Abstract
Lesion bypass is an important mechanism to overcome replication blockage by DNA damage. Translesion synthesis requires a DNA polymerase (Pol). Human Pol ι encoded by the RAD30B gene is a recently identified DNA polymerase that shares sequence similarity to Pol η. To investigate whether human Pol ι plays a role in lesion bypass we examined the response of this polymerase to several types of DNA damage in vitro. Surprisingly, 8-oxoguanine significantly blocked human Pol ι. Nevertheless, translesion DNA synthesis opposite 8-oxoguanine was observed with increasing concentrations of purified human Pol ι, resulting in predominant C and less frequent A incorporation opposite the lesion. Opposite a template abasic site human Pol ι efficiently incorporated a G, less frequently a T and even less frequently an A. Opposite an AAF-adducted guanine, human Pol ι was able to incorporate predominantly a C. In both cases, however, further DNA synthesis was not observed. Purified human Pol ι responded to a template TT (6–4) photoproduct by inserting predominantly an A opposite the 3′ T of the lesion before aborting DNA synthesis. In contrast, human Pol ι was largely unresponsive to a template TT cis-syn cyclobutane dimer. These results suggest a role for human Pol ι in DNA lesion bypass.
INTRODUCTION
Lesion bypass is a ubiquitous biological process in response to unrepaired DNA damage during replication. Depending on the outcome, lesion bypass is divided into error-free bypass and error-prone bypass. Error-free lesion bypass predominantly incorporates the correct nucleotide opposite the DNA damage, whereas error-prone lesion bypass frequently results in mutations opposite the damage. In Escherichia coli DNA polymerase (Pol) V, composed of the UmuC′2D protein complex, is involved in an error-prone lesion bypass mechanism (1,2). In eukaryotes a damage-induced mutagenesis pathway, originally discovered in the yeast Saccharomyces cerevisiae (3,4), constitutes an important error-prone lesion bypass mechanism (5–7). In this mutagenesis pathway DNA Pol ζ (the REV3–REV7 protein complex) is thought to be involved in the translesion DNA synthesis step (8–10). More recently the UmuC superfamily of proteins has been recognized and is thought to be important for lesion bypass (11,12). The prototypic members of this superfamily include E.coli UmuC, E.coli DinB, yeast Rev1 and yeast Rad30 (11,12).
Pol η, encoded by the RAD30 gene in yeast and the XPV gene in humans, was originally discovered as a eukaryotic DNA polymerase for error-free bypass of TT dimers in DNA (13–15). The critical role of Pol η in response to UV radiation has been unequivocally established by the phenotypes of the human hereditary disease xeroderma pigmentosum variant (XPV). XPV patients show sun sensitivity and a predisposition to skin cancer (16), as a result of reduced UV resistance and enhanced UV mutability of the XPV cells (17). It is believed that the error-free bypass activity of Pol η opposite TT dimers and perhaps other cyclobutane pyrimidine dimers significantly contributes to the suppression of UV-induced cytotoxicity and mutagenesis in human cells. However, recent in vitro studies have demonstrated that additional lesions can be bypassed by Pol η (18–23). Furthermore, error-prone translesion synthesis has also been observed with Pol η (19–21).
RAD30B is another human homolog of the yeast RAD30 gene (11). The human RAD30B gene codes for the ninth DNA polymerase, Pol ι (24–26). Human Pol ι is an unprecedented error-prone DNA polymerase (24–26). Two unique properties of this enzyme differentiate it from all other DNA polymerases known: (i) violation of the Watson–Crick base pairing rule opposite template T with preferential G, rather than A, incorporation (24–26); (ii) the T stop that frequently aborts DNA synthesis opposite a template T (24). These remarkable features suggest that human Pol ι may play a specialized function in human biology. On the other hand, since human Pol ι belongs to the UmuC superfamily of proteins, it is possible that this polymerase may also play a role in DNA lesion bypass. We have investigated this possibility by asking how human Pol ι would respond to DNA lesions in vitro. In this report we demonstrate efficient nucleotide insertion by human Pol ι opposite a template AP site and show different responses of human Pol ι to a template 8-oxoguanine, AAF-adducted guanine, TT cis-syn cyclobutane dimer and TT (6–4) photoproduct.
