/ml) (Sigma-Aldrich) was added. After incubation for various times at 37°C, cells were lysed and proteins were precipitated with ice-cold trichloroacetic acid (10%) for 1 h. Precipitates were washed with acetone, air dried, and solubilized with 1% SDS in 0.1 M NaOH. Acid-precipitable counts were measured by liquid scintillation counting to quantitate the incorporation of 35S-labeled amino acids into proteins.
SDS-PAGE and immunoblotting.
SDS-PAGE was performed with 10% acrylamide gels as described previously (51). Viral proteins were detected with Coomassie brilliant blue R-250. Gels loaded with radiolabeled proteins were dried on filter paper and visualized by phosphorimaging with the Storm system. For immunoblotting, proteins were transferred to nitrocellulose with a Mini-TransBlot system (Bio-Rad, Hercules, Calif.). μ1-specific mouse MAb 10F6 (1:1,000 dilution) and mouse-specific goat IgG conjugated to alkaline phosphatase (1:2,000 dilution) (Sigma-Aldrich) were used as primary and secondary antibodies, respectively. Antibody binding was detected with colorimetric reagents p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (Bio-Rad).
RESULTS
MAbs 4A3 and 10F6 detect the μ1 conformational change in vitro and at early times p.i.
To further explore the conformational change in μ1 that accompanies the ISVP→ISVP* transition, we assessed the capacity of four MAbs specific for the δ region of μ1 (70) to immunoprecipitate purified T1L ISVPs or ISVP*s in vitro. MAbs 10H2 and 8H6 precipitated similar numbers of ISVPs and ISVP*s, suggesting that their epitopes are similarly exposed and accessible in both particles (Fig. 1a). In contrast, MAbs 4A3 and 10F6 precipitated much smaller numbers of ISVPs than of ISVP*s (only 1 in 100 to 1 in 50 as many) (Fig. 1a), suggesting that their epitopes are accessible in ISVP*s but masked in ISVPs.
The capacity of MAbs 4A3 and 10F6 to recognize the δ region of μ1 in ISVP*s but not in ISVPs in vitro led us to hypothesize that they may be useful for detecting the μ1 conformational change during cell entry. To test this hypothesis, we allowed T1L ISVPs to adsorb to Mv1Lu cells at 4°C and later immunostained the samples with 10H2, 8H6, 4A3, or 10F6 (Fig. 1b to e). Before adsorption, we treated the cells with CHX to abolish protein synthesis and to ensure that only proteins in infecting particles were later detected by IF microscopy. When cells kept at 4°C after adsorption were stained with 10H2 (Fig. 1b) or 8H6 (Fig. 1c), we observed punctate staining concentrated at cell margins, suggesting that the MAbs were bound to ISVPs at the cell surface. We obtained similar results with σ1-specific MAb 5C6 (70) (data not shown). In contrast, we observed little staining with 4A3 (Fig. 1d) or 10F6 (Fig. 1e). These findings are consistent with the immunoprecipitation results (Fig. 1a) and provide further evidence that 10H2 and 8H6 recognize the μ1 conformer in ISVPs, whereas 4A3 and 10F6 do so only poorly.
When ISVP-adsorbed cells were warmed to 37°C and immunostained with 10H2 (Fig. 1b) or 8H6 (Fig. 1c) at 2 h p.i., punctate staining was still observed, suggesting that the MAbs were bound to the δ region of μ1 in viral particles before or after uptake from the cell surface. Interestingly, some diffuse cytoplasmic and nuclear staining also was seen in many cells. More striking was the dramatic induction of staining by 4A3 (Fig. 1d) or 10F6 (Fig. 1e) at 2 h p.i. This staining was largely diffuse and distributed throughout the cytoplasm and nucleus, although some punctate staining also was noted. We conclude from these results that μ1 changes conformation at early times p.i., such that a large proportion of its δ region can be newly recognized by 4A3 and 10F6. We also conclude that much of this conformationally altered protein region is no longer particle bound, since its staining is diffuse and not confined to punctate spots. In addition, the presence of 4A3 and 10F6 staining in the cytoplasm and nucleus strongly suggested that a substantial fraction of the δ fragment had successfully penetrated the cellular membrane barrier.
Anticore serum detects changes in particle structure in vitro and at early times p.i.
In the course of the present experiments, we found that a polyclonal antiserum raised against reovirus cores (15) also differs in its capacity to recognize ISVPs and ISVP*s. This antiserum precipitated similar numbers of ISVP*s and cores but only small numbers of ISVPs in vitro (1 in 50 to 1 in 25 as many ISVPs as ISVP*s or cores) (Fig. 2a). These findings suggested that core epitopes are largely masked in ISVPs but become more accessible upon changes in particle structure during the ISVP→ISVP* and ISVP*→core transitions. Anticore serum therefore may be another useful reagent for detecting changes in particle structure during cell entry. To test this possibility, we allowed T1L ISVPs to adsorb to CHX-treated Mv1Lu cells as described above and later immunostained the samples with anticore serum (Fig. 2b and c). Little staining was observed in cells kept at 4°C, confirming that anticore serum binds poorly to ISVPs at the cell surface (Fig. 2b). When the cells were warmed to 37°C and immunostained with anticore serum, however, a dramatic induction of staining was seen. This staining was exclusively punctate in nature (Fig. 2b and c) and colocalized with staining for core protein λ2 by λ2-specific MAb 7F4 (Fig. 2c). We conclude that infecting ISVPs undergo changes in particle structure at early times p.i., such that they can be newly recognized by antibodies in anticore serum.
Changes in particle structure, including the μ1 conformational change, precede viral protein synthesis.
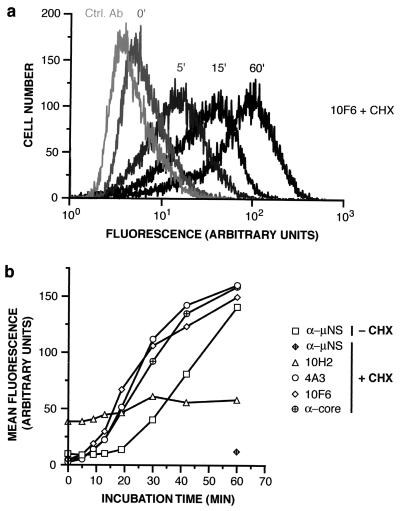
We hypothesized that the structural changes exhibited by ISVPs within 2 h p.i. may be linked to productive cell entry. As a test, we compared the kinetics of changes detected by MAb 4A3 or 10F6 or by anticore serum with those of a marker for productive cell entry, the synthesis of viral nonstructural protein μNS. T1L ISVPs were allowed to adsorb to CHX-treated or untreated Mv1Lu cells at 4°C, and cells were prepared for immunostaining either immediately or after incubation at 37°C for various times. Untreated samples were immunostained with anti-μNS serum (10). All samples were then analyzed by flow cytometry. Induction of staining with 4A3, 10F6, or anticore serum in CHX-treated cells occurred rapidly and with similar kinetics (0 to 10 min p.i.) (Fig. 3), suggesting that the μ1 conformational change and particle disassembly are contemporaneous. The induction of μNS synthesis in untreated cells lagged the structural changes by ∼20 min (Fig. 3b), consistent with the hypothesis that structural changes are associated with penetration of viral particles into the cytoplasm and/or derepression of transcriptase complexes within the particles.
