Abstract
The title compound, C8H11N2O3 +·Cl−, was synthesized as an intermediate in the development of a new sugar sensor. The structure displays N—H⋯Cl and O—H⋯O hydrogen bonding, as well as weak O—H⋯Cl interactions and π–π stacking (3.298 Å). There are two formula units in the asymmetric unit.
Related literature
For related literature, see: James et al. (1995 ▶).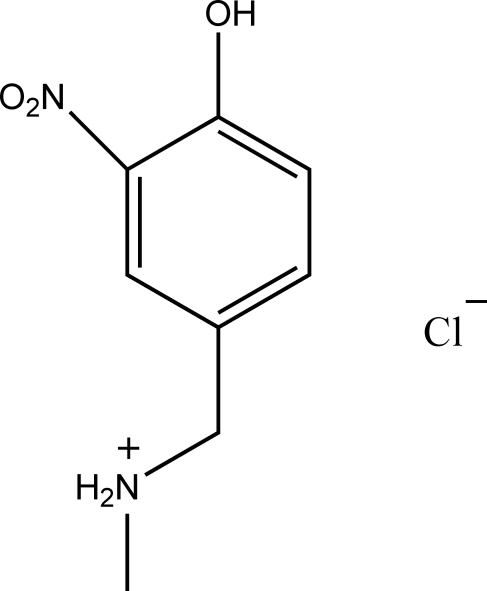
Experimental
Crystal data
C8H11N2O3 +·Cl−
M r = 218.64
Triclinic,
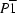
a = 7.7650 (2) Å
b = 10.5922 (3) Å
c = 13.5987 (4) Å
α = 70.262 (1)°
β = 78.368 (1)°
γ = 76.459 (1)°
V = 1014.27 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.36 mm−1
T = 173 (2) K
0.48 × 0.39 × 0.36 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: none
11798 measured reflections
4901 independent reflections
4057 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.033
wR(F 2) = 0.094
S = 1.06
4901 reflections
257 parameters
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT-Plus (Bruker, 1999 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: Mercury (Macrae et al., 2006 ▶) and WinGX (Farrugia, 1999 ▶); software used to prepare material for publication: SHELXTL and PLATON (Spek, 2003 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680800473X/hg2362sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800473X/hg2362Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2A—H2A⋯Cl1A | 0.92 | 2.23 | 3.1301 (11) | 167 |
| N2A—H2B⋯Cl1B | 0.92 | 2.18 | 3.0898 (11) | 173 |
| O1A—H1A⋯O2A | 0.84 | 1.89 | 2.5917 (14) | 140 |
| O1A—H1A⋯Cl1Bi | 0.84 | 2.87 | 3.3918 (10) | 122 |
| N2B—H2C⋯Cl1Aii | 0.92 | 2.17 | 3.0775 (11) | 168 |
| N2B—H2D⋯Cl1Biii | 0.92 | 2.26 | 3.1671 (10) | 168 |
| O1B—H1B⋯O2B | 0.84 | 1.88 | 2.5860 (14) | 141 |
Symmetry codes: (i) 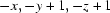 ; (ii)
; (ii) 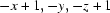 ; (iii)
; (iii) 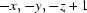 .
.
Acknowledgments
We thank Dr Manuel Fernandes of the Jan Boeyens Structural Chemistry Laboratory at the University of the Witwatersrand for his assistance in the acquisition of the crystallographic data. Aspen Pharmacare is acknowledged for their financial support.
supplementary crystallographic information
Comment
The title compound, (I), was synthesized as an intermediate in the development of a new sugar sensor (James et al., 1995). The compound itself is also novel and is being reported for the first time.