MATERIALS AND METHODS
Materials
N-acetoxy-N-2-acetylaminofluorene (AAAF) was obtained from the Midwest Research Institute (Kansas City, MO). The yeast rad30 deletion mutant strain BY4741rad30Δ (MATa his3 leu2 met15 ura3 rad30Δ) was purchased from Research Genetics (Huntsville, AL). Human Pol ι was expressed in the yeast rad30 deletion mutant cells and purified to apparent homogeneity as previously described (24). Human Pol η, Pol κ and Pol β and yeast Pol η were expressed in yeast cells and purified to apparent or near homogeneity (19,20,27,28).
Damaged DNA templates
A 30mer DNA template containing a site-specific 8-oxoguanine was synthesized by automated DNA phosphoramidite methods by Operon (Alameda, CA). Its sequence is 5′-GGATGGACTGCAGGATCCGGAGGCCGCGCG-3′, where the position of the 8-oxoguanine is underlined. DNA templates containing a site-specific tetrahydrofuran (AP site analog) were also synthesized by Operon. Their sequences are 5′-GCGCGCTTCTGGCCAATXCTAGACGGTAGG-3′ (template AP), 5′-GAAGGGATCCTTAAGACTXTAACCGGTCTTCGCGCG-3′ (template AP-T), 5′-GAAGGGATCCTTAAGACAXTAACCGGTCTTCGCGCG-3′ (template AP-A), 5′-GAAGGGATCCTTAAGACGXTAACCGGTCTTCGCGCG-3′ (template AP-G) and 5′-GAAGGGATCCTTAAGACCXTAACCGGTCTTCGCGCG-3′ (template AP-C), where X designates the AP site analog. A 49 nt DNA template containing a site-specific cis-syn TT dimer or a TT (6–4) photoproduct was prepared as previously described (29). Its sequence is 5′-AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT-3′, where the modified TT is underlined.
To prepare AAF-adducted DNA template, 2 nmol oligonucleotide 5′-CCTTCTTCTTCATACAAGCTTACTTCTTCC-3′ was incubated with 200 nmol AAAF at 37°C in the dark for 3 h in 100 µl of TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA) containing 20% ethanol. AAAF predominantly modified the unique G in the DNA. Free AAAF was removed by extracting the reaction mixture five times with water-saturated ether. Then, damaged and undamaged oligonucleotides were separated by electrophoresis on a 20% denaturing polyacrylamide gel. AAF-modified DNA migrated slower on the gel and was sliced out of the gel. The gel slices were soaked in 150 µl of water at room temperature for 4 h. Finally, AAF-damaged DNA was recovered using a GenElute DNA spin column (Sigma, St Louis).
DNA lesion bypass assays
Lesion bypass assays were performed in standard DNA polymerase reactions using various damaged DNA templates as indicated. The standard DNA polymerase reaction (10 µl) contained 25 mM potassium phosphate (pH 7.0), 5 mM MgCl2, 5 mM dithiothreitol, 100 µg/ml bovine serum albumin, 10% glycerol, 50 µM dNTPs (dATP, dCTP, dTTP and dGTP individually or together as indicated), 50 fmol DNA substrate containing a 32P-labeled primer and purified DNA Pol ι. After incubation at 30°C for 10 min reactions were terminated with 7 µl of a stop solution (20 mM EDTA, 95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol). The reaction products were resolved on a 20% polyacrylamide gel containing 8 M urea and visualized by autoradiography. Primer extension was quantitated by scanning densitometry of the autoradiogram using the SigmaGel software (Sigma) for analysis.
RESULTS
Translesion synthesis of 8-oxoguanine by human Pol ι
8-Oxoguanine is a major form of oxidative damage in DNA. To examine whether human Pol ι can bypass 8-oxoguanine we first purified this DNA polymerase to apparent homogeneity (24) and then performed DNA synthesis in vitro using an 8-oxoguanine template. The 30mer DNA template contained a site-specific 8-oxoguanine residue and a 32P-labeled 17mer primer annealed right before the lesion (Fig. 1A). As shown in Figure 1B (lanes 6–10), purified human Pol ι was able to bypass the template 8-oxoguanine. Compared to the undamaged control template (Fig. 1B, lanes 1–5), however, significant inhibition of human Pol ι activity by the template 8-oxoguanine was evident (Fig. 1C). On the undamaged DNA template human Pol ι extended the primer up to 5 nt to stop opposite a template T (Fig. 1B, lanes 1–5), a Pol ι phenomenon known as the T stop (24). On the 8-oxoguanine template human Pol ι incorporated 1 nt opposite the lesion and then extended only 1 nt further without reaching the template T (Fig. 1B, lanes 6–10). These results show that human Pol ι is capable of translesion synthesis opposite a template 8-oxoguanine with low efficiency.