FIG. 3.
Kinetics of changes in particle structure and onset of viral protein synthesis within infected cells. ISVPs were allowed to adsorb to untreated or CHX-pretreated Mv1Lu cells in suspension at 4°C, and cells were fixed and permeabilized at different times p.i. at 37°C. CHX-treated cells were immunostained with μ1-specific MAb 10H2, 4A3, or 10F6 or with anticore serum (α-core), followed by goat anti-mouse or goat anti-rabbit IgG conjugated to Alexa 488. Both untreated and CHX-treated cells were immunostained with anti-μNS serum (α-μNS), followed by goat anti-rabbit IgG conjugated to Alexa 488. Cells were analyzed by flow cytometry. (a) Representative histograms. Ctrl. Ab, isotype-matched control antibody. Numbers above each histogram indicate the time in minutes p.i. (b) The geometric mean of each histogram was used as a measure of antibody binding, after subtraction of the value obtained with control antibody. Averages from two independent experiments are shown.
IF microscopy evidence for the release of δ and σ1 from viral particles during cell entry.
To characterize the particles detected by anticore serum at early times p.i. in the absence of protein synthesis, we infected CHX-treated Mv1Lu cells with T1L ISVPs and later immunostained the samples with anticore serum together with MAbs specific for the δ region of μ1 or σ1. At 2 h p.i., we observed colocalization in only a few punctate spots with anticore serum and δ-specific MAb 10H2 (Fig. 4a), 4A3 (Fig. 4b), or 10F6 (data not shown), suggesting that most of the particles recognized by anticore serum contained little δ. In addition, the punctate staining detected with anticore serum largely did not colocalize with that detected with σ1-specific MAb 5C6, suggesting that the particles also contained little σ1 (Fig. 4c). We considered it possible that the particles resembling cores might have been generated by proteolysis of ISVPs or ISVP*s that failed to penetrate the cytoplasm and were instead trapped within hydrolytic compartments of the endocytic pathway. To test this possibility, we coimmunostained ISVP-infected cells with anticore serum and antibodies specific for the late endosomal and lysosomal proteins LAMP-1 and LAMP-2 (11, 44). At 2 h p.i., little colocalization was observed between the particles and LAMP-1-positive (data not shown) or LAMP-2-positive (Fig. 4d) compartments, leading us to conclude that particles that resemble cores and that are produced during cell entry are not associated with late endosomes or lysosomes and likely are localized to the cytoplasm (see Discussion for other results).
Immunoprecipitation evidence for the release of δ and σ1 from viral particles during cell entry.
We used immunoprecipitation as a further test of whether particles resembling cores are generated from infecting ISVPs at early times p.i. [35S]methionine-cysteine-labeled ISVPs were allowed to adsorb to CHX-treated Mv1Lu cells at 4°C, and the cells were lysed with nonionic detergent either immediately or after incubation at 37°C for 2 h. Postnuclear cell extracts were incubated with anticore serum or λ2-specific MAb 7F4, and antibody-protein complexes were affinity purified and analyzed by SDS-PAGE. We found that anticore serum precipitated about 10-fold more particles from the 2-h cell extract than from the 0-h cell extract (Fig. 4e and data not shown), consistent with its increased immunostaining of cells incubated for 2 h at 37°C relative to those kept at 4°C (Fig. 2b). A more modest (about twofold) increase was observed with 7F4 (Fig. 4e and data not shown). Densitometric quantitation of core proteins and δ (Fig. 4f) and visual comparison of σ1 levels in Fig. 4e further indicated that particles precipitated from the 2-h cell extract by anticore serum or 7F4 contained much less δ and σ1 than purified ISVPs, whereas particles precipitated from the 0-h cell extract resembled ISVPs in protein composition (Fig. 4f). These results provide additional evidence that both δ and σ1 are lost from viral particles during cell entry.
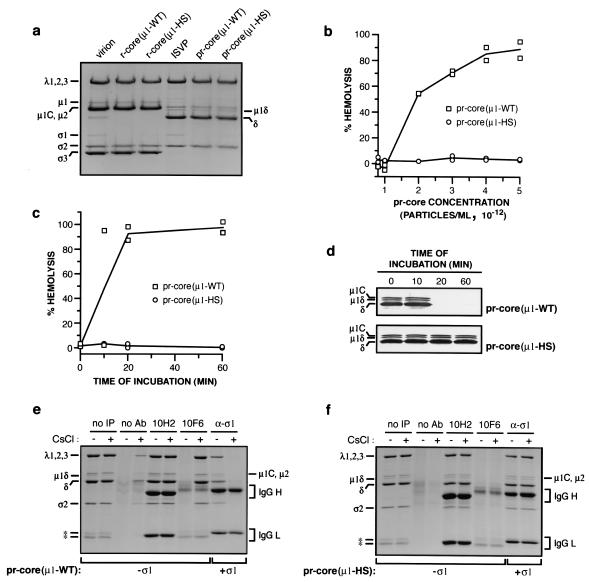
ISVP-like particles derived from recoated cores containing mutant μ1-HS are defective in hemolysis and do not undergo the ISVP→ISVP* transition at 37°C in vitro.
Having observed structural changes that precede viral protein synthesis at early times p.i. with ISVPs, we sought further evidence that these changes are linked to productive cell entry. To this end, we obtained and characterized hyperstable ISVPs that convert to ISVP*s only poorly at physiological temperatures in vitro. Previous work provided clues for engineering such particles. ISVPs derived from ethanol-resistant viral clones containing mutations in μ1 (e.g., A319E) and ISVP-like particles derived from recoated cores containing μ1 with the P344L and L359F mutations exhibit quantitative defects in in vitro membrane permeabilization (15, 35, 71; K. Chandran and M. L. Nibert, unpublished data). Because virus-induced permeabilization of erythrocytes requires ISVP*-like particles (13), we reasoned that these mutations may act by slowing the ISVP→ISVP* transition and may cause a more severe defect if combined in the same protein. Accordingly, we engineered μ1-HS together with wild-type σ1 and generated recoated cores containing μ1-HS or μ1-WT [r-cores(μ1-HS) and r-cores(μ1-WT), respectively). r-cores(μ1-HS) resembled r-cores(μ1-WT) in protein composition (Fig. 5a), three-dimensional structure determined by scanning electron cryomicroscopy (12) (data not shown), and capacity to convert to ISVP-like particles [pr-cores(μ1-HS)] upon chymotrypsin treatment (Fig. 5a), indicating that the mutations in μ1-HS caused no gross defects in viral structure.
FIG. 5.