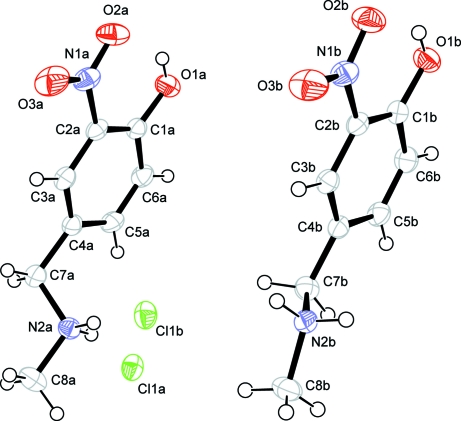
The structure consists of two molecules in the asymmetric unit (Figure 1). The cation consists of a planar nitro phenol ring with a methylaminomethyl group in the para position with respect to the hydroxy group (O1) on the ring. The methylammonium groups attached to the methylene carbon (C7) deviate from the plane of the ring with a torsion angle of -121.52 (13)° for C3A—C4A—C7A—N2A and -46.81 (16)° for C3B—C4B—C7B—N2B.
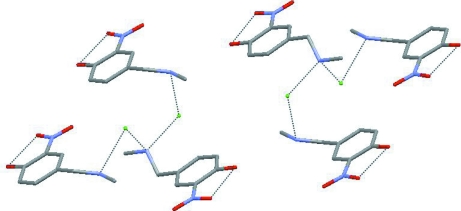
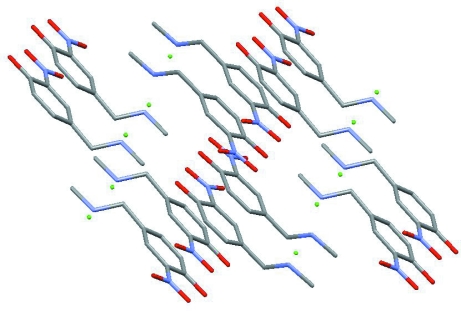
The structure exhibits both intermolecular (N1—H1···Cl) and intramolecular (O1—H1···O2) hydrogen bonding interactions (Table 1, Figure 2). The chloride ions act as hydrogen bond acceptors between adjacent molecules. Weak interactions are also observed between O1—H1···Cl1. These interactions, with a bond length of 2.87Å (O1A—H1A···Cl1Bi), are more likely weak Van der Waals interactions rather than true hydrogen bonds. See Table 1 for a full list of all hydrogen bond interactions. An interdigitated, layered structure is observed with the aromatic groups π–π stacking above each other and the methylaminomethyl group interacting with the chloride ions in hydrogen bonded layers (Figure 3).
Experimental
4-Chloromethyl-2-nitrophenol, 3.8 g (20 mmol), was dissolved in DMF (30 ml). To this was added triethylamine (3 ml) followed by 40% methylamine in H2O (5 ml, 58 mmol). The reaction was heated to 333 K and left to stir overnight. The solvent was removed under vacuum to afford an orange solid, which was recrystallized from methanol at room temperature. Yield 3.49 g (80%). Decomposition point 373–375 K.
1H-NMR (400 MHz, D2O): p.p.m. = 0.00 (TMS), 2.62 (s, 3H, CH3), 4.12 (s, 2H, CH2), 7.14 (d, J = 8.5 Hz, 1H, H5), 7.60 (d, J = 8.5 Hz, 1H, H6), 8.13 (s, 1H, H3).13C-NMR(100 MHz, D2O): p.p.m. = 0.00 (TMS), 32.58 (CH3), 51.50 (CH2),121.30 (C6), 123.50 (C4), 127.75 (C3), 136.86 (C2), 139.04 (C5), 154.76 (C1).
Refinement
Hydrogen atoms were located in the difference map then positioned geometrically, and allowed to ride on their respective parent atoms, with bond lengths of 0.99Å (CH2), 0.98Å (CH3), 0.95Å (CH), 0.98Å (NH2) or 0.84Å (OH). Isotropic displacement parameters for these atoms were set equal to 1.2 (CH2, CH and NH2), or 1.5 (CH3 and OH) times Ueq of the parent atom.
Figures
Fig. 1.
The asymmetric unit showing ellipsoids at the 50% probability level and the numbering scheme employed.
Fig. 2.
Diagram of the inter- and intramolecular hydrogen bonding. Hydrogen atoms have been omitted for clarity.
Fig. 3.
Depiction of the packing. Hydrogen atoms have been omitted for clarity.