Figure 1.

Translesion DNA synthesis opposite a template 8-oxoguanine (8-oxoG) by human Pol ι. (A) The DNA template for translesion synthesis. The 17mer primer was labeled with 32P at its 5′-end as indicated by an asterisk and annealed right before the lesion. (B) DNA polymerase assays were performed with increasing amounts of purified human Pol ι using DNA templates without (lanes 1–5) or with (lanes 6–10) a site-specific 8-oxoguanine. DNA size markers in nt are indicated in the middle. (C) Quantitation of the results in (B).
To identify the nucleotide incorporated opposite 8-oxoguanine we performed DNA synthesis assays with only one deoxyribonucleoside triphosphate: dATP, dCTP, dGTP or dTTP individually. As shown in Figure 2 (lanes 6–10), human Pol ι extended 44% of the primers using dCTP, 22% of the primers using dATP and 14% of the primers using dGTP opposite the template 8-oxoguanine. In comparison, human Pol ι misincorporated T more frequently than A opposite the undamaged template G (Fig. 2, lanes 1–5). These results indicate that 8-oxoguanine significantly alters the coding property of the template G during DNA synthesis by human Pol ι and that the probability of misincorporating A opposite 8-oxoguanine by human Pol ι is significantly increased.
Figure 2.

Specificity of nucleotide incorporation by human Pol ι opposite the template 8-oxoguanine. DNA polymerase assays were performed with purified human Pol ι (19 ng, 238 fmol) using 50 fmol of DNA templates without (lanes 1–5) or with (lanes 6–10) a site-specific 8-oxoguanine. The 8-oxoguanine DNA template containing a 32P-labeled 17mer primer annealed right before the lesion is shown in Figure 1A. Polymerase reactions were carried out in the presence of each deoxyribonucleoside triphosphate dATP (A), dCTP (C), dTTP (T) and dGTP (G) individually or all four dNTPs (N4). Quantitation of extended primers is shown at the bottom of the gel. DNA size markers in nt are indicated on the left.
8-Oxoguanine is not a significant blocker of many DNA polymerases tested (20,30). However, our results in Figure 1B would suggest that 8-oxoguanine may significantly block human Pol ι. To confirm this conclusion we annealed a 32P-labeled 15mer primer 2 nt before the template 8-oxoguanine (Fig. 3) and performed primer extension assays. Undamaged templates with identical sequence were used as controls to determine the effect of 8-oxoguanine on primer extension. As shown in Figure 3 (compare lanes 1 and 2 with lanes 3 and 4), the majority of DNA synthesis by human Pol ι stopped right before the template 8-oxoguanine, yielding the 17mer DNA band as the major product. In contrast, a major DNA synthesis stop right before the template 8-oxoguanine (17mer band) was not observed with human Pol β, human Pol η, yeast Pol η and human Pol κ (Fig. 3, lanes 5–20). These results show that template 8-oxoguanine significantly and uniquely blocks human Pol ι during DNA synthesis.
Figure 3.
Effect of a template 8-oxoguanine on DNA synthesis by various DNA polymerases. A 32P-labeled 15mer primer was annealed 2 nt before the template 8-oxoguanine as indicated or annealed to the undamaged control template of the same sequence. Using the annealed templates without (8-oxoG, –) or with (8-oxoG, +) the template 8-oxoguanine, DNA polymerase assays were performed with increasing amounts of purified human Pol ι (lanes 1–4), human Pol β (lanes 5–8), human Pol η (lanes 9–12), yeast Pol η (lanes 13–16) and human Pol κ (lanes 17–20). Products extended to opposite the template 8-oxoguanine are indicated by the arrowheads. DNA size markers in nt are indicated at the sides.