Capacities of ISVP-like particles derived from recoated cores to mediate the lysis of erythrocytes and to undergo ISVP→ISVP* change in vitro. (a) Purified virions, r-cores(μ1-WT), or r-cores(μ1-HS) (8 × 1012 particles/ml) were converted to their respective ISVPs or ISVP-like particles [pr-cores(μ1-WT) or pr-cores(μ1-HS)] by digestion with chymotrypsin for 15 min at 37°C. Viral capsid proteins were resolved by SDS-PAGE and visualized by Coomassie brilliant blue R-250 staining. Positions of viral proteins are indicated at the left and right. (b) Capacities of nonpurified pr-cores(μ1-WT) and pr-cores(μ1-HS) to mediate hemolysis, determined as previously described for ISVPs (13). Washed citrated bovine calf erythrocytes (3% [vol/vol]) were incubated with increasing concentrations of viral particles in virion buffer for 1 h at 37°C, and the extent of hemoglobin release into the supernatant was measured relative to that in controls containing only virion buffer and erythrocytes (0%) or water and erythrocytes (100%). Values from two independent experiments are shown. (c) Kinetics of hemolysis mediated by pr-cores(μ1-WT) and pr-cores(μ1-HS). Erythrocytes were incubated with viral particles (4 × 1012 per ml) for different times at 37°C, and the extent of hemolysis was determined as described for panel b. Values from two independent experiments are shown. (d) Conformational status of protein μ1. Aliquots (10 μl) from the hemolysis reactions in panel c were incubated with trypsin (100 μg/ml) for 30 min at 4°C, digestion was stopped by the addition of soybean trypsin inhibitor (300 μg/ml), and viral capsid proteins were resolved by SDS-PAGE. Protein μ1 was visualized by immunoblotting with μ1-specific MAb 10F6. A portion of each immunoblot containing the region of interest is shown. Positions of viral proteins are indicated at the left. (e and f) Capacities of pr-cores±σ1(μ1-WT) and pr-cores±σ1(μ1-HS) to undergo conversion to ISVP*-like particles. pr-cores(μ1-WT) (e) and pr-cores(μ1-HS) (f) (4 × 1012 per ml) were incubated with 300 mM NaCl (−) or CsCl (+) for 20 min at 37°C to inhibit or promote conversion to ISVP*-like particles, respectively (see the text for details). Viral particles were immunoprecipitated with protein A-conjugated magnetic beads alone (no Ab) or beads bound to 10H2 or 10F6. pr-cores+σ1(μ1-WT) (e) andpr-cores+σ1(μ1-HS) (f) were prepared and incubated with NaCl or CsCl as described above and immunoprecipitated with σ1-specific MAb 5C6 (α-σ1). Viral capsid proteins bound to beads were resolved by SDS-PAGE and visualized by Coomassie brilliant blue R-250 staining. Positions of viral proteins are indicated at the left and right. IgG H, IgG heavy chain; IgG L, IgG light chain; no IP, no immunoprecipitation (see the legend to Fig. 4). Asterisks indicate positions of protease bands.
We next monitored the capacity of pr-cores(μ1-WT) and pr-cores(μ1-HS) to lyse erthyrocytes at 37°C. pr-cores(μ1-WT) induced hemolysis at concentrations above 1012 particles per ml, whereas no hemolysis was detected with pr-cores(μ1-HS), even at 5- to 10-fold-higher concentrations of viral particles (Fig. 5b and data not shown). Assessment of the conformational status of μ1-WT and μ1-HS within hemolysis reactions by protease treatment at 4°C confirmed that the onset of hemolysis was temporally correlated with the μ1→μ1* change, as described previously (13), and provided evidence that pr-cores(μ1-HS) failed to induce hemolysis because they did not undergo the μ1→μ1* change under the conditions used (Fig. 5c and d).
We also used μ1-specific MAbs to assess the capacity of pr-cores(μ1-HS) to undergo the μ1→μ1* change. pr-cores(μ1-WT) and pr-cores(μ1-HS) were incubated under conditions that favored either the maintenance of ISVPs or the conversion to ISVP*s. Samples were then immunoprecipitated with MAb 10H2 or 10F6 and resolved by SDS-PAGE and Coomassie brilliant blue R-250 staining. pr-cores(μ1-WT) preincubated with NaCl were recognized by 10H2 but not by 10F6, whereas pr-cores(μ1-WT) preincubated with CsCl were recognized by both MAbs, consistent with a Cs+-accelerated μ1→μ1* change in viral particles (13) (Fig. 5e). pr-cores(μ1-HS) preincubated with NaCl produced a pattern of MAb recognition similar to that produced by pr-cores(μ1-WT), providing additional evidence that particle-bound μ1-HS resembles μ1-WT in overall protein conformation (Fig. 5f). In contrast to pr-cores(μ1-WT), however, pr-cores(μ1-HS) preincubated with CsCl were recognized by 10H2 but not by 10F6, supporting the conclusion that they undergo the μ1→μ1* change very slowly at 37°C (Fig. 5f).
In native ISVPs, the μ1→μ1* change is accompanied by σ1 release from particles (13) (Fig. 5f). To test whether recoated particles containing μ1-HS exhibited σ1 release even in the absence of a detectable μ1→μ1* change, we preincubated pr-cores+σ1(μ1-WT) and pr-cores+σ1(μ1-HS) with NaCl or CsCl, immunoprecipitated the samples with σ1-specific MAb 5C6, and analyzed the precipitated proteins as described above (Fig. 5e and f). pr-cores+σ1(μ1-WT) preincubated with NaCl were precipitated by 5C6, whereas pr-cores+σ1(μ1-WT) preincubated with CsCl were not, indicating the release of σ1 in association with the μ1→μ1* change, as expected (Fig. 5e). In contrast, pr-cores+σ1(μ1-HS) preincubated with either NaCl or CsCl were precipitated by 5C6, indicating that they had not lost σ1 and providing further evidence that σ1 release from particles requires the μ1→μ1* change (Fig. 5f).
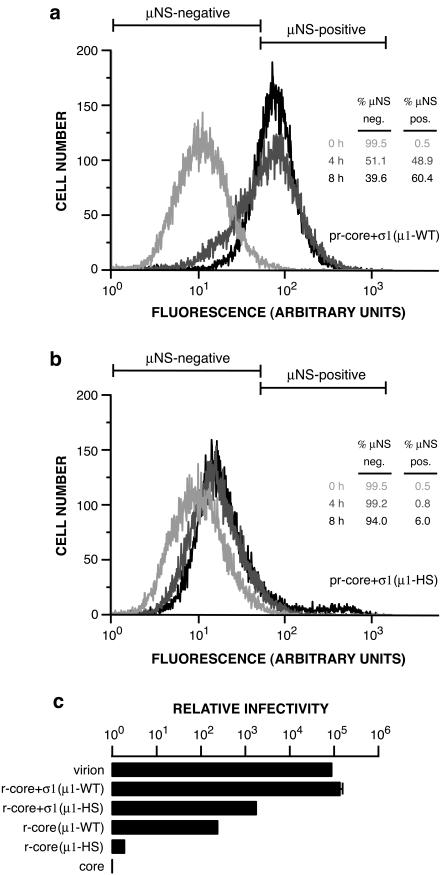
pr-cores+σ1(μ1-HS) are defective in cell entry.