Crystal data
| C8H11N2O3+·Cl– | Z = 4 |
| Mr = 218.64 | F000 = 456 |
| Triclinic, P1 | Dx = 1.432 Mg m−3Dm = 1.432 Mg m−3Dm measured by not measured |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 7.7650 (2) Å | Cell parameters from 6008 reflections |
| b = 10.5922 (3) Å | θ = 4.6–28.4º |
| c = 13.5987 (4) Å | µ = 0.36 mm−1 |
| α = 70.262 (1)º | T = 173 (2) K |
| β = 78.368 (1)º | Block, orange |
| γ = 76.459 (1)º | 0.48 × 0.39 × 0.36 mm |
| V = 1014.27 (5) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 4057 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.034 |
| Monochromator: graphite | θmax = 28.0º |
| T = 173(2) K | θmin = 1.6º |
| φ and ω scans | h = −10→10 |
| Absorption correction: none | k = −13→13 |
| 11798 measured reflections | l = −17→16 |
| 4901 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.033 | H-atom parameters constrained |
| wR(F2) = 0.094 | w = 1/[σ2(Fo2) + (0.0542P)2 + 0.0174P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 4901 reflections | Δρmax = 0.24 e Å−3 |
| 257 parameters | Δρmin = −0.31 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1A | 0.07924 (17) | 0.64421 (12) | 0.39067 (10) | 0.0280 (3) | |
| C2A | −0.05629 (16) | 0.60295 (12) | 0.36161 (10) | 0.0266 (3) | |
| C3A | −0.02230 (17) | 0.54096 (12) | 0.28266 (10) | 0.0277 (3) | |
| H3A | −0.1168 | 0.5129 | 0.2652 | 0.033* | |
| C4A | 0.14689 (17) | 0.52009 (12) | 0.22990 (10) | 0.0272 (3) | |
| C5A | 0.28403 (17) | 0.55999 (13) | 0.25897 (11) | 0.0311 (3) | |
| H5A | 0.4022 | 0.5452 | 0.2238 | 0.037* | |
| C6A | 0.25051 (17) | 0.61995 (13) | 0.33725 (11) | 0.0318 (3) | |
| H6A | 0.3462 | 0.6456 | 0.3556 | 0.038* | |
| C7A | 0.17964 (18) | 0.46001 (13) | 0.14103 (11) | 0.0328 (3) | |
| H7A | 0.0658 | 0.4412 | 0.1318 | 0.039* | |
| H7B | 0.2224 | 0.5269 | 0.0749 | 0.039* | |
| C8A | 0.3333 (2) | 0.26316 (15) | 0.07972 (12) | 0.0408 (3) | |
| H8A | 0.2200 | 0.2359 | 0.0819 | 0.061* | |
| H8B | 0.4277 | 0.1822 | 0.0934 | 0.061* | |
| H8C | 0.3646 | 0.3263 | 0.0100 | 0.061* | |
| N1A | −0.23762 (14) | 0.62436 (11) | 0.41284 (10) | 0.0339 (3) | |
| N2A | 0.31439 (14) | 0.33107 (10) | 0.16086 (8) | 0.0270 (2) | |
| H2A | 0.4233 | 0.3503 | 0.1621 | 0.032* | |
| H2B | 0.2807 | 0.2726 | 0.2259 | 0.032* | |
| O1A | 0.06011 (13) | 0.70500 (10) | 0.46491 (8) | 0.0388 (2) | |
| H1A | −0.0465 | 0.7123 | 0.4939 | 0.058* | |
