Abstract
The RNA-dependent RNA polymerase of hepatitis C virus (HCV) is the catalytic subunit of the viral RNA amplification machinery and is an appealing target for the development of new therapeutic agents against HCV infection. Nonnucleoside inhibitors based on a benzimidazole scaffold have been recently reported. Compounds of this class are efficient inhibitors of HCV RNA replication in cell culture, thus providing attractive candidates for further development. Here we report the detailed analysis of the mechanism of action of selected benzimidazole inhibitors. Kinetic data and binding experiments indicated that these compounds act as allosteric inhibitors that block the activity of the polymerase prior to the elongation step. Escape mutations that confer resistance to these compounds map to proline 495, a residue located on the surface of the polymerase thumb domain and away from the active site. Substitution of this residue is sufficient to make the HCV enzyme and replicons resistant to the inhibitors. Interestingly, proline 495 lies in a recently identified noncatalytic GTP-binding site, thus validating it as a potential allosteric site that can be targeted by small-molecule inhibitors of HCV polymerase.
Hepatitis C virus (HCV) is the causative agent of themajority of chronic liver disease throughout the world. More than 170 million individuals are estimated to be infected with this virus (27). The size of the HCV epidemic and the limited efficacy of current therapy (based on the use of alpha interferon) have stimulated intense research efforts towards the development of antiviral drugs that are both better tolerated and more effective. The most widely established strategy for developing novel anti-HCV therapeutics aims at the identification of low-molecular-weight inhibitors of essential HCV enzymes.
RNA-dependent RNA polymerase (RdRP) activity, carried out by the NS5B protein, is essential for virus replication (13) and has no functional equivalent in uninfected mammalian cells. It is thus likely that specific inhibitors of this enzyme can be found that block HCV replication with negligible associated toxicity. The NS5B RdRP has been expressed in a variety of recombinant forms (2, 4). The production of highly soluble forms of the enzyme (12, 24), devoid of the C-terminal membrane anchoring domain (23), has allowed considerable progress toward the determination of the enzyme's three-dimensional structure and mechanism of action. The crystal structure of NS5B revealed a classical “right hand” shape, showing the characteristic fingers, palm, and thumb subdomains (1, 7, 14). More recently, the three-dimensional structure of the HCV polymerase was solved in complex with RNA (20) as well as in a complex with nucleoside triphosphates (6). Three distinct nucleotide-binding sites were observed in the catalytic center of HCV RdRP whose geometry was remarkably similar to that observed in the initiation complex of the RNA phage Φ6 RdRP (8), strengthening the proposal that the two enzymes initiate replication de novo by similar mechanisms. An unexpected result of this study was the observation of a GTP-binding site on the enzyme surface at the interface between the finger and thumb domains, 30 Å away from the polymerase catalytic center (6). This previously unidentified GTP pocket was proposed to be a potential allosteric regulatory site that could modulate alternative interactions between the two domains during the conformational change of the enzyme required for efficient initiation. The presence of a unique nucleotide-binding site away from the enzyme catalytic center could potentially provide an attractive target for allosteric inhibitors of the HCV polymerase reaction.
A number of structurally diverse nonnucleoside inhibitors (NNI) of the HCV polymerase have now been reported (10). Among these, two promising compound series that share a common benzimidazole scaffold have been described (P.-L. Beaulieu, G. Fazal, J. Gillard, G. Kukolj, and V. Austel, July 2002, World Intellectual Property Organization; H. Hashimoto, K. Mizutani, and A. Yoshida, Dec. 2001, World Intellectual Property Organization). Interestingly, an orally bioavailable benzimidazole analogue (JTK-003) is currently under investigation in early clinical trials (18). We have synthesized two benzimidazole-containing inhibitors of the HCV RdRP that are representative of each series. We show that these compounds act as allosteric inhibitors that block the activity of the polymerase prior to the polymerization step. By taking advantage of the recently developed subgenomic replication system (15), we demonstrate that at least one compound of this class is able to interfere with the replication of the HCV RNA in cell culture. Replicon clones that are resistant to inhibition were selected that allowed the identification of the possible inhibitor interaction site on the enzyme. This site, which we show to be common to the two compounds tested, corresponds to the previously identified surface GTP-binding site and thereby validates its relevance as a target for allosteric inhibitors of the HCV polymerase.