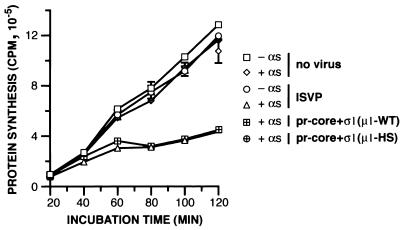
Because pr-cores±σ1(μ1-HS) are defective in undergoing the ISVP→ISVP* transition and in mediating membrane permeabilization in vitro, we predicted that they also would be defective in productive cell entry. To test this prediction, we allowed pr-cores+σ1(μ1-WT) or pr-cores+σ1(μ1-HS) to adsorb to Mv1Lu cells at 4°C and then fixed and immunostained the cells with anti-μNS serum either immediately or after incubation at 37°C for 4 or 8 h. The stained cells were analyzed by flow cytometry to measure viral protein synthesis. A large increase in anti-μNS serum staining was seen in cells infected with pr-cores+σ1(μ1-WT) by 4 h p.i., with an even larger increase by 8 h p.i. (Fig. 6a). In contrast, only low levels of μNS synthesis were apparent by 4 or 8 h p.i. in cells infected with pr-cores+σ1(μ1-HS) (Fig. 6b). These results demonstrated that the levels of productive cell entry and/or induction of viral protein synthesis achieved by pr-cores+σ1(μ1-HS) were much lower than those achieved by pr-cores+σ1(μ1-WT) at the same times.
FIG. 6.
Capacities of r-cores+σ1(μ1-WT), r-cores+σ1(μ1-HS), and their respective pr-cores+σ1 to synthesize viral proteins and produce viral progeny within cells. (a and b) pr-cores+σ1(μ1-WT) (a) or pr-cores+σ1(μ1-HS) (b) were allowed to adsorb to Mv1Lu cells in 60-mm dishes at 4°C, and cells were suspended, washed, and fixed at 0, 4, or 8 h p.i. at 37°C. Cells were permeabilized and immunostained with anti-μNS serum, followed by goat anti-mouse IgG conjugated to Alexa 488, and analyzed by flow cytometry. Protein μNS-negative (neg.) and μNS-positive (pos.) regions for each set of histograms were set so that 99.5% of cells fixed at 0 h were designated μNS negative. The tables (insets) indicate the percentages of μNS-negative and μNS-positive cells at each time point. (c) Infectivities of purified virions, r-cores+σ1(μ1-WT), r-cores+σ1(μ1-HS), r-cores(μ1-WT), and r-cores(μ1-HS) in L929 cells relative to their parent cores (infectivity set to 1), as determined by a plaque assay. Averages and standard deviations from three trials are shown.
To determine whether viral particles containing μ1-HS have reduced infectivity, as the protein synthesis findings would infer, we examined the capacities of pr-cores±σ1(μ1-WT) and pr-cores±σ1(μ1-HS) to form plaques on L929 cell monolayers. In each case, we normalized the infectivity value to reflect the efficiency of plaque formation on a per-particle basis. pr-cores+σ1(μ1-WT) resembled native ISVPs in relative infectivity, with an enhancement over parent cores in excess of 100,000 (Fig. 6c). In contrast, pr-cores+σ1(μ1-HS) were only 0.01 times as infectious as pr-cores+σ1(μ1-WT) (Fig. 6c). This difference was also reproducible with r-cores lacking σ1 (Fig. 6c). These findings indicate that viral particles containing μ1-HS have greatly reduced infectivity, likely because the mutations in μ1 compromise their capacity to mediate productive cell entry.
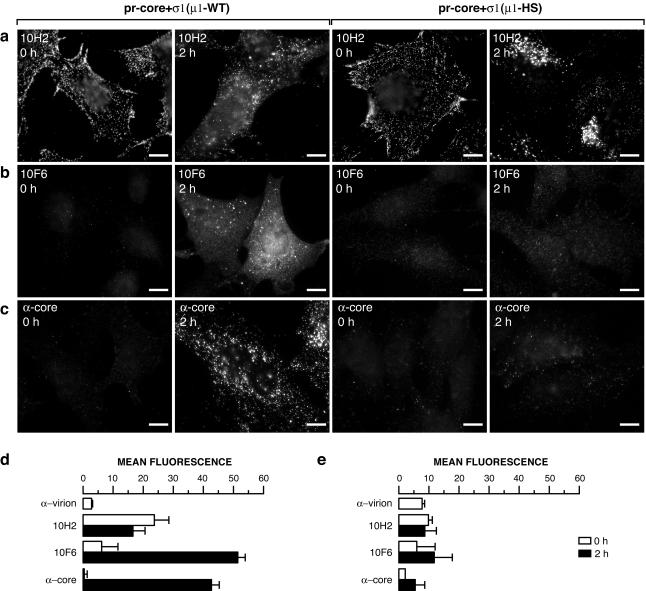
pr-cores+σ1(μ1-HS) do not undergo the μ1 conformational change and conversion to particles resembling cores by 2 h p.i.
In an effort to pinpoint the defect of pr-cores+σ1(μ1-HS) in cell entry, we allowed pr-cores+σ1(μ1-WT) or pr-cores+σ1(μ1-HS) to adsorb to CHX-treated Mv1Lu cells; later, we immunostained the cells with MAb 10H2 or 10F6 or anticore serum and analyzed the staining patterns by IF microscopy (Fig. 7a to c) or flow cytometry (Fig. 7d and e). In cells adsorbed to either particle type and then kept at 4°C, we observed a pattern of staining with all three antibodies similar to that in ISVP-adsorbed cells. Specifically, 10H2 showed punctate staining concentrated at cell margins, indicating that both pr-cores+σ1(μ1-WT) and pr-cores+σ1(μ1-HS) were bound to Mv1Lu cells and detected by 10H2 at the cell surface (Fig. 7a). The capacity of both particle types to bind to cells was confirmed by flow cytometric quantitation of cell-associated fluorescence following immunostaining with 10H2 or a virion-specific antiserum (Fig. 7d and e). In contrast to 10H2, 10F6 (Fig. 7b, d, and e) and anticore serum (Fig. 7c to e) showed little staining, consistent with the limited capacities of these antibodies to immunoprecipitate pr-cores+σ1(μ1-WT) and pr-cores+σ1(μ1-HS) in vitro (Fig. 5e and f and data not shown for anticore serum).
FIG. 7.
Induction of μ1 conformational change and generation of particles resembling cores within cells infected with pr-cores+σ1(μ1-WT) or pr-cores+σ1(μ1-HS). (a to c) pr-cores+σ1(μ1-WT) or pr-cores+σ1(μ1-HS) were allowed to adsorb to CHX-pretreated Mv1Lu cells at 4°C, and cells were fixed at 0 or 2 h p.i. at 37°C. Cells were permeabilized and immunostained with μ1-specific MAb 10H2 (a) or 10F6 (b) or with anticore serum (α-core) (c), followed by goat anti-mouse IgG or goat anti-rabbit IgG conjugated to Alexa 488. Bars, 10 μm. (d and e) Suspension cultures of CHX-pretreated Mv1Lu cells were infected and immunostained as described above and analyzed by flow cytometry. The geometric mean of each histogram was used as a measure of antibody binding, after subtraction of the value obtained with control antibody. Averages and standard deviations from three independent experiments are shown.
Qualitatively different 10H2 staining patterns between pr-cores+σ1(μ1-WT) and pr-cores+σ1(μ1-HS) were observed in particle-adsorbed cells warmed to 37°C and fixed at 2 h p.i. In cells infected with pr-cores+σ1(μ1-WT), 10H2 showed a combination of punctate and diffuse staining (Fig. 7a) resembling that in ISVP-infected cells (Fig. 1b). In contrast, 10H2 staining in cells infected with pr-cores+σ1(μ1-HS) was almost entirely punctate and was markedly concentrated in perinuclear regions (Fig. 7a). Similar punctate staining, concentrated in perinuclear regions, was observed with σ1-specific MAb 5C6 as well (data not shown). These findings suggested different intracellular fates for particles containing μ1-WT and μ1-HS.