| O2A | −0.27172 (14) | 0.68749 (10) | 0.47848 (9) | 0.0457 (3) | |
| O3A | −0.35065 (13) | 0.58060 (12) | 0.39075 (10) | 0.0518 (3) | |
| C1B | 0.08090 (17) | 0.14787 (12) | 0.89457 (10) | 0.0271 (3) | |
| C2B | −0.03119 (15) | 0.10203 (12) | 0.85044 (10) | 0.0248 (3) | |
| C3B | 0.03195 (16) | 0.04521 (12) | 0.76886 (10) | 0.0256 (3) | |
| H3B | −0.0483 | 0.0164 | 0.7401 | 0.031* | |
| C4B | 0.21073 (16) | 0.03066 (12) | 0.72966 (10) | 0.0251 (3) | |
| C5B | 0.32453 (17) | 0.07415 (13) | 0.77435 (11) | 0.0294 (3) | |
| H5B | 0.4486 | 0.0636 | 0.7488 | 0.035* | |
| C6B | 0.26146 (17) | 0.13157 (13) | 0.85399 (11) | 0.0317 (3) | |
| H6B | 0.3423 | 0.1608 | 0.8821 | 0.038* | |
| C7B | 0.28683 (17) | −0.02588 (12) | 0.63923 (10) | 0.0285 (3) | |
| H7C | 0.2538 | 0.0440 | 0.5729 | 0.034* | |
| H7D | 0.4189 | −0.0455 | 0.6340 | 0.034* | |
| C8B | 0.3304 (2) | −0.22269 (14) | 0.57491 (11) | 0.0383 (3) | |
| H8D | 0.3227 | −0.1618 | 0.5028 | 0.057* | |
| H8E | 0.2844 | −0.3051 | 0.5841 | 0.057* | |
| H8F | 0.4554 | −0.2478 | 0.5879 | 0.057* | |
| N1B | −0.22054 (14) | 0.11494 (11) | 0.88843 (9) | 0.0311 (3) | |
| N2B | 0.22290 (13) | −0.15229 (10) | 0.65039 (9) | 0.0260 (2) | |
| H2C | 0.2293 | −0.2100 | 0.7180 | 0.031* | |
| H2D | 0.1052 | −0.1306 | 0.6391 | 0.031* | |
| O1B | 0.03022 (13) | 0.20461 (10) | 0.97277 (8) | 0.0357 (2) | |
| H1B | −0.0805 | 0.2095 | 0.9912 | 0.054* | |
| O2B | −0.28171 (13) | 0.17175 (11) | 0.95706 (8) | 0.0424 (3) | |
| O3B | −0.31379 (13) | 0.06993 (12) | 0.85167 (10) | 0.0492 (3) | |
| Cl1A | 0.70902 (4) | 0.37097 (3) | 0.13787 (3) | 0.03519 (10) | |
| Cl1B | 0.18542 (4) | 0.12266 (3) | 0.36875 (2) | 0.03163 (10) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1A | 0.0284 (6) | 0.0254 (6) | 0.0266 (7) | −0.0009 (5) | −0.0051 (5) | −0.0051 (5) |
| C2A | 0.0210 (6) | 0.0246 (6) | 0.0276 (7) | −0.0023 (5) | −0.0007 (5) | −0.0024 (5) |
| C3A | 0.0258 (6) | 0.0239 (6) | 0.0309 (7) | −0.0044 (5) | −0.0065 (5) | −0.0039 (5) |
| C4A | 0.0286 (6) | 0.0240 (6) | 0.0247 (6) | −0.0008 (5) | −0.0042 (5) | −0.0041 (5) |
| C5A | 0.0213 (6) | 0.0327 (7) | 0.0350 (7) | −0.0018 (5) | −0.0002 (5) | −0.0087 (6) |
| C6A | 0.0238 (6) | 0.0334 (7) | 0.0394 (8) | −0.0037 (5) | −0.0074 (6) | −0.0119 (6) |
| C7A | 0.0336 (7) | 0.0331 (7) | 0.0283 (7) | 0.0017 (6) | −0.0063 (6) | −0.0088 (6) |
| C8A | 0.0502 (9) | 0.0437 (8) | 0.0341 (8) | −0.0092 (7) | −0.0013 (7) | −0.0208 (7) |
