Abstract
The hemagglutinin (HA) and neuraminidase (NA) genes of H7 avian influenza virus (AIV) isolated between 1994 and 2002 from live-bird markets (LBMs) in the northeastern United States and from three outbreaks in commercial poultry have been characterized. Phylogenetic analysis of the HA and NA genes demonstrates that the isolates from commercial poultry were closely related to the viruses circulating in the LBMs. Also, since 1994, two distinguishing genetic features have appeared in this AIV lineage: a deletion of 17 amino acids in the NA protein stalk region and a deletion of 8 amino acids in the HA1 protein which is putatively in part of the receptor binding site. Furthermore, analysis of the HA cleavage site amino acid sequence, a marker for pathogenicity in chickens and turkeys, shows a progression toward a cleavage site sequence that fulfills the molecular criteria for highly pathogenic AIV.
Avian influenza virus (AIV), a type A influenza virus, especially in its highly pathogenic (HP) form, can cause a devastating disease in poultry, resulting in severe economic losses. Of the 15 hemagglutinin (HA) subtypes of influenza virus, H5 and H7 are considered to be the most important in gallinaceous poultry, as only these subtypes have been associated with HP AIV (reviewed in reference 1). Importantly, HP AIV is considered exotic to the United States; however, low-pathogenic avian influenza H7N2 subtype viruses have been routinely isolated from the live-bird market (LBM) system in the northeastern United States since 1994 (13, 14) and are believed to be linked to two outbreaks in commercial poultry in Pennsylvania, the first from 1997 to 1998 (22) and the second from 2001 to 2002, and to an outbreak of H7N2 AIV in the spring of 2002 in Virginia, West Virginia, and North Carolina.
This study characterizes the HA and neuraminidase (NA) genes from H7N2 AIV isolates obtained from the premises of LBMs and poultry farms or isolated from birds at LBMs and poultry farms in the northeastern United States between 1994 and 2002. The full coding regions of the HA gene of 54 H7N2 isolates and only the HA1 region of an additional 4 isolates were included as well as 45 NA subtype 2 genes. LBM isolates obtained from 1994 to 1998, initially characterized by Suarez et al. (17), were also included in the study with the addition of the full coding sequences for the HA and the NA genes.
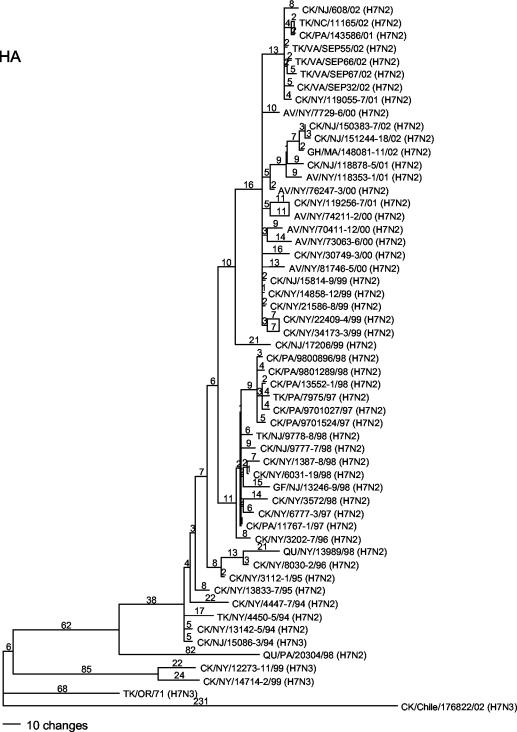
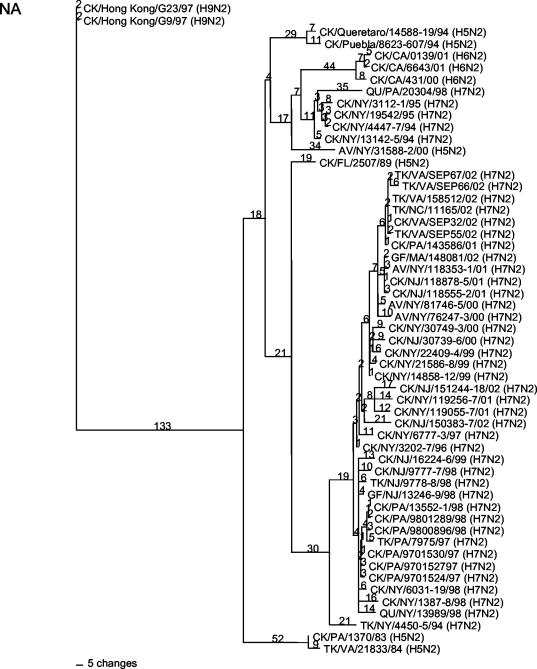
Phylogenetic analysis was performed with PAUP 4.0b10 (Sinauer Associates, Inc., Sunderland, Mass.) with the maximum parsimony tree-building method by heuristic search with 500 bootstrap replicates. Phylogenetic analysis of the HA gene of the H7N2 viruses revealed a close relationship, probably representing a single lineage and a chronological assortment among the isolates (Fig. 1). The isolates from the AIV outbreaks in Pennsylvania (from both 1997 to 1998 and 2001 to 2002) and the outbreak in Virginia, West Virginia, and North Carolina in 2002 were clearly derived from the LBM H7N2 lineage. This relationship is supported by the NA phylogenetics also, although the early NA genes assort into several groups (Fig. 2).