To test for the intracellular μ1 conformational change, infected cells warmed to 37°C and fixed at 2 h p.i. were immunostained with 10F6. A dramatic induction of 10F6 staining was observed by both IF microscopy (Fig. 7b) and flow cytometry (Fig. 7d) in cells infected with pr-cores+σ1(μ1-WT), indicating that these particles undergo the μ1 conformational change shortly after infection, as was also found with ISVPs (Fig. 1e). In contrast, little increase in 10F6 staining was seen in cells infected with pr-cores+σ1(μ1-HS), suggesting that the latter particles do not undergo the μ1 conformational change to an appreciable degree by 2 h p.i. (Fig. 7b and e).
In cells infected with pr-cores+σ1(μ1-WT), 10F6 showed a combination of diffuse and punctate staining (Fig. 7b), as was also found with ISVPs (Fig. 1e). The diffuse staining provided evidence for the disassembly of δ from particles and suggested that particles resembling cores are generated in cells infected with pr-cores+σ1(μ1-WT). Two further observations supported that conclusion. First, a large induction in immunostaining with anticore serum was observed by both IF microscopy and flow cytometry (Fig. 7c and d). Second, coimmunostaining with λ2-specific MAb 7F4 and either μ1-specific MAb 10H2 or σ1-specific polyclonal antiserum showed many punctate spots containing core protein λ2 but neither the δ region of μ1 (Fig. 8a) nor σ1 (data not shown). In contrast to the results obtained with pr-cores+σ1(μ1-WT), little induction of immunostaining with anticore serum was seen in cells infected with pr-cores+σ1(μ1-HS) (Fig. 7c and e), and few punctate spots containing core proteins but lacking δ (Fig. 8b) or σ1 (data not shown) were seen. Taken together, these results provided evidence that few pr-cores+σ1(μ1-HS) undergo the μ1 conformational change or conversion to particles resembling cores by 2 h p.i.
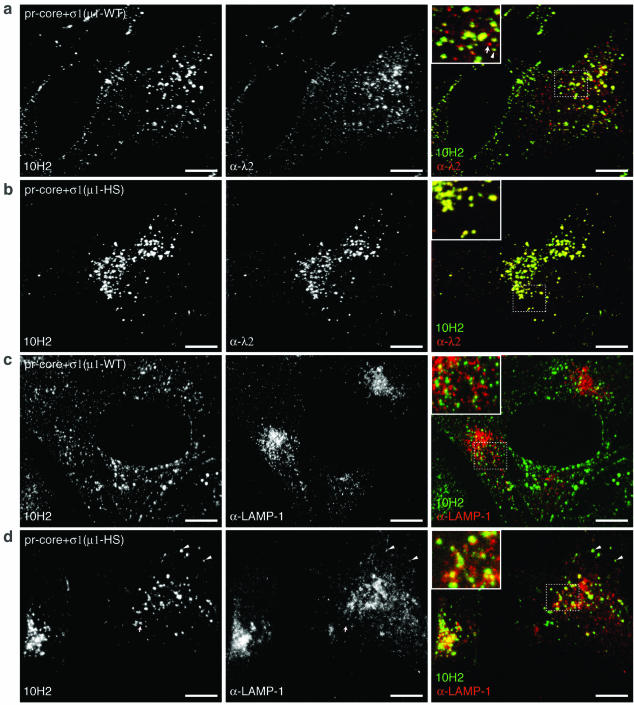
FIG. 8.
Localization of viral particles, the δ region of μ1, and LAMP-1-positive vacuoles in cells infected with pr-core+σ1(μ1-WT) or pr-core+σ1(μ1-HS). pr-cores+σ1(μ1-WT) (a and c) or pr-cores+σ1(μ1-HS) (b and d) were allowed to adsorb to CHX-pretreated Mv1Lu cells at 4°C, and cells were fixed at 2 h p.i. at 37°C. (a and b) Cells were permeabilized and immunostained with λ2-specific MAb 7F4 (α-λ2), followed by goat anti-mouse IgG conjugated to Alexa 594 and then by μ1-specific MAb 10H2 directly conjugated to Alexa 488. The arrow in the panel a inset indicates a punctate spot detected by 7F4 but not by 10H2. The arrowhead indicates a punctate spot detected by both 7F4 and 10H2. (c and d) Cells were permeabilized and coimmunostained with a LAMP-1-specific polyclonal antiserum (α-LAMP-1) and 10H2, followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. Arrowheads in panel d indicate punctate spots that were detected by 10H2 and that colocalized with LAMP-1-positive structures. Images were captured by spinning-disk confocal microscopy and processed as described in Materials and Methods. Bars, 20 μm.
ISVP-like particles accumulate within LAMP-positive endocytic compartments in cells infected with pr-cores+σ1(μ1-HS).
The marked concentration of pr-cores+σ1(μ1-HS) in perinuclear regions (Fig. 7a and 8b) led us to suspect that these particles are defective in penetrating into the cytoplasm and consequently are trapped within the endocytic pathway. To test this possibility, we infected CHX-pretreated Mv1Lu cells with pr-cores+σ1(μ1-WT) or pr-cores+σ1(μ1-HS) for 2 h at 37°C and later coimmunostained the samples with μ-specific MAb 10H2 and LAMP-1-specific antiserum (11) (Fig. 8c and d). Confocal microscopy revealed limited colocalization between 10H2 staining and anti-LAMP-1 staining in cells infected with pr-cores+σ1(μ1-WT) (Fig. 8c). Much more extensive colocalization of 10H2 staining and anti-LAMP-1 staining was observed in cells infected with pr-cores+σ1(μ1-HS) (Fig. 8d) [approximately three times the area of colocalization for pr-cores+σ1(μ1-HS) as for pr-cores+σ1(μ1-WT)]. Furthermore, colocalization of staining for 10H2 and anti-σ1 serum in perinuclear regions was essentially complete in cells infected with pr-cores+σ1(μ1-HS) (data not shown). In summary, these findings strongly suggested that ISVP-like particles containing μ1 and σ1 accumulated within LAMP-positive compartments (i.e., late endosomes and/or lysosomes) in infections with pr-cores+σ1(μ1-HS).
pr-cores+σ1(μ1-HS) are defective in cellular membrane permeabilization during entry.
The behaviors of pr-cores+σ1(μ1-HS) described above consistently suggested that they are defective in membrane penetration during cell entry. As a further test of this conclusion, we assessed the capacity of pr-cores+σ1(μ1-HS) to mediate cytoplasmic delivery of the ribonucleotoxin α-sarcin in trans, a property previously shown to be associated with productive entry by reovirus and rotavirus particles (41, 46). Inhibition of protein synthesis was used to monitor the extent of α-sarcin entry (41). In control series, incubation of L929 cells with ISVPs alone or α-sarcin alone had little effect on protein synthesis, even after 2 h at 37°C (Fig. 9). However, incubation with ISVPs plus α-sarcin led to a shutoff of protein synthesis within 30 to 60 min p.i. at 37°C, consistent with the entry of toxin into the cytoplasm along with penetrating viral particles (Fig. 9). As expected, pr-cores+σ1(μ1-WT) resembled ISVPs in their capacity to potentiate α-sarcin intoxication of cells (Fig. 9). In contrast, pr-cores+σ1(μ1-HS) caused little increase in cellular intoxication by 2 h p.i. (Fig. 9), supporting the conclusion that they are defective in membrane penetration during cell entry.