| N1A | 0.0250 (6) | 0.0310 (6) | 0.0382 (7) | −0.0029 (5) | 0.0015 (5) | −0.0055 (5) |
| N2A | 0.0277 (5) | 0.0285 (5) | 0.0241 (5) | −0.0061 (4) | −0.0002 (4) | −0.0083 (4) |
| O1A | 0.0370 (5) | 0.0461 (6) | 0.0373 (6) | −0.0035 (5) | −0.0040 (5) | −0.0213 (5) |
| O2A | 0.0373 (6) | 0.0484 (6) | 0.0471 (7) | −0.0046 (5) | 0.0121 (5) | −0.0213 (5) |
| O3A | 0.0232 (5) | 0.0635 (7) | 0.0721 (8) | −0.0122 (5) | 0.0004 (5) | −0.0258 (6) |
| C1B | 0.0277 (6) | 0.0275 (6) | 0.0248 (6) | −0.0024 (5) | −0.0058 (5) | −0.0066 (5) |
| C2B | 0.0207 (6) | 0.0238 (6) | 0.0269 (6) | −0.0030 (5) | −0.0032 (5) | −0.0045 (5) |
| C3B | 0.0236 (6) | 0.0245 (6) | 0.0287 (7) | −0.0049 (5) | −0.0061 (5) | −0.0065 (5) |
| C4B | 0.0244 (6) | 0.0231 (6) | 0.0251 (6) | −0.0025 (5) | −0.0046 (5) | −0.0044 (5) |
| C5B | 0.0211 (6) | 0.0338 (7) | 0.0320 (7) | −0.0029 (5) | −0.0052 (5) | −0.0085 (6) |
| C6B | 0.0258 (6) | 0.0382 (7) | 0.0353 (7) | −0.0063 (5) | −0.0100 (6) | −0.0128 (6) |
| C7B | 0.0292 (6) | 0.0286 (6) | 0.0264 (7) | −0.0069 (5) | −0.0011 (5) | −0.0072 (5) |
| C8B | 0.0434 (8) | 0.0395 (8) | 0.0332 (8) | −0.0029 (6) | 0.0012 (6) | −0.0193 (6) |
| N1B | 0.0233 (5) | 0.0322 (6) | 0.0359 (7) | −0.0050 (5) | −0.0010 (5) | −0.0098 (5) |
| N2B | 0.0232 (5) | 0.0282 (5) | 0.0255 (5) | −0.0020 (4) | −0.0036 (4) | −0.0084 (4) |
| O1B | 0.0335 (5) | 0.0462 (6) | 0.0327 (5) | −0.0062 (4) | −0.0038 (4) | −0.0200 (4) |
| O2B | 0.0322 (5) | 0.0554 (6) | 0.0411 (6) | −0.0068 (5) | 0.0061 (4) | −0.0237 (5) |
| O3B | 0.0251 (5) | 0.0671 (7) | 0.0690 (8) | −0.0133 (5) | −0.0015 (5) | −0.0376 (6) |
| Cl1A | 0.02968 (17) | 0.0419 (2) | 0.02772 (18) | −0.00866 (14) | −0.00213 (13) | −0.00206 (14) |
| Cl1B | 0.02849 (17) | 0.03788 (18) | 0.02609 (18) | −0.00808 (13) | −0.00452 (13) | −0.00475 (13) |
Geometric parameters (Å, °)
| C1A—O1A | 1.3378 (15) | C1B—O1B | 1.3413 (15) |
| C1A—C6A | 1.3924 (18) | C1B—C6B | 1.3925 (18) |
| C1A—C2A | 1.3981 (18) | C1B—C2B | 1.3996 (17) |
| C2A—C3A | 1.3908 (18) | C2B—C3B | 1.3882 (17) |
| C2A—N1A | 1.4425 (16) | C2B—N1B | 1.4473 (15) |
| C3A—C4A | 1.3713 (18) | C3B—C4B | 1.3755 (17) |
| C3A—H3A | 0.9500 | C3B—H3B | 0.9500 |
| C4A—C5A | 1.4008 (18) | C4B—C5B | 1.3994 (17) |
| C4A—C7A | 1.5000 (18) | C4B—C7B | 1.5030 (17) |
| C5A—C6A | 1.3682 (19) | C5B—C6B | 1.3691 (18) |
| C5A—H5A | 0.9500 | C5B—H5B | 0.9500 |
| C6A—H6A | 0.9500 | C6B—H6B | 0.9500 |