FIG. 1.
Phylogenetic tree of the HA gene of H7 subtype AIVs isolated in the Americas (with subtypes noted on the tree). The tree was generated by the maximum parsimony method with PAUP 4.0b10 (heuristic search and 500 bootstrap replicates) and is rooted with CK/Chile/176822/02. AV, avian; CK, chicken; DK, duck; GF, guinea fowl; QU, quail; TK, turkey. The names of the states where the viruses were isolated are represented by their standard two-letter postal codes.
FIG. 2.
Phylogenetic tree of the NA (subtype N2) gene of AIVs isolated in North America (with subtypes noted on the tree). Groupings of H7N2 viruses isolated in the northeastern United States between 1994 and 2002 are shown. The tree was generated by the maximum parsimony method with PAUP 4.0b10 (heuristic search and 500 bootstrap replicates) and is not rooted. AV, avian; CK, chicken; GF, guinea fowl; QU, quail; TK, turkey. The names of the states where the viruses were isolated are represented by their standard two-letter postal codes.
A 24-nucleotide (nt) deletion resulting in an eight-amino-acid deletion from residues 212 to 219 in the HA1 segment, first reported by Suarez et al. (17), was present in all isolates from the LBM lineage obtained since 1996 and in all isolates from AIV outbreaks in commercial poultry in Pennsylvania, Virginia, and North Carolina. Based on the structure of the H3 HA gene (20), this deletion removes five of six consecutive amino acids in part of the receptor binding site. The nucleotide sequence similarity between viruses with and without the deletion is high. Therefore, the HA gene with this deletion arose from the H7 subtype viruses circulating in the LBMs at the time and was not a separate introduction of a different HA gene with a deletion.
Four different NA stalk lengths were observed during the sample period. Isolates with a full-length protein were only seen prior to 1996. The largest deletion was in TK/NY/4450-5/94, which had a stalk deletion from nt 142 to 211, resulting in a loss of 23 amino acids. Another isolate, QU/PA/20304/98, had a stalk deletion from nt 142 to 199. A fourth group, which was phylogenetically distinct and which contained most of the N2 isolates obtained since 1997 from the LBMs and commercial poultry outbreaks, had a 51-nt deletion between positions 164 and 211 (17 amino acids) and was phylogenetically distinct from earlier N2 genes. Based on the deletions and sequence variation, it appears that four separate NA genes were introduced between 1994 and 2002 by reassortment into the LBM H7 virus lineage, with a single gene lineage becoming dominant after 1997.