FIG. 9.
Capacities of ISVPs, pr-cores+σ1(μ1-WT), and pr-cores+σ1(μ1-HS) to potentiate α-sarcin (αs) intoxication of L929 cells. ISVPs, pr-cores+σ1(μ1-WT), or pr-cores+σ1(μ1-HS) were allowed to adsorb to L929 cells in 24-well plates at 4°C, and cells were washed and incubated in methionine-free growth medium containing [35S]methionine-cysteine and with or without α-sarcin (50 μg/ml) for different times at 37°C. Proteins were then precipitated with trichloroacetic acid, and acid-precipitable radioactivity was measured by scintillation counting. Averages and standard deviations from three trials are shown.
DISCUSSION
Changes that characterize the ISVP→ISVP* transition in vitro.
This article is a follow-up to an earlier report on structural changes in ISVPs that precede membrane permeabilization in vitro (13). To the published data, we added new in vitro evidence from immunoprecipitations with purified particles, corroborating the conformational change in putative membrane penetration protein μ1 and the release of adhesin σ1 that accompany the ISVP→ISVP* transition. Immunoprecipitations also revealed an increase in the exposure of core epitopes in ISVP*s. We made use of the core recoating approach (15, 16) to show that particles containing a hyperstable mutant form of μ1 exhibit none of the structural and functional changes that characterize the ISVP→ISVP* transition in vitro. The broad in vitro effects of this single type of mutant μ1 provided new genetic evidence that μ1 plays a key role in the cascade of programmed disassembly events and their functional consequences.
Changes in ISVPs that accompany cell entry.
In this study, we demonstrated that features of the ISVP→ISVP* transition in vitro attend cell entry. Structural features included the conformational change in μ1, exposure of core epitopes, and shedding of σ1. A further structural change, release of the δ fragment of μ1, is discussed below. Functional features of the ISVP→ISVP* transition—membrane permeabilization and transcriptase activation—were also shown to accompany cell entry. Membrane permeabilization was indicated by the cytoplasmic localization of both particles resembling cores and free δ in CHX-treated cells, as well as by cellular intoxication following α-sarcin coentry in the absence of CHX. Recent demonstrations of entering particles, resembling cores, being recruited to cytoplasmic inclusions of reovirus nonstructural protein μNS have provided further evidence for the cytoplasmic localization of these particles (T. J. Broering, J. Kim, C. L. Miller, M. L. Nibert, and J. S. L. Parker, submitted for publication). Transcriptase activation was indicated by the onset of viral protein synthesis in cells not treated with CHX. As in the in vitro experiments, the consistently “inert” behavior of recoated particles containing hyperstable μ1 established a genetic link between the structural and functional changes that accompanied cell entry by particles containing μ1-WT.
Hypothesis that ISVP*-like particles are a transient cell entry intermediate.
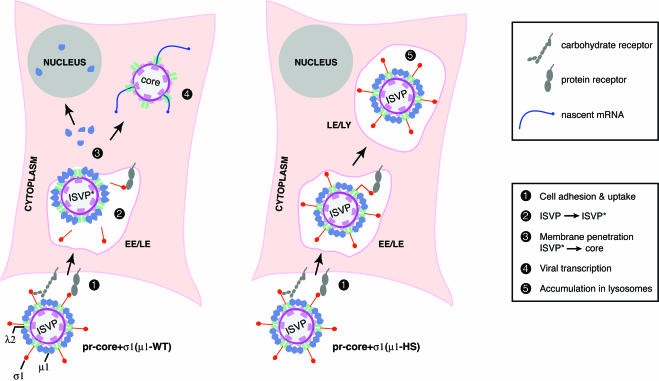
We detected few particles that corresponded directly to ISVP*s inside infected cells. Nevertheless, previous and present findings indicated that ISVP*s are generated and are a necessary entry intermediate (Fig. 10). (i) Conversion of ISVPs to cores in vitro, with the attendant disassembly of σ1 and μ1, must proceed through an ISVP* intermediate (13). (ii) ISVP*s, but not ISVPs or cores, can permeabilize erythrocyte membranes in vitro (13). (iii) Recoated particles containing mutant μ1 defective in ISVP→ISVP* conversion in vitro undergo intracellular changes in particle structure only at low levels and are defective in membrane penetration and productive cell entry (Fig. 5 to 9) (K. Chandran, A. L. Odegard, M. A. Agosto, K. S. Myers, and M. L. Nibert, unpublished data). A probable explanation for the absence of large numbers of ISVP*-like particles in infected cells is that they are quickly converted to a “downstream” particle form, i.e., cores, and therefore do not accumulate (Fig. 10) (see below for further discussion). This hypothesis suggests that ISVP→ISVP* conversion is a rate-limiting step in cell entry by wild-type ISVPs.
FIG. 10.
Updated model for reovirus entry. Whether productive infection by ISVPs can be initiated by penetration at the plasma membrane remains unresolved and is not illustrated here. Wild-type ISVPs and pr-cores+σ1(μ1-WT) are converted to ISVP*-like particles following cell adhesion and/or uptake. This step is characterized by conformational changes in μ1 and λ1, release of σ1 from viral particles, and derepression of core transcriptases. ISVP*-like particles initiate membrane penetration by inserting viral protein sequences into a cellular lipid bilayer. During or following membrane penetration, protein μ1 is removed from ISVP*s by the action of an unknown cellular factor(s), yielding particles that resemble cores. The δ fragment of released μ1 localizes to the cytoplasm and nucleus. Particles within the cytoplasm initiate viral mRNA synthesis. Unlike their μ1-WT-containing counterparts, pr-cores+σ1(μ1-HS) fail to be converted to ISVP*-like particles, to penetrate the cytoplasm, and to undergo the release of μ1. These mutant ISVP-like particles accumulate within late endosomes or lysosomes (LE/LY). EE, early endosomes.
Accumulation of viral particles in LAMP-positive compartments.
In most cells infected with recoated particles containing hyperstable μ1, features of the ISVP→ISVP* transition or productive cell entry were not observed, and ISVP-like particles localized extensively to LAMP-positive vacuoles. We postulate that because these particles failed to penetrate from the plasma membrane or an early endocytic compartment, they were passed along the endocytic pathway to later, LAMP-containing compartments, i.e., late endosomes or lysosomes (11, 44) (Fig. 10). Alternatively, the particles may normally penetrate from a LAMP-positive vacuole and accumulate there if defective in penetration. Yet another possibility is that one or more of the mutations in hyperstable μ1 affects trafficking of the particles to particular subcellular locations. For example, some of the entry-linked structural changes may signal specific trafficking events within cells. Work with other mutant forms of μ1 is needed to address these possibilities, but the present study suggests that analysis of the subcellular localization of particles may be a key element in future studies of membrane penetration by reoviruses.
Nature of the μ1 conformational change during the ISVP→ISVP* transition.