| C7A—N2A | 1.4928 (16) | C7B—N2B | 1.4852 (15) |
| C7A—H7A | 0.9900 | C7B—H7C | 0.9900 |
| C7A—H7B | 0.9900 | C7B—H7D | 0.9900 |
| C8A—N2A | 1.4759 (16) | C8B—N2B | 1.4788 (16) |
| C8A—H8A | 0.9800 | C8B—H8D | 0.9800 |
| C8A—H8B | 0.9800 | C8B—H8E | 0.9800 |
| C8A—H8C | 0.9800 | C8B—H8F | 0.9800 |
| N1A—O3A | 1.2109 (15) | N1B—O3B | 1.2125 (14) |
| N1A—O2A | 1.2406 (15) | N1B—O2B | 1.2350 (14) |
| N2A—H2A | 0.9200 | N2B—H2C | 0.9200 |
| N2A—H2B | 0.9200 | N2B—H2D | 0.9200 |
| O1A—H1A | 0.8400 | O1B—H1B | 0.8400 |
| O1A—C1A—C6A | 116.81 (12) | O1B—C1B—C6B | 117.37 (11) |
| O1A—C1A—C2A | 126.21 (12) | O1B—C1B—C2B | 125.88 (12) |
| C6A—C1A—C2A | 116.98 (12) | C6B—C1B—C2B | 116.74 (11) |
| C3A—C2A—C1A | 121.68 (12) | C3B—C2B—C1B | 122.28 (11) |
| C3A—C2A—N1A | 117.71 (11) | C3B—C2B—N1B | 117.56 (11) |
| C1A—C2A—N1A | 120.61 (11) | C1B—C2B—N1B | 120.15 (11) |
| C4A—C3A—C2A | 120.34 (12) | C4B—C3B—C2B | 119.98 (11) |
| C4A—C3A—H3A | 119.8 | C4B—C3B—H3B | 120.0 |
| C2A—C3A—H3A | 119.8 | C2B—C3B—H3B | 120.0 |
| C3A—C4A—C5A | 118.47 (12) | C3B—C4B—C5B | 118.22 (11) |
| C3A—C4A—C7A | 119.74 (12) | C3B—C4B—C7B | 122.67 (11) |
| C5A—C4A—C7A | 121.76 (12) | C5B—C4B—C7B | 119.08 (11) |
| C6A—C5A—C4A | 121.07 (12) | C6B—C5B—C4B | 121.63 (12) |
| C6A—C5A—H5A | 119.5 | C6B—C5B—H5B | 119.2 |
| C4A—C5A—H5A | 119.5 | C4B—C5B—H5B | 119.2 |
| C5A—C6A—C1A | 121.45 (12) | C5B—C6B—C1B | 121.13 (12) |
| C5A—C6A—H6A | 119.3 | C5B—C6B—H6B | 119.4 |
| C1A—C6A—H6A | 119.3 | C1B—C6B—H6B | 119.4 |
| N2A—C7A—C4A | 111.77 (10) | N2B—C7B—C4B | 113.02 (10) |
| N2A—C7A—H7A | 109.3 | N2B—C7B—H7C | 109.0 |
| C4A—C7A—H7A | 109.3 | C4B—C7B—H7C | 109.0 |
| N2A—C7A—H7B | 109.3 | N2B—C7B—H7D | 109.0 |
| C4A—C7A—H7B | 109.3 | C4B—C7B—H7D | 109.0 |
| H7A—C7A—H7B | 107.9 | H7C—C7B—H7D | 107.8 |
| N2A—C8A—H8A | 109.5 | N2B—C8B—H8D | 109.5 |
| N2A—C8A—H8B | 109.5 | N2B—C8B—H8E | 109.5 |
| H8A—C8A—H8B | 109.5 | H8D—C8B—H8E | 109.5 |
| N2A—C8A—H8C | 109.5 | N2B—C8B—H8F | 109.5 |
| H8A—C8A—H8C | 109.5 | H8D—C8B—H8F | 109.5 |
| H8B—C8A—H8C | 109.5 | H8E—C8B—H8F | 109.5 |
| O3A—N1A—O2A | 122.35 (12) | O3B—N1B—O2B | 122.23 (11) |
| O3A—N1A—C2A | 119.40 (12) | O3B—N1B—C2B | 118.92 (11) |
| O2A—N1A—C2A | 118.25 (11) | O2B—N1B—C2B | 118.84 (10) |
| C8A—N2A—C7A | 112.24 (11) | C8B—N2B—C7B | 111.57 (10) |
| C8A—N2A—H2A | 109.2 | C8B—N2B—H2C | 109.3 |
| C7A—N2A—H2A | 109.2 | C7B—N2B—H2C | 109.3 |
| C8A—N2A—H2B | 109.2 | C8B—N2B—H2D | 109.3 |