Efficient error-prone nucleotide insertion opposite a template AP site by human Pol ι
AP sites are significant spontaneous and induced DNA lesions. Recently we observed that human Pol ι incorporated 1 nt opposite a template AP site more efficiently than opposite a template T (24). Kinetic analyses indicated that human Pol ι is catalytically most inefficient opposite a template T, but most efficient opposite a template A (24). To compare nucleotide incorporation opposite a template AP site versus a template A we labeled a 17mer primer at its 5′-end with 32P and annealed it to a DNA template right before a template A (template FY3) or an AP site (template AP) (Fig. 4). Primer extension was then performed with increasing amounts of purified human Pol ι. As shown in Figure 4 (compare lanes 1–3 with lanes 4–6), nucleotide incorporation opposite the template AP site was nearly as efficient as opposite the template A. These results show that human Pol ι is able to incorporate 1 nt opposite a template AP site very efficiently.
Figure 4.

Efficient nucleotide insertion by human Pol ι opposite a template AP site. A 32P-labeled 17mer primer was annealed right before an A of the undamaged template FY3 or annealed right before the AP site (designated by X) of the template AP as shown. Polymerase assays were performed with increasing amounts of human Pol ι using 50 fmol undamaged template (FY3, lanes 1–3) and the AP site-containing template (template AP, lanes 4–6). Quantitation of extended primers is shown at the bottom of the gel. DNA size markers in nt are indicated on the right.
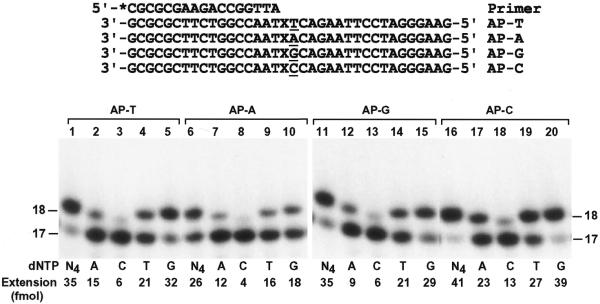
To identify the nucleotide incorporated by human Pol ι opposite the template AP site we performed lesion bypass assays with only one deoxyribonucleoside triphosphate. Furthermore, to examine whether nucleotide incorporation opposite the AP site by human Pol ι is influenced by the sequence context we synthesized four DNA templates that differed by 1 nt 5′ to the AP site (Fig. 5). A 17mer primer was labeled at its 5′-end with 32P and annealed to the templates, right before the template AP site (Fig. 5). Then the primed DNA templates were incubated with human Pol ι for primer extension assays. As shown in Figure 5, nucleotide incorporation opposite the AP site by human Pol ι was most efficient when the template base 5′ to the lesion was a C and most inefficient when this template base was an A (Fig. 5, compare lanes 1, 6, 11 and 16). Nucleotide incorporation opposite the AP site by human Pol ι followed the order, from most frequent to least frequent: G > T > A > C. These results show that human Pol ι predominantly incorporates G opposite a template AP site, whose further extension is blocked by the lesion.
Figure 5.
Specificity of nucleotide incorporation opposite the template AP site. A 32P-labeled 17mer primer was annealed right before the template AP site of four templates as shown. The templates differed by one base (underlined) 5′ to the AP site. DNA polymerase assays were then performed with 3.7 ng (46 fmol) human Pol ι using 50 fmol of the various AP site-containing templates as indicated. Polymerase reactions were carried out with dATP (A), dCTP (C), dTTP (T) and dGTP (G) individually or all four dNTPs (N4). Quantitation of extended primers is shown at the bottom of the gels. DNA size markers in nt are indicated on the sides.
Error-free nucleotide insertion opposite AAF-adducted guanine by human Pol ι
Unlike 8-oxoguanine and AP sites, AAF adducts are bulky lesions in DNA. AAF-adducted guanines (AAF-G) are the major AAF lesions in DNA, which block many DNA polymerases (20,31). To determine how human Pol ι would respond to the bulky AAF-guanine we performed polymerase assays using a DNA template containing a site-specific AAF-G. A 32P-labeled 17mer primer was annealed to the template right before the AAF-adducted guanine (Fig. 6). As shown in Figure 6 (lane 1), human Pol ι incorporated 1 nt opposite the template AAF-G. However, human Pol ι was unable to extend the DNA synthesis further. To identify the incorporated nucleotide, we performed a polymerase assay in the presence of only one deoxyribonucleoside triphosphate. As shown in Figure 6 (lanes 2–5), human Pol ι predominantly incorporated a C opposite the AAF-G. These results show that human Pol ι is able to accurately insert 1 nt opposite an AAF-adducted guanine.