MATERIALS AND METHODS
Compound synthesis.
Compound A (2-[4-({4′-chloro-4-[(4-hydroxypiperidin-1-yl) carbonyl]-1,1′-biphenyl-2-yl}methoxy)-2-fluorophenyl]-1-cyclohexyl-1H-benzimidazole-5-carboxylic acid) and compound B (N-{[1-cyclohexyl-2-(3-furyl)-1H-benzimidazol-5-yl]carbonyl}-5-hydroxy-l-tryptophan) were synthesized as previously described (Hashimoto et al., World Intellectual Property Organization; Beaulieu et al., World Intellectual Property Organization).
Plasmids.
pHCVNeo17.B (25) encodes an HCV replicon identical to I377neo/NS3-3′/wt (15) but containing the adaptive mutations E176G in NS3 and a AAA triplet (coding for K) insertion after the GTG triplet, coding for V67 in NS5A. All other replicon plasmids were derived from pHCVNeo17.B and contain the following mutations: pHCVNeo17.BR1 and pHCVNeo17.BR2, replacement of CCG codon for P495 in NS5B with CTG (coding for L) or GCG (coding for A), respectively; pHCVNeo17.D, replacement of ATC codon for I585 in NS5B with GTC (coding for T); pHCVNeo17.DR2, replacement of CCG codon for P495 with CTG (coding for L) and of ATC codon for I585 with GTC (coding for T).
pT7-NS5BΔC55 contains the HCV-BK sequence coding for the NS5B protein lacking 55 C-terminal residues (residues 1 to 536) in the pT7-7 expression vector. pT7-GB/NS5BΔC23 encodes a GB virus B (GBV-B) NS5B protein lacking 23 C-terminal residues (residues 1 to 567).
NS5B expression and purification.
Expression of the HCV and GBV-B NS5B proteins in Escherichia coli BL21(DE3) and purification of the proteins were carried out as described previously (5).
Polymerase assays.
Primer-dependent assays were performed with either the heteropolymeric RNA template Dcoh (4) or the homopolymeric template-primer couple poly(A)-oligo(U)18 as previously described (24). Compounds were dissolved and diluted in dimethyl sulfoxide. Unless otherwise specified, compounds, polymerase, and template RNA were incubated at room temperature (RT) for 25 min before the addition of nucleoside triphosphates (NTPs). Alternatively, compounds were added to the preformed polymerase-template complex (15 min at RT) and incubated at RT for 10 min before the addition of NTPs. Elongation proceeded for 1 h at RT and the activity was measured as acid-insoluble radioactivity. Fifty percent inhibitory concentration (IC50) values were calculated by using a three-parameter logistic equation, and inhibition data were fitted by use of Kaleidagraph software.
Kinetic parameters were calculated from a least-square fit of initial rates as a function of substrate concentration, assuming Michaelis-Menten kinetics.
Inhibition mechanisms were determined by performing substrate titration experiments. In the single-turnover experiments, elongation reactions were started by the addition of nucleotides and 50 ng of heparin per μl.
Polymerase-inhibitor binding.
The polymerase-inhibitor complex was monitored essentially as previously described (21). Polymerase and compound (10 μM each) were mixed in 60 μl of incubation buffer (20 mM Tris-HCl [pH 7.5], 3 mM dithiothreitol, 100 mM NaCl, 0.03% n-octyl-β-d-glucopyranoside,10% glycerol) with or without 15 μM poly(A)-oligo(U)18. After a 10-min incubation at RT, the mixture was applied to a gel filtration G-25 spin column (Pharmacia) prewashed with incubation buffer. The eluate, containing the protein-inhibitor complex and the unbound protein, was recovered by centrifugation for 2 min at 1,450 × g. The eluting protein was quantified by Bradford assay (Bio-Rad), and the inhibitor was quantified by mass spectrometry as follows. The column eluate (0.4 μl) was injected into a reverse-phase C18 column coupled online with an ion trap mass spectrometer (LCQ DECA; Thermoquest, San Jose, Calif.) operated with selected reaction monitoring. The flow from the column was split 1:10 towards the electrospray ionization (ESI) inlet of the ion trap mass spectrometer and the diode array detector. All spectra were acquired at unit resolution and 0.3% mass accuracy. The inhibitor was quantitated from a five-point calibration curve.