The in vitro immunoprecipitation results identified new reagents for tracking the μ1 conformational change both in vitro and in cells. MAbs 4A3 and 10F6 displayed a greater affinity for μ1* (in ISVP*s) than for μ1 (in ISVPs), whereas MAbs 10H2 and 8H6 bound to the two conformers with similar affinities (Fig. 1). Immunoblotting and competition ELISA experiments previously indicated that all four MAbs bind to the δ region of μ1 (residues 43 to 581) and that 4A3, 10H2, and 8H6 specifically bind to the β-barrel upper domain (residues 312 to 506) (34, 70). In contrast, the 10F6 epitope is contained within more C-terminal sequences of δ that form part of an α-helical “neck” region (residues 507 to 581) (34, 70). We postulate that 4A3 and 10F6 detect refolding and/or exposure of sequences in the upper parts of the μ1 trimer during the μ1→μ1* change. Such refolded or exposed sequences may participate directly in membrane permeabilization or may form part of a cascade of conformational changes that mobilize membrane-seeking regions from other parts of μ1 (see below).
The inert behavior of recoated particles containing hyperstable μ1 also provided clues to the nature of the μ1→μ1* change. μ1-HS contains mutations at two positions in the upper domain (residues 319 and 359) that, when separately present in native or recoated particles, confer mildly enhanced thermostability and reduced propensity to undergo the ISVP→ISVP* transition (35, 71; K. S. Myers and M. L. Nibert, unpublished data). Analysis of μ1 sequence changes in panels of thermostable viral mutants revealed that many of these changes are located in the δ region at intra- or intertrimer interfaces between the upper domains, suggesting that the μ1→μ1* change involves some degree of trimer dissociation (35, 71; J. K. Middleton, M. A. Agosto, K. S. Myers, J. Yin, and M. L. Nibert, unpublished data). A recent study suggested that such dissociation in upper portions of the trimer may precede wider rearrangements in μ1 involved in membrane permeabilization, including release of the N-myristoylated N-terminal peptide μ1N (A. L. Odegard, K. Chandran, M. Ehrlich, T. Kirchhausen, and M. L. Nibert, unpublished data).
Properties of the δ fragment released from entering particles.
We observed by IF microscopy that the δ fragment of μ1 shed from entering particles localized to the cytoplasm and nucleus of cells at early times p.i. (Fig. 1). Immunoprecipitations of particles from cytoplasmic lysates revealed that a large proportion of δ within the lysates remained intact but was indeed no longer particle bound (Fig. 4). The shed form of δ may resemble that in the μ1* conformer because it is strongly recognized by MAbs 4A3 and 10F6 (Fig. 1). At odds with that hypothesis, however, is the finding that the released form of δ is less strongly recognized by MAbs 10H2 and 8H6, which immunoprecipitate similar numbers of ISVPs and ISVP*s in vitro (Fig. 1). Thus, the released δ might represent a novel conformer. To investigate this possibility, we will isolate δ from extracts of CHX-treated cells for further analyses. We will also attempt to identify cellular factors that may be involved in the removal and potential refolding of μ1 regions during cell entry. A related question is whether any proportion or region of the δ fragment is embedded in the lipid bilayer during membrane penetration and, if so, how it may be induced to exit the bilayer to assume the diffusely distributed form seen by IF microscopy. The postentry fates of the μ1N and φ fragments of μ1 (51, 53) will also be critical to ascertain.
The accumulation of large amounts of free δ within the cytoplasm and nucleus and its persistence for several hours p.i. suggest that this protein may have some postentry function(s) or effect(s). One possibility is that it needs simply to be removed from particles for subsequent steps in replication to proceed efficiently. For example, the removal of δ may permit more efficient targeting of the resulting core to a μNS inclusion in which RNA synthesis and assembly of new particles can occur (Broering et al., submitted). Another possibility is that the μ1* conformer in entering particles is toxic, perhaps due to its enhanced hydrophobicity (13), and must be refolded or degraded to prevent cell injury. In fact, several studies have indicated that the M2 genome segment encoding μ1 is a genetic determinant of strain differences in virus-induced apoptosis (58, 69) and that the generation of ISVP-like particles, but not viral protein synthesis, is required for apoptosis in some cell lines (19, 21). Thus, an interesting hypothesis is that a μ1 conformer either present in or released from infecting particles may play a positive or negative role in apoptosis. The released σ1 protein may also have some postentry function(s) or effect(s), but its predominant localization was not extensively analyzed in this study.
Membrane penetration mechanism.
Passage of large nonenveloped particles across the cellular membrane barrier during entry is commonly thought to require localized disruption of the plasma membrane or lysis of an endocytic compartment (5, 8, 41). A “reverse budding” model in which particle translocation is mechanistically coupled to disassembly and membrane insertion of viral capsid proteins also has been proposed (66). Definitive evidence for any mechanism is nonetheless lacking. The capacity of reovirus particles to support the inhibition of cellular protein synthesis by α-sarcin (46) (Fig. 9) likely indicates that toxin molecules enter the cytoplasm along with or in the wake of a penetrating viral particle. Similar findings obtained with α-sarcin or other toxins have been reported for rotavirus and adenovirus (29, 41, 56). Because α-sarcin is a 17-kDa protein, these findings are consistent with a membrane breach or rupture mechanism for cell entry by these viruses. However, although α-sarcin coentry clearly indicates that membrane leakage of such proportion can accompany cell entry, it does not necessarily represent the mechanism by which viral particles productively penetrate the membrane barrier (47). Isolation and characterization of membrane-associated penetration intermediates may be required to resolve this question.
Acknowledgments
We thank Elaine Freimont and Jason Dinoso for skillful technical assistance and laboratory management. Many thanks also are due to other members of our laboratory for helpful discussions and to Kim Myers for comments on a preliminary version of this article. We are very grateful to Hidde Ploegh for use of his flow cytometry facility and to Ya Chen for cryoscanning electron microscopy.
This work was supported by NIH grants K08 AI52209 (to J.S.L.P.), R01 GM36548 (to T.K.), and R01 AI46440 (to M.L.N.). K.C. was additionally supported by a predoctoral fellowship from the Howard Hughes Medical Institute and a Fields postdoctoral fellowship made available to the Department of Microbiology and Molecular Genetics through the generosity of Ruth Peedin Fields. M.E. was additionally supported by a Dorit fellowship from Harvard Medical School. The confocal microscope was made available through a gift from the Perkin Fund.