Figure 6.

Nucleotide incorporation opposite a template AAF-guanine by human Pol ι. A 32P-labeled 17mer primer was annealed right before the template AAF-G as shown. DNA polymerase assays were then performed with 37 ng (463 fmol) purified human Pol ι using dATP (A), dCTP (C), dTTP (T) and dGTP (G) individually or all four dNTPs (N4). DNA size markers in nt are indicated on the right.
Response of human Pol ι to a TT dimer and a TT (6–4) photoproduct
Human Pol ι and human Pol η are two homologs of the yeast Rad30 protein (11,15,32). Since Pol η is capable of error-free bypass opposite a TT cis-syn cyclobutane dimer (13–15) it is of particular interest to determine whether human Pol ι could bypass a TT dimer. To do this a 15mer primer was labeled at its 5′-end with 32P and annealed to a 49mer DNA template right before the site-specific TT dimer (Fig. 7A). Primer extension was then performed with purified human Pol ι. As shown in Figure 7A (lane 1), only 1.8% of the primers were extended by 1 nt opposite the TT dimer by human Pol ι in 10 min at 30°C. In contrast, 58% of the primers were extended by 1 nt opposite the 3′ T of the TT (6–4) photoproduct by human Pol ι (Fig. 7A, lane 2). Lowering human Pol ι by 4-fold in the reaction (9 ng, 113 fmol), 50% of the primers were also extended by 1 nt opposite the 3′ T of the TT (6–4) photoproduct in 10 min at 30°C (data not shown). To identify the incorporated nucleotide opposite the TT (6–4) photoproduct we performed a polymerase assay in the presence of only one deoxyribonucleoside triphosphate. As shown in Figure 7A (lanes 3–6), human Pol ι predominantly incorporated an A opposite the 3′ T of the TT (6–4) photoproduct. These results show that the TT cis-syn cyclobutane dimer is a strong blocker of human Pol ι and that human Pol ι is able to predominantly insert the correct nucleotide opposite the 3′ T of the TT (6–4) photoproduct.
Figure 7.

DNA synthesis by human Pol ι on templates containing UV photoproducts. (A) A 32P-labeled 15mer primer was annealed to a DNA template containing a TT dimer or a TT (6–4) photoproduct right before the modified TT sequence. DNA polymerase assays were then performed with 37 ng (463 fmol) purified human Pol ι and 50 fmol DNA using dATP (A), dCTP (C), dTTP (T) and dGTP (G) individually or all four dNTPs (N4). Lane 1, DNA synthesis on the template containing a TT dimer; lanes 2–6, DNA synthesis on the template containing a TT (6–4) photoproduct. (B) Translesion synthesis opposite the TT dimer was extended to 30 min and the reaction temperature shifted to 37°C, using 2-fold higher concentrations (100 µM) of a single dNTP or all four dNTPs as indicated. Human Pol ι in the reaction was increased to 50 ng (625 fmol). Quantitation of extended primers is shown at the bottom of the gel. DNA size markers in nt are indicated on the right.
Most recently, Johnson et al. (26) concluded that purified human Pol ι cannot insert a nucleotide opposite a template TT dimer, whereas Tissier et al. (33) reported that purified human Pol ι is able to perform limited lesion bypass opposite a template TT dimer with T or G incorporation opposite the 3′ T of the lesion. These conflicting results may be largely attributed to the different reaction conditions used. To examine this possibility we performed lesion bypass assays with purified human Pol ι under similar conditions to those used by Tissier et al. (33), in which dNTP concentration was increased from 50 to 100 µM and the reaction was extended from 10 to 30 min at 37 rather than 30°C. Additionally, we increased human Pol ι to 50 ng (625 fmol) in the reaction. As shown in Figure 7B (lane 1), under these conditions of extended reaction and large excess of human Pol ι only a small fraction (23%) of the primers were extended by 1 nt opposite the 3′ T of the TT dimer. Under these conditions T was found to be predominantly incorporated by human Pol ι opposite the 3′ T of the TT dimer (Fig. 7B, lane 4). Less frequently G was also incorporated opposite the lesion (Fig. 7B, lane 5). The correct A was rarely incorporated opposite the lesion (Fig. 7B, lane 2). These results confirm our conclusion that human Pol ι alone is largely unresponsive to a template TT dimer in vitro.
DISCUSSION
Human Pol ι is a unique and extraordinarily error-prone DNA polymerase (24–26). As a member of the UmuC superfamily of proteins human Pol ι is suspected to play a role in DNA lesion bypass during replication. In this study we have examined the ability of purified human Pol ι in translesion DNA synthesis opposite a template 8-oxoguanine, AP site, AAF-adducted guanine, TT (6–4) photoproduct and TT dimer. We found that human Pol ι incorporated nucleotides opposite the first four DNA lesions, thus supporting a role for this DNA polymerase in lesion bypass.
8-Oxoguanine, a major product of oxidative damage in DNA, has long been recognized as a miscoding lesion (30). This lesion directs both C and A incorporation during DNA synthesis by a polymerase (30). The probability of C versus A insertion opposite 8-oxoguanine depends on the specific DNA polymerase. For example, while yeast Pol η predominantly incorporates C opposite 8-oxoguanine (20), human Pol κ inserts A more frequently (28). Consistent with the miscoding property of 8-oxoguanine, human Pol ι incorporates C predominantly, but it also incorporates A and G with decreasing frequencies. Thus, with respect to the specificity of base selection opposite a template 8-oxoguanine, human Pol ι behaves similarly to yeast Pol η, but differently from human Pol η, which incorporates A and C opposite the lesion with similar efficiencies (19). However, to our surprise human Pol ι is significantly blocked by a template 8-oxoguanine during DNA synthesis. Yeast Pol η and other DNA polymerases studied are not significantly blocked by 8-oxoguanine (Fig. 3; 20,30). The inhibitory effect of 8-oxoguanine on human Pol ι occurs at two steps: (i) nucleotide incorporation opposite the lesion; (ii) extension after bypassing the lesion by 1 nt (+2 nt extension). Our Pol ι results demonstrate that the inability of 8-oxoguanine to block a DNA polymerase is not universally true.
Unlike 8-oxoguanine, an AP site in DNA is non-coding. Since an AP site can derive from A, T, C or G, nucleotide incorporation opposite the AP site would be error-prone. In response to the template AP site human Pol ι is able to incorporate predominantly a G opposite the lesion. Additionally, T and A could also be incorporated by human Pol ι with decreasing probabilities. Again, this behavior of human Pol ι is similar to yeast Pol η (20), but different from human Pol η, which most frequently incorporates A opposite the template AP site (19). Nucleotide incorporation by human Pol ι opposite the template AP site is remarkably efficient, more efficient than nucleotide incorporation opposite the template T (24). Following nucleotide incorporation, however, the extension step is blocked by the template AP site. It is conceivable that following human Pol ι action the blocked primer end may be extended by Pol ζ in cells. Indeed, yeast Pol ζ is able to extend the blocked primer end following 1 nt incorporation opposite the template AP site by yeast Pol η in vitro (20). Consistent with the notion that Pol ι plays a role in AP site bypass in humans, major G incorporation opposite AP sites was observed in a study using a shuttle vector that had been replicated in human lymphoblastoid cells (34).
Human Pol ι is able to insert a C opposite a bulky AAF-adducted guanine, but is unable to extend DNA synthesis further. It is possible that the combined actions of human Pol ι and Pol ζ may be able to achieve true bypass of the template AAF-guanine in cells. Supporting this model, we have observed that purified yeast Pol ζ is able to extend a primer with a 3′ C annealed opposite a template AAF-guanine adduct (D.Guo and Z.Wang, unpublished results). If this model proves to be correct in human cells, Pol ι would contribute to error-free bypass of AAF adducts in DNA.
Human Pol ι is largely unresponsive to a template TT dimer. This property clearly separates human Pol ι from Pol η. A hallmark of Pol η is its ability to perform efficient error-free bypass opposite template TT dimers (13–15). In fact, without functional Pol η (encoded by the XPV gene in humans) XPV disease is manifested in humans (15,32), which is characterized by sun sensitivity and a predisposition to skin cancer (16). The dramatic phenotype of XPV patients suggests that error-free bypass of TT dimers and perhaps additional cyclobutane pyrimidine dimer (CPD) lesions is mainly contributed by Pol η and could not be substituted by other DNA polymerases in vivo. The inability of human Pol ι to efficiently respond to a TT dimer supports this conclusion.
Recently it was discovered that human Pol ι predominantly incorporates a G opposite the template T (24–26), thus violating the Watson–Crick base pairing rule. In the present study we surprisingly found that human Pol ι predominantly incorporates an A opposite the 3′ T of a template TT (6–4) photoproduct. Most recently, a similar observation was also reported by Johnson et al. (26) and a slight preference for A incorporation opposite the 3′ T of a template TT (6–4) photoproduct was also noticed by Tissier et al. (33). Using the same DNA template without the TT (6–4) photoproduct, preferential G incorporation by human Pol ι opposite this undamaged template T was observed (24). We postulate that human Pol ι may have evolved to contain a loose and flexible pocket at its active site. Undamaged template would fit the pocket loosely without the stringent geometry constraints for highly accurate Watson–Crick base pairings, resulting in extraordinarily low fidelity DNA synthesis and the extreme template T-primer G instead of T-A base pairing. The loose pocket of Pol ι, however, would be able to fit some damaged templates, leading to nucleotide incorporation opposite the lesion. In the case of TT (6–4) photoproduct the bulky lesion may be able to fit into the active site of human Pol ι, constraining the otherwise loose pocket into a restrictive conformation to allow A pairing with the 3′ T of the lesion in the template. Supporting such a loose active site model, the lesion bypass polymerases Pol η and Pol κ are associated with extraordinarily low fidelity of DNA synthesis when copying undamaged DNA templates (27,35,36). Since human Pol ι is unable to effectively insert a nucleotide opposite a TT dimer, the loose pocket of the polymerase likely retains certain specificity as to what lesions can fit into it.
Human Pol ι inserts 1 nt opposite the 3′ T of TT (6–4) photoproduct much more efficiently than opposite the 3′ T of TT cis-syn cyclobutane dimer. Thus, Pol ι may play an important role in response to TT (6–4) photoproduct in human cells. It is conceivable that following A incorporation by human Pol ι opposite the 3′ T of the TT (6–4) photoproduct, the 5′ T of the lesion may subsequently be bypassed by human Pol ζ. Supporting this model, when the primer 3′-end is an A and annealed opposite to the 3′ T of the TT (6–4) photoproduct, this primer is effectively extended to the end of the template by purified yeast Pol ζ (D.Guo and Z.Wang, unpublished results). Most recently, in vitro bypass of a template TT (6–4) photoproduct by the combined activities of human Pol ι and yeast Pol ζ has also been observed by Johnson et al. (26).
It has now been shown that human Pol η (14,15,18,19,21–23), Pol κ (28,37) and Pol ι (26,33; this study) can respond to DNA lesions in an error-free or an error-prone manner, depending on the specific lesion. Some lesions, such as AP sites and AAF adducts, may be responded to by all three DNA polymerases. Hence, it is very likely that some DNA lesion bypass in vivo may be overlapped by several mechanisms. Such functional redundancy would supply the cell with a highly efficient system for lesion bypass during DNA replication. Escherichia coli does not contain Pol η and Pol ι and the yeast S.cerevisiae does not contain Pol ι and DinB. It should be noted that the yeast Trf4 protein was recently named DNA polymerase κ (38). However, this polymerase is involved in sister chromatid cohesion (38) and is not related to the human Pol κ encoded by the DINB1 gene. Humans contain Pol ζ (7,39,40), Pol η, Pol κ (the DINB protein) and Pol ι. Hence, it appears that humans have evolved to possess a more versatile and perhaps more efficient system for lesion bypass in response to unrepaired DNA damage during replication. Lesion bypass is not merely a survival mechanism. When error-prone lesion bypass is manifested, mutations are accompanied by survival. Mutations under genotoxic pressure are essential building blocks for evolution and adaptation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a THRI grant from the Tobacco and Health Research Institute of the University of Kentucky (Z.W.), a New Investigator Award in Toxicology from Burroughs Wellcome Fund (Z.W.) and a NIH grant CA40463 (J.-S.T.).
References
- 1.Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA and SSB and is specialized for translesion replication. J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- 3.Lemontt J.F. (1971) Mutants of yeast defective in mutation induction by ultraviolet light. Genetics, 68, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemontt J.F. (1977) Pathways of ultraviolet mutability in Saccharomyces cerevisiae