Tissue culture, replication analysis selection, and sequencing of resistant replicons.
Huh-7 and HBI10A cells were cultured as previously described (25). Transient transfections by electroporation of in vitro-transcribed RNAs were performed using cells that are highly competent for HCV replication, obtained by curing HBI10A cells of the endogenous replicons with human alpha interferon 2b as described previously (25). The effect of compounds on viral replication was monitored by cell enzyme-linked immunosorbent assay (cell-ELISA) (25) or by in situ RNase protection assay (isRPA) (9). Clones resistant to compound A were selected as previously described (25). HBI10A cells were plated at 3 × 103/cm2 and cultured in the presence of 1 mg of G418 per ml and increasing concentrations of compound A, from 1.6 to 4 μM. Approximately 15 days after beginning selection, small colonies of cells resistant to the inhibitor and the antibiotic became visible and were isolated. Replicon RNAs extracted from resistant clones were retrotranscribed, amplified by PCR, and sequenced by automated sequencing.
Transient-transfection assays were performed as described previously (25). Replication efficiency was determined 96 h after transfection by cell-ELISA and was expressed as the ratio between the absorbance value of the sample transfected with a given RNA and the absorbance value of mock-transfected cells. The values were normalized to the transfection efficiency measured by cell-ELISA 24 h after transfection. Each experiment was performed in triplicate, and average absorbance values were used for calculations.
RESULTS
Effect of benzimidazole-based inhibitors on NS5B polymerase activity.
Compounds A and B were chosen as representative examples of benzimidazole-based inhibitors of the HCV polymerase (Beaulieu et al., World Intellectual Property Organization; Hashimoto et al., World Intellectual Property Organization). Both compounds were confirmed to strongly inhibit HCV RdRP activity in a dose-dependent manner. We measured similar IC50 values of about 0.25 μM using either homopolymeric poly(A)-oligo(U)18 template-primer or heteromeric RNA (Table 1; see below). The inhibitor potency was independent of the form of recombinant polymerase used, as both full-length and C-terminally truncated NS5BΔC21 and ΔC55 enzymes were inhibited, with similar IC50 values (data not shown). Compounds A and B were highly selective against HCV polymerase and did not inhibit the closely related GBV-B RdRP (Table 1). In assays using poly(A)-oligo(U)18 as template-primer, both compounds appeared to be noncompetitive with respect to UTP (Table 1). Moreover, increasing amounts of template-primer RNA did not affect the inhibition potency, suggesting that RNA binding does not interfere with the enzyme-compound interaction. Consistent with their structural similarities, the two compounds appeared clearly competitive with each other when tested in direct competition assays (not shown). Taken together, these results suggest that both the compounds interact with the polymerase at a site distinct from the catalytic center.
TABLE 1.
Potency, selectivity, and mechanism of inhibition of compounds A and B
Ki and Kii values were derived from a replot of slopes and 1/Vmax, respectively, of double-reciprocal plots.
NA, not active at 50 μM.
Data are means ± standard deviations.
Order of addition.
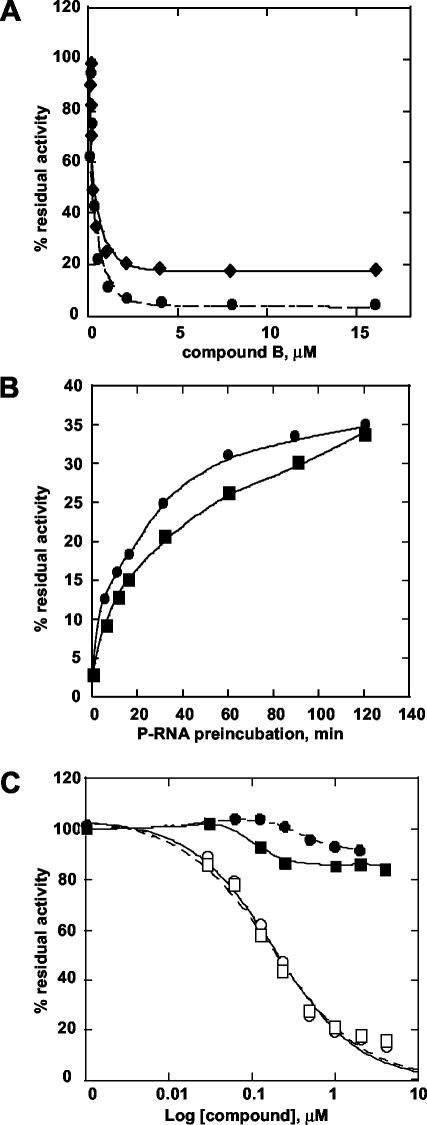