REFERENCES
- 1.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, A. K., and A. J. Shatkin. 1970. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J. Virol. 6:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 4.Bass, D. M., D. Bodkin, R. Dambrauskas, J. S. Trier, B. N. Fields, and J. L. Wolf. 1990. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 64:1830-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, R., P. Seth, M. C. Willingham, and I. Pastan. 1986. pH dependent lysis of liposomes by adenovirus. Biochemistry 25:2231-2237. [DOI] [PubMed] [Google Scholar]
- 6.Bodkin, D. K., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borsa, J., T. P. Copps, M. D. Sargent, D. G. Long, and J. D. Chapman. 1973. New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J. Virol. 11:552-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsa, J., B. D. Morash, M. D. Sargent, T. P. Copps, P. A. Lievaart, and J. G. Szekely. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161-170. [DOI] [PubMed] [Google Scholar]
- 9.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 10.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson, S. R., and M. Fukuda. 1989. Structure of human lysosomal membrane glycoprotein 1. Assignment of disulfide bonds and visualization of its domain arrangement. J. Biol. Chem. 264:20526-20531. [PubMed] [Google Scholar]
- 12.Centonze, V. E., Y. Chen, T. F. Severson, G. G. Borisy, and M. L. Nibert. 1995. Visualization of individual reovirus particles by low-temperature, high-resolution scanning microscopy. J. Struct. Biol. 115:215-225. [DOI] [PubMed] [Google Scholar]
- 13.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of reovirus penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandran, K., and M. L. Nibert. 1998. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J. Virol. 72:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandran, K., X. Zhang, N. H. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 75:5335-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappell, J. D., E. S. Barton, T. H. Smith, G. S. Baer, D. T. Duong, M. L. Nibert, and T. S. Dermody. 1998. Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J. Virol. 72:8205-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell, J. D., A. E. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke, P., S. M. Meintzer, L. Moffitt, and K. L. Tyler. 2003. Two distinct phases of virus-induced NF-κB regulation enhance TRAIL-mediated apoptosis in virus-infected cells. J. Biol. Chem. 278:18092-18100. [DOI] [PubMed] [Google Scholar]
- 20.Cleveland, D. R., H. Zarbl, and S. Millward. 1986. Reovirus guanylyltransferase is L2 gene product λ2. J. Virol. 60:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly, J. L., and T. S. Dermody. 2002. Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 76:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218-228. [DOI] [PubMed] [Google Scholar]
- 23.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drayna, D., and B. N. Fields. 1982. Genetic studies on the mechanism of chemical and physical inactivation of reovirus. J. Gen. Virol. 63:149-159. [DOI] [PubMed] [Google Scholar]
- 25.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 27.Farsetta, D. L., K. Chandran, and M. L. Nibert. 2000. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J. Biol. Chem. 275:39693-39701. [DOI] [PubMed] [Google Scholar]
- 28.Fausnaugh, J., and A. J. Shatkin. 1990. Active site localization in a viral mRNA capping enzyme. J. Biol. Chem. 265:7669-7672. [PubMed] [Google Scholar]
- 29.FitzGerald, D. J. P., R. Padmanabhan, I. Pastan, and M. C. Willingham. 1983. Adenovirus-induced release of epidermal growth factor and Pseudomonas toxin into the cytosol of KB cells during receptor-mediated endocytosis. Cell 32:607-617. [DOI] [PubMed] [Google Scholar]
- 30.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J. Biol. Chem. 251:5043-5053. [PubMed] [Google Scholar]
- 32.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 33.Hazelton, P. R., and K. M. Coombs. 1995. The reovirus mutant tsA279 has temperature-sensitive lesions in the M2 and L2 genes: the M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology 207:46-58. [DOI] [PubMed] [Google Scholar]
- 34.Hooper, J. W. 1996. Ph.D. thesis. Harvard University, Cambridge, Mass.
- 35.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joklik, W. K. 1972. Studies on the effect of chymotrypsin on reovirions. Virology 49:700-715. [DOI] [PubMed] [Google Scholar]
- 37.Joklik, W. K., and M. R. Roner. 1995. What reassorts when reovirus genome segments reassort? J. Biol. Chem. 270:4181-4184. [DOI] [PubMed] [Google Scholar]
- 38.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Protein σ1 is the reovirus cell attachment protein. Virology 108:156-163. [DOI] [PubMed] [Google Scholar]
- 39.Legesse-Miller, A., R. H. Massol, and T. Kirchhausen. 2003. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14:1953-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liprandi, F., Z. Moros, M. Gerder, J. E. Ludert, F. H. Pujol, M. C. Ruiz, F. Michelangeli, A. Charpilienne, and J. Cohen. 1997. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor α-sarcin. Virology 237:430-438. [DOI] [PubMed] [Google Scholar]
- 42.Loh, P. C., and A. J. Shatkin. 1968. Structural proteins of reovirus. J. Virol. 2:1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mane, S. M., L. Marzella, D. F. Bainton, V. K. Holt, Y. Cha, J. E. Hildreth, and J. T. August. 1989. Purification and characterization of human lysosomal membrane glycoproteins. Arch. Biochem. Biophys. 268:360-378. [DOI] [PubMed] [Google Scholar]
- 45.Mao, Z. X., and W. K. Joklik. 1991. Isolation and enzymatic characterization of protein λ2, the reovirus guanylyltransferase. Virology 185:377-386. [DOI] [PubMed] [Google Scholar]
- 46.Martinez, C. G., R. Guinea, J. Benavente, and L. Carrasco. 1996. The entry of reovirus into L cells is dependent on vacuolar proton-ATPase activity. J. Virol. 70:576-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meier, O., K. Boucke, S. V. Hammer, S. Keller, R. P. Stidwill, S. Hemmi, and U. F. Greber. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 158:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metcalf, P., M. Cyrklaff, and M. Adrian. 1991. The three-dimensional structure of reovirus obtained by cryo-electron microscopy. EMBO J. 10:3129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Middleton, J. K., T. F. Severson, K. Chandran, A. L. Gillian, J. Yin, and M. L. Nibert. 2002. Thermostability of reovirus disassembly intermediates (infectious subvirion particles) correlates with genetic, biochemical, and thermodynamic properties of major surface protein μ1. J. Virol. 76:1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano, M. Y., K. Boucke, M. Suomalainen, R. P. Stidwill, and U. F. Greber. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 53.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noble, S., and M. L. Nibert. 1997. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 71:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker, J. S., and C. R. Parrish. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74:1919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 58.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shatkin, A. J., and A. J. LaFiandra. 1972. Transcription by infectious subviral particles of reovirus. J. Virol. 10:698-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanisms of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 61.Silverstein, S. C., M. Schonberg, D. H. Levin, and G. Acs. 1970. The reovirus replicative cycle: conservation of parental RNA and protein. Proc. Natl. Acad. Sci. USA 67:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skehel, J. J., and W. K. Joklik. 1969. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 39:822-831. [DOI] [PubMed] [Google Scholar]
- 63.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 64.Strong, J. E., G. Leone, R. Duncan, R. K. Sharma, and P. W. K. Lee. 1991. Biochemical and biophysical characterization of the reovirus cell attachment protein σ1: evidence that it is a homotrimer. Virology 184:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tihova, M., K. A. Dryden, A. R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]
- 67.Tosteson, M. T., M. L. Nibert, and B. N. Fields. 1993. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. USA 90:10549-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyler, K. L. 2001. Mammalian reoviruses, p. 1729-1745. In D. M. Knipe, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 69.Tyler, K. L., M. K. Squier, A. L. Brown, B. Pike, D. Willis, S. M. Oberhaus, T. S. Dermody, and J. J. Cohen. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J. Virol. 70:7984-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wessner, D. R., and B. N. Fields. 1993. Isolation and genetic characterization of ethanol-resistant reovirus mutants. J. Virol. 67:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White, C. K., and H. J. Zweerink. 1976. Studies on the structure of reovirus cores: selective removal of polypeptide λ2. Virology 70:171-180. [DOI] [PubMed] [Google Scholar]
- 73.Yamakawa, M., Y. Furuichi, and A. J. Shatkin. 1982. Reovirus transcriptase and capping enzymes are active in intact virions. Virology 118:157-168. [DOI] [PubMed] [Google Scholar]