Although the HA1 stalk deletion is unique to this lineage, deletions in the NA stalk region have been observed in other AIVs isolated from poultry; however, the positions and lengths of the deletions vary among isolates (3, 21). NA stalk deletions have been shown to reduce the enzymatic activity of the protein (10) and, presumably, adversely affect the spread of the virus to uninfected cells. Compensatory changes in the HA gene have been described, including decreases in receptor binding affinity arising from increased glycosylation near the receptor binding site (11, 19). The presence of an eight-amino-acid deletion in a region of the HA1 segment that putatively corresponds to the receptor binding site (based on the structure of the H3 HA gene) (20) has been reported only in this lineage (17, 18) and may represent a novel compensatory mutation for the NA stalk deletion. The impact this deletion may have on HA binding needs to be investigated further. Interestingly, both the HA and NA stalk deletions were first observed in isolates obtained in 1996.
Compared to several other viruses in poultry, the LBM-lineage H7N2 viruses have circulated for a relatively long period of time in chickens and turkeys without becoming HP. In some previous reports, low-pathogenic H5 or H7 AIV circulated in commercial poultry for at most 2 years after they were first detected before mutating into the HP form (2, 3, 5-7). The HA cleavage site sequence has been well characterized as an indicator of pathogenicity, where multiple basic amino acids are associated with high pathogenicity (4, 9). Throughout the 8 years during which H7 influenza viruses have been consistently isolated from the LBMs, five different HA cleavage site sequences have been observed (Table 1). The earliest isolates (from 1994 to 1998) had two basic amino acids; in 1998 isolates with three basic amino acids were observed and became the predominant sequence, and then in early 2002 isolates with four basic amino acids were observed. It has not yet been determined if the sequence with four basic amino acids will become the predominant sequence. In H7 subtype viruses, five or more basic amino acids would likely result in a phenotype for high pathogenicity (15). Importantly, all of the LBM-lineage H7N2 isolates that have been characterized, even those with four basic amino acids, have been of low pathogenicity in chickens (data not shown).
TABLE 1.
HA cleavage site amino acid sequences observed in the AIV isolates included in this study
| HA cleavage site sequence | Year observed | No. of isolates |
|---|---|---|
| P-E-N-P-K-T-R/Ga | 1994-1995, 1999 | 6 |
| P-E-N-P-K-P-R/Ga | 1995-1999, 2002 | 23 |
| P-E-K-P-K-P-R/G | 1998-2002 | 30 |
| P-E-K-P-K-T-R/G | 1995, 1998 | 2 |
| P-E-K-P-K-K-R/G | 2002 | 3 |
The sequence was originally reported by Suarez et al. (17).
Finally, reassortment in the matrix and nonstructural genes has a similar pattern to that of the HA and NA genes. Previously, it was shown that the matrix and nonstructural genes were reassorting fairly often in viruses isolated before 1997 (17). However, both genes became stably associated in the H7N2 lineage after 1997, with no further evidence of reassortment (data not shown). The stable combination of these four gene segments after 1997 suggests that this constellation of genes offers some selective advantage to the virus in this environment, particularly when AIVs of as many as nine different subtypes other than H7N2 have been isolated in the LBM system since 1994 (12). Isolates of non-H5 and non-H7 subtypes from the same time period have not been well characterized by sequencing.
Because the H7N2 viruses have continued to circulate in the LBM system for more than 8 years with consistent monitoring, the LBMs provide a unique opportunity to observe the evolution and ecology of the influenza virus in a nonnatural avian host. The LBMs also provide a somewhat unique environment for the evolution of AIV through the poor bio-security that results from housing multiple avian species (and sometimes mammalian species) in close contact and from a constant influx of new and susceptible birds. The markets likely provide a direct route for AIV transmission to poultry from waterfowl, which are one of the natural host reservoirs of type A influenza virus (8, 16).
Nucleotide sequence accession numbers.
The nucleotide sequences for the HA gene and the NA gene have been deposited in the GenBank database under accession numbers AY240877 to AY240925 and AY254105 to AY254149, respectively.
Acknowledgments
We thank Suzanne DeBlois, Chang-won Lee, and the U.S. Department of Agriculture, Agricultural Research Service (USDA-ARS), South Atlantic Area sequencing facility for technical assistance with this work.
This work was supported by USDA-ARS Current Research Information System project 6612-32000-039.
REFERENCES
- 1.Alexander, D. J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3-13. [DOI] [PubMed] [Google Scholar]
- 2.Banks, J., E. C. Speidel, J. W. McCauley, and D. J. Alexander. 2000. Phylogenetic analysis of H7 haemagglutinin subtype influenza A viruses. Arch. Virol. 145:1047-1058. [DOI] [PubMed] [Google Scholar]
- 3.Banks, J., E. S. Speidel, E. Moore, L. Plowright, A. Piccirillo, I. Capua, P. Cordioli, A. Fioretti, and D. J. Alexander. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963-973. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, F. X., W. Garten, H. D. Klenk, and R. Rott. 1981. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology 113:725-735. [DOI] [PubMed] [Google Scholar]
- 5.Buisch, W. W., A. E. Hall, and H. A. McDaniel. 1984. 1983-1984 Lethal avian influenza, p. 430-445. In Proceedings of the 88th Annual Meeting of the U.S. Animal Health Association. U.S. Animal Health Association, Richmond, Va.
- 6.Garcia, M., J. M. Crawford, J. W. Latimer, E. Rivera-Cruz, and M. L. Perdue. 1996. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J. Gen. Virol. 77:1493-1504. [DOI] [PubMed] [Google Scholar]
- 7.Horimoto, T., E. Rivera, J. Pearson, D. Senne, S. Krauss, Y. Kawaoka, and R. G. Webster. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213:223-230. [DOI] [PubMed] [Google Scholar]
- 8.Kawaoka, Y., T. M. Chambers, W. L. Sladen, and R. G. Webster. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 163:247-250. [DOI] [PubMed] [Google Scholar]
- 9.Klenk, H. D., and R. Rott. 1988. The molecular biology of influenza virus pathogenicity. Adv. Virus Res. 34:247-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo, G., J. Chung, and P. Palese. 1993. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res. 29:141-153. [DOI] [PubMed] [Google Scholar]
- 11.Matrosovich, M., N. Zhou, Y. Kawaoka, and R. Webster. 1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 73:1146-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panigrahy, B., D. A. Senne, and J. C. Pedersen. 2002. Avian influenza virus subtypes inside and outside the live bird markets, 1993-2000: a spatial and temporal relationship. Avian Dis. 46:298-307. [DOI] [PubMed] [Google Scholar]
- 13.Pomeroy, B. S. E. 1994. Avian influenza, p. 512-523. In Proceedings of the 98th Annual Meeting of the U.S. Animal Health Association. U.S. Animal Health Association, Richmond, Va.
- 14.Senne, D. 1997. NVSL report on avian influenza, p. 497-498. In Proceedings of the 101st Annual Meeting of the U.S. Animal Health Association. U.S. Animal Health Association, Richmond, Va.
- 15.Senne, D. A., B. Panigrahy, Y. Kawaoka, J. E. Pearson, J. Suss, M. Lipkind, H. Kida, and R. G. Webster. 1996. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40:425-437. [PubMed] [Google Scholar]
- 16.Slemons, R. D., D. C. Johnson, J. S. Oborn, and F. Hayes. 1974. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 18:119-124. [PubMed] [Google Scholar]
- 17.Suarez, D. L., M. Garcia, J. Latimer, D. Senne, and M. Perdue. 1999. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the northeast United States. J. Virol. 73:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suarez, D. L., E. Spackman, and D. Senne. Update on molecular epidemiology of H1, H5 and H7 influenza infection in poultry in North America. Avian Dis. Suppl., in press. [DOI] [PubMed]
- 19.Wagner, R., T. Wolff, A. Herwig, S. Pleschka, and H. D. Klenk. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 74:6316-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weis, W., J. H. Brown, S. Cusack, J. C. Paulson, J. J. Skehel, and D. C. Wiley. 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333:426-431. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, N. N., K. F. Shortridge, E. C. Claas, S. L. Krauss, and R. G. Webster. 1999. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J. Virol. 73:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler, A. F., S. Davison, H. Acland, and R. J. Eckroade. 1999. Characterization of H7N2 (non-pathogenic) avian influenza virus infections in commercial layer, in Pennsylvania, 1997-98. Avian Dis. 43:142-149. [PubMed] [Google Scholar]