We observed that the order of reagent addition in the RdRP reaction affected the shapes of the inhibition curves. Inhibition experiments were performed by either preincubating the enzyme with the RNA template prior to addition of the inhibitors or by omitting the preincubation step (for compound B, see Fig. 1A; for compound A, data not shown). Similar IC50 values were measured in both cases, but complete inhibition of the enzymatic activity could not be attained if the inhibitors were added to a preformed enzyme-RNA complex. In this case, a significant fraction of the polymerase activity was not inhibited, even at very high compound concentrations. A possible explanation for this finding is that the fraction of the enzyme engaged with the RNA in a preelongation complex is protected from the action of the inhibitor. In line with this interpretation, when the NS5B polymerase and the RNA template were preincubated for increasing times before the addition of compounds, at a concentration 15-fold above the IC50 values, the percentage of residual activity increased as a function of preincubation time (Fig. 1B). Interestingly, polymerase activity in the absence of inhibitor also increased with the enzyme-RNA preincubation time (data not shown), likely reflecting the formation of a productive preelongation complex.
FIG. 1.
Inhibition by compounds A and B. (A) Dependence of inhibition curves on polymerase-RNA (P-RNA) complex formation. Increasing amounts of compound B (30 nM to 16 μM) were added to polymerase and heteropolymeric RNA (Dcoh) that were (♦) or were not (•) preincubated for 15 min at RT. (B) Effect of P-RNA preincubation time. NS5BΔC55 and Dcoh template were preincubated from 0 to 120 min before the addition of 4 μM compound A (▪) or compound B (•). The residual activity was expressed as the percentage of that obtained at each time point in the absence of inhibitor. (C) Inhibition curves for compound A and compound B under single-cycle conditions. NS5BΔC55 and the Dcoh template (20 nM) were preincubated for 15 min at RT before the addition of compounds from 30 nMto 4 μM. Elongation was started by the addition of nucleotide mixture (5 μM each plus 4 μCi of [3H]UTP) with (▪, compound A; •, compound B) or without (□, compound A; ○, compound B) heparin (50 ng/μl).
Inhibition under single-cycle conditions.
The RdRP assays described above were performed under continuous polymerization conditions whereby the polymerase performs multiple sequential rounds of processive RNA synthesis. The inhibited activity after preincubation of enzyme and template might therefore result from those polymerase molecules that dissociated from the template during the reaction and were thus susceptible to inhibition. In order to assess directly whether compounds A and B could no longer interfere with the enzymatic activity of polymerase molecules already engaged in a preelongation complex, we measured inhibition under conditions that favored single-cycle RNA synthesis. The inhibitors were added to preformed polymerase-RNA complexes and the elongation reaction was started by the addition of nucleotides and heparin (Fig. 1C). Heparin functions as a trapping agent to titrate free enzyme as well as polymerase molecules that dissociate from the template after completion of a processive round of RNA synthesis (3). Under these conditions, we measured only the activity of those polymerase molecules that were engaged in a productive preelongation complex prior to the addition of the trapping agent. As shown in Fig. 1C, both compounds failed to inhibit polymerase activity under these conditions, suggesting their inability to act on preformed enzyme-RNA complexes.
Polymerase-inhibitor interaction.
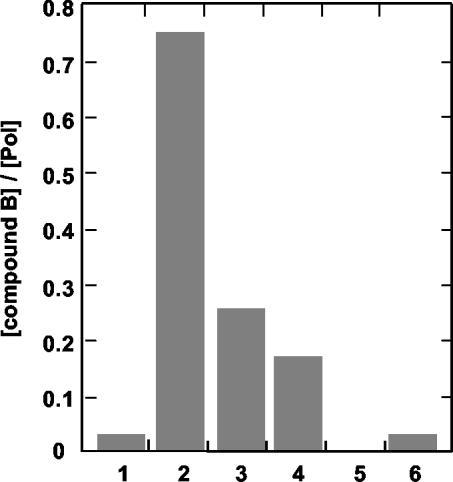
To further investigate the mechanism of inhibition, we performed experiments aimed at the direct characterization of the polymerase-inhibitor interaction. Following preincubation, polymerase-inhibitor complexes were separated from the unbound compound through gel filtration columns. The concentrations of enzyme and inhibitor in the complex were measured by Bradford assay and mass spectrometry, respectively. Due to the low solubility of compound A, these experiments were performed only with compound B. As shown in Fig. 2 (bars 1 and 2), compound B eluted with the enzyme at almost stoichiometric concentrations when incubation was performed in the presence of poly(A)-oligo(U)18 RNA. As expected, compound B did not associate with GBV-B NS5B (Fig. 2, bar 5) or with RNA alone (not shown). This finding suggests that compound B interacts with the purified enzyme in the presence of template RNA. However, when the inhibitor was added after prolonged preincubation of the polymerase with template RNA, the amount of compound eluting with the enzyme decreased with longer preincubation times, reaching about 20% of the initial value after 60 min (Fig. 2, bars 3 and 4).
FIG. 2.
Polymerase-inhibitor binding. Compound B and polymerases (bars 1 to 4, NS5BΔC55; bar 5, GB/NS5BΔC23; bar 6, NS5BΔC55-P495L) were incubated in the absence (bar 1) or presence (bars 2, 5, and 6) of 15 μM poly(A)-oligo(U)18. Alternatively, NS5BΔC55 and poly(A)-oligo(U)18 were preincubated for 30 (bar 3) or 60 (bar 4) min before the addition of compound B. Polymerase-inhibitor complexes were separated by gel exclusion chromatography, and the eluted polymerase and compound were quantitated as described in Materials and Methods. Data are reported as molar ratios between eluting compound and polymerase.
This and the previous findings suggest that although the initial interaction of the enzyme with the template RNA appears to be essential for compound B binding, the polymerase-RNA complex undergoes a slow conformational change to a form of the enzyme that is no longer susceptible to inhibition.
Antiviral activity and selection of resistant mutants.
The effect of compounds A and B on the replication of HCV subgenomic replicons was determined by using Huh-7 clone HBI10A (19) and was monitored by measuring replicon RNA by isRPA and NS3 protein by cell-ELISA (9, 25). Incubation with compound A resulted in a dose-dependent reduction of both viral RNA and NS3 protein synthesis, with an IC50 of about 0.35 μM in both assays (Table 2). Conversely, compound B up to 10 μM showed no inhibition of viral replication (not shown). Compound A was nontoxic and had no effect on cell growth rate up to 10 μM (not shown), indicating that its direct effect is on viral replication. Taking advantage of the expression of neomycin resistance, we cultured HBI10A cells in the presence of G418 and compound A in order to select inhibitor-resistant replicon variants. Selection yielded several resistant clones that duplicated at the same rate as parental cells and expressed HCV RNA and proteins at comparable levels. More importantly, the IC50 values for compound A on all the selected clones were at least 10-fold higher than that on parental cells (Table 2). The NS5B sequences of four resistant clones were determined. Remarkably, all shared replacement of proline 495 with leucine (P495L) or alanine (P495A), and in addition, clone 10AI14 contained replacement of isoleucine 585 with threonine (I585T) (Table 2). These clones were as sensitive to alpha interferon and the nucleoside inhibitor 2′-C-methyl-adenosine (9) as the parent cells were, indicating that resistance was specific for compound A (not shown). In order to assess their relevance for resistance, the NS5B mutations were segregated in the pHCVNeo17.B replicon. This replicon harbors two adaptive mutations that enhance replication efficiency (25). Among the resulting replicons, pHCVNeo17.BR1 and pHCVNeo17.BR2, bearing mutations P495L and P495A, respectively, were clearly resistant to compound A but replicated less efficiently than the selected clones (Table 3). The IC50 values measured for these replicons were similar to those observed in the resistant cells, indicating that substitutions of P495 were sufficient to confer the resistance phenotype of the selected clones. Conversely, pHCVNeo17.D, bearing an I585T substitution, was still sensitive to inhibition by compound A and even showed enhanced replication efficiency with respect to the parent replicon. Interestingly, a replicon containing both P495L and I585T (pHCVNeo17.DR1) replicated more efficiently than pHCVNeo17.BR1 and was resistant to inhibition by compound A, suggesting that the I585T substitution was irrelevant for resistance but partially compensated the replication defect due to P495L.
TABLE 2.
Effect of compound A on parental and resistant clones
| Clone | IC50 (μM) for compound Aa
|
NS5B mutation(s) | |
|---|---|---|---|
| Cell-ELISA | isRPA | ||
| HBI10A | 0.35 ± 0.15 | 0.30 ± 0.2 | |
| 10AI1 | >5 | >5 | P495L |
| 10AI2 | >5 | >5 | P495L |
| 10AI12 | >5 | >5 | P495A |
| 10AI14 | >5 | >5 | P495L, 1585T |
Data are means ± standard deviations.
TABLE 3.
Resistant replicons and enzyme
| Replicon or enzyme | NS5B mutation(s) | Replication efficiency (arbitrary units)a | IC50 (μM)
|
||
|---|---|---|---|---|---|
| Compound A | Compound B | ||||
| pHCVNEO17 replicons
|
|||||
| B | 14.2 | 0.32 | |||
| BR1 | P495L | 3.1 | >5 | ||
| BR2 | P495A | 10.9 | 2.2 | ||
| D | I585T | >20 | 0.4 | ||
| DR1 | P495L, I585T | 10.6 | >5 | ||
| Enzyme ΔC55 | |||||
| P495L | >20 | 16 ± 0.8 | |||
Replication efficiency was determined by cell-ELISA.
Mechanism of resistance of mutant polymerase.
To support the genetic evidence, we introduced the P495L mutation in the NS5BΔC55 protein. The mutant protein showed substantially reduced susceptibility to inhibition by both compounds A and B (Table 3). Conversely, comparable kinetic parameters were measured for the poly(A)-oligo(U)18 RNA for wild-type and mutant proteins (kcat/Km = 15,830 and 9,649 s−1 M−1, respectively), indicating that the P495L mutation did not significantly affect polymerase activity.
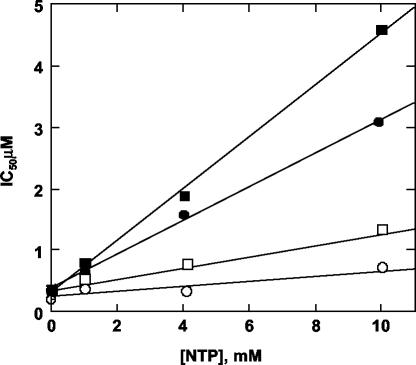
In order to assess whether the P495 mutations conferred resistance by impairing the interaction with the inhibitors, we measured by mass spectrometry the amount of compound B eluting with the purified P495L mutant protein from gel filtration columns. As shown in Fig. 2 (bar 6), compound B was hardly detectable, indicating that the mutant enzyme had a reduced affinity for the inhibitors. This result confirmed that compounds A and B inhibited polymerase activity by binding the enzyme at the same or overlapping sites. Interestingly, P495 is part of a specific noncatalytic GTP-binding site recently identified by X-ray crystallography on the surface of the enzyme, at the interface between the finger and thumb domains (6). Thus, we verified whether GTP was specifically able to interfere with inhibition by compounds A and B by measuring potencies at increasing GTP concentrations. As shown in Fig. 3, the IC50 values increased with increasing GTP concentrations, while they were marginally affected by even high concentrations of CTP. The GTP concentration that produced doubling of the IC50 values was very high (about 1 mM), in agreement with the low-affinity nature of the surface GTP-binding site (5), and did not significantly affect the RdRP activity in our assay (not shown).
FIG. 3.
Effect of GTP and CTP on inhibition potency. The IC50 values for inhibition of NS5BΔC55 on Dcoh RNA were measured in the presence of 5 μM and 1, 4, and 10 mM GTP (•, compound A; ▪, compound B) or CTP (○, compound A; □, compound B). Reactions were carried out as described in Materials and Methods in the presence of 15 mM MgCl2, 10 μM UTP, 2 μCi of [3H]UTP, 250 μM ATP, and 250 μM CTP or GTP.
DISCUSSION
Although they are derived from independent studies, both HCV polymerase NNIs used in this study share a common cyclohexyl-benzimidazole scaffold that might constitute the active center of the molecule. In light of this similarity, we thought that these compounds may inhibit the HCV RdRP through a common mechanism, interacting at the same site of the enzyme. As expected on the basis of their chemical structures, these compounds were found to be noncompetitive with nucleotide substrates. Interestingly, inhibition of RNA synthesis by compounds A and B was not observed in single-turnover experiments, indicating that both are unable to affect the actively elongating enzyme. Their inability to act during the elongation phase excludes the idea that inhibition might be due to alteration of the NS5B processivity by inducing premature dissociation of the enzyme or by altering its translocation along the RNA product.
As is the case for other polymerases, the HCV RdRP catalyzes RNA synthesis through an ordered stepwise mechanism, with RNA template binding occurring first. Each step presumably involves conformational changes of the enzyme leading to the proper positioning of template, the growing RNA chain, and incoming nucleotides in the catalytic center. Our data support a model in which the benzimidazole-containing NNIs act at a step prior to the formation of a productive polymerase-RNA complex. Interestingly, though these compounds do not prevent interaction with the RNA template, prolonged incubation of the enzyme with RNA abolishes the interaction of the inhibitors. We propose, therefore, that by interacting with the enzyme in the polymerase-RNA complex, the compounds might effect a slow conformational transition preceding nucleotide binding that is required for the formation of a productive preelongation complex. However, once the conformational transition has happened, the polymerase may no longer be sensitive to inhibition. This possibility is supported by the observation that when the compounds are added to preformed polymerase-RNA complexes, there is residual enzyme activity even at a saturating inhibitor concentration. This activity may correspond to the fraction of enzyme that has undergone the conformational change and is therefore no longer susceptible to inhibition. The time course experiments shown in Fig. 1 and 2 are in line with the existence of an intrinsically slow conformational change that occurs within a polymerase-RNA complex and leads to the formation of a productive preelongation complex. The existence of isomerization steps within the enzyme-template-primer complex is well documented for the human immunodeficiency virus (HIV) reverse transcriptase (28), and additional studies would be required to obtain a more direct proof that this is also the case for the HCV RdRP.
As for HIV, the high mutational frequency of HCV is expected to favor the generation of drug-resistant mutants upon long-term treatment with inhibitors of viral enzymes. Thus, resistance studies using tissue culture systems are considered crucial to optimize the resistance profiles of inhibitors and can contribute important information for understanding the mechanism of inhibition. In the absence of a suitable in vitro infection model, we took advantage of the recently developed subgenomic replication system (15) to select for replicon clones harboring resistant mutations. This approach has already been successfully used to select mutants resistant to an inhibitor of the viral NS3-4A serine protease (25) and has now allowed us to identify the putative region where compounds A and B interact with the polymerase enzyme. Remarkably, all the selected replicons contained mutations of proline 495 in NS5B, which we demonstrated to be responsible for the acquired resistance to inhibition by both the compounds. Proline 495 in NS5B is conserved in >99% of natural HCV isolates of all strains, suggesting a significant role for the region where P495 lies during HCV replication and possibly explaining the lower replication efficiency of replicons in which its substitutions were segregated. Supplementary mutations might have emerged to compensate for the replication defect in the selected cell clones. In fact, an additional mutation in NS5B, I585T, partially restored the replication ability of replicons containing P495 substitutions. The I585T mutation was able to enhance replication capability even in the absence of the P495 substitution, therefore excluding a direct effect on polymerase enzymatic activity. Interestingly, P495 has been recently identified by X-ray crystallography as one of the key residues involved in the interaction with a noncatalytic GTP molecule on the NS5B surface (6), leading to the speculation that the binding site for benzimidazole-based inhibitors at least partially overlaps with the surface GTP-binding site (Fig. 4). The reduced ability of the mutant enzyme to interact with compound B and the effect of high GTP concentrations on the potency of both compounds A and B strengthen this hypothesis. This model, however, will ultimately require confirmation by structural studies of the polymerase in complex with RNA template and inhibitors.
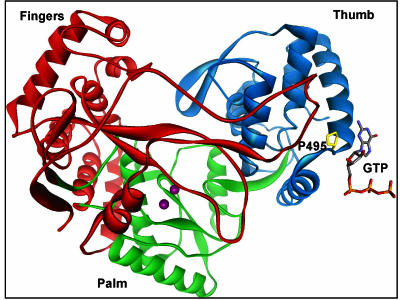
FIG. 4.
Location of GTP-binding site and P495. Top view of the NS5BΔC55 polymerase with finger, palm, and thumb domains in red, green, and blue, respectively. A stick representation shows the noncatalytic GTP in the surface site interacting with P495 (yellow). Divalent Mn2+ metal ions in the catalytic center are displayed as violet spheres.
Though specific, GTP binding at the surface site appears to have no consequence on in vitro polymerase activity. In our experiments, while GTP strongly stimulated de novo activity of the NS5B polymerase, high GTP concentrations only very modestly enhanced the overall polymerase efficiency on heteromeric templates, which is different from what was previously reported by others (16). We believe that these effects are not exerted by binding at the surface site, as mutations of residues in the surface GTP-binding site neither alter the GTP response of activity nor significantly affect the de novo efficiency of NS5B (L. Tomei and A. Biroccio, unpublished observations). The latter result is in line with recently published observations (22) that point to the initiating-NTP site in the catalytic cavity of the enzyme, not the surface site, as the site at which GTP binding plays a regulatory role for de novo activity of NS5B. Whether or not GTP binding at the surface site could be of biological significance per se, our finding clearly points to this site as an allosteric pocket on the enzyme surface that may be targeted by small-molecule inhibitors of HCV polymerase activity. Moreover, the observation that replicons carrying P495 substitutions do not replicate efficiently suggests that this region of the molecule might play a key function during viral replication through, for example, the interaction with other viral and/or cellular factors.
The relevance of the thumb domain surface as a target for allosteric inhibitors of the HCV polymerase is becoming increasingly evident. Two series of NNIs of the NS5B enzyme have recently been shown by X-ray crystallography to interact in a hydrophobic pocket at the base of the thumb domain (17, 26; R. A. Love and X. Yu, May 2001, European Patent Office). This site does not overlap with the surface GTP-binding site and is almost 15 Å from it. In contrast to HIV reverse transcriptase, for which all known NNIs have been shown to bind to the same site of the enzyme (11), HCV NS5B apparently contains multiple regions of the thumb domain that are potential targets for allosteric inhibitors. This is of particular relevance in consideration of the possible requirement for a combination therapy regimen based on the use of multiple NNIs.
Acknowledgments
L. Tomei and S. Altamura contributed equally to this work.
We are grateful to Uwe Koch for continuous helpful discussions and suggestions. We also thank Simona Ponzi for the chemical synthesis of the compounds used in this work.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Al, R. H., Y. Xie, Y. Wang, and C. H. Hagedorn. 1998. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 53:141-149. [DOI] [PubMed] [Google Scholar]
- 3.Bambara, R. A., P. J. Fay, and L. M. Mallaber. 1995. Methods of analyzing processivity. Methods Enzymol. 262:270-280. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Biroccio, A., J. Hamm, I. Incitti, R. De Francesco, and L. Tomei. 2002. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3688-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 10.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 11.Ding J., K. Das, Y. Hsiou, W. Zhang, E. Arnold, P. N. S. Yadav, and S. H. Hughes. 1997. Structural studies on HIV-1 reverse transcriptase and implications for drug design, p. 41-82. In P. Veerapandian, (ed.), Structure-based drug design. Marcel Dekker, Inc., New York, N.Y.
- 12.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 16.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 17.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McHutchison, J. G., and K. Patel. 2002. Future therapy of hepatitis C. Hepatology 36:S245-S252. [DOI] [PubMed] [Google Scholar]
- 19.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 20.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 21.Orsatti, L., S. Di Marco, C. Volpari, A. Vannini, P. Neddermann, and F. Bonelli. 2002. Determination of the stoichiometry of noncovalent complexes using reverse-phase high-performance liquid chromatography coupled with electrospray ion trap mass spectrometry. Anal. Biochem. 309:11-18. [DOI] [PubMed] [Google Scholar]
- 22.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 24.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 25.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, R. C. Bethell, and M. N. James. 2003. Nonnucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase: crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 27.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 28.Wohrl, B. M., R. Krebs, R. S. Goody, and T. Restle. 1999. Refined model for primer/template binding by HIV-1 reverse transcriptase: pre-steady-state kinetic analyses of primer/template binding and nucleotide incorporation events distinguish between different binding modes depending on the nature of the nucleic acid substrate. J. Mol. Biol. 292:333-344. [DOI] [PubMed] [Google Scholar]