| C7A—N2A—H2B | 109.2 | C7B—N2B—H2D | 109.3 |
| H2A—N2A—H2B | 107.9 | H2C—N2B—H2D | 108.0 |
| C1A—O1A—H1A | 109.5 | C1B—O1B—H1B | 109.5 |
| O1A—C1A—C2A—C3A | 179.66 (12) | O1B—C1B—C2B—C3B | 179.51 (12) |
| C6A—C1A—C2A—C3A | −0.25 (19) | C6B—C1B—C2B—C3B | −1.17 (19) |
| O1A—C1A—C2A—N1A | 0.0 (2) | O1B—C1B—C2B—N1B | 0.5 (2) |
| C6A—C1A—C2A—N1A | −179.92 (11) | C6B—C1B—C2B—N1B | 179.83 (11) |
| C1A—C2A—C3A—C4A | −0.80 (19) | C1B—C2B—C3B—C4B | 0.92 (19) |
| N1A—C2A—C3A—C4A | 178.87 (11) | N1B—C2B—C3B—C4B | 179.94 (11) |
| C2A—C3A—C4A—C5A | 1.30 (18) | C2B—C3B—C4B—C5B | 0.13 (18) |
| C2A—C3A—C4A—C7A | −176.71 (11) | C2B—C3B—C4B—C7B | −178.01 (11) |
| C3A—C4A—C5A—C6A | −0.77 (19) | C3B—C4B—C5B—C6B | −0.89 (19) |
| C7A—C4A—C5A—C6A | 177.19 (12) | C7B—C4B—C5B—C6B | 177.32 (12) |
| C4A—C5A—C6A—C1A | −0.3 (2) | C4B—C5B—C6B—C1B | 0.6 (2) |
| O1A—C1A—C6A—C5A | −179.13 (12) | O1B—C1B—C6B—C5B | 179.78 (12) |
| C2A—C1A—C6A—C5A | 0.8 (2) | C2B—C1B—C6B—C5B | 0.4 (2) |
| C3A—C4A—C7A—N2A | −121.52 (13) | C3B—C4B—C7B—N2B | −46.81 (16) |
| C5A—C4A—C7A—N2A | 60.54 (16) | C5B—C4B—C7B—N2B | 135.07 (12) |
| C3A—C2A—N1A—O3A | 4.67 (18) | C3B—C2B—N1B—O3B | 3.65 (18) |
| C1A—C2A—N1A—O3A | −175.66 (12) | C1B—C2B—N1B—O3B | −177.30 (12) |
| C3A—C2A—N1A—O2A | −175.38 (12) | C3B—C2B—N1B—O2B | −176.18 (11) |
| C1A—C2A—N1A—O2A | 4.30 (18) | C1B—C2B—N1B—O2B | 2.86 (18) |
| C4A—C7A—N2A—C8A | 173.76 (12) | C4B—C7B—N2B—C8B | −166.60 (11) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2A—H2A···Cl1A | 0.92 | 2.23 | 3.1301 (11) | 167 |
| N2A—H2B···Cl1B | 0.92 | 2.18 | 3.0898 (11) | 173 |
| O1A—H1A···O2A | 0.84 | 1.89 | 2.5917 (14) | 140 |
| O1A—H1A···Cl1Bi | 0.84 | 2.87 | 3.3918 (10) | 122 |
| N2B—H2C···Cl1Aii | 0.92 | 2.17 | 3.0775 (11) | 168 |
| N2B—H2D···Cl1Biii | 0.92 | 2.26 | 3.1671 (10) | 168 |
| O1B—H1B···O2B | 0.84 | 1.88 | 2.5860 (14) | 141 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x+1, −y, −z+1; (iii) −x, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2362).
References
- Bruker (1999). SAINT-Plus Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- James, T. D., Sandanayake, K. R. A. S., Iuguchi, R. & Shinkai, S. (1995). J. Am. Chem. Soc.117, 8982–8987.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680800473X/hg2362sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800473X/hg2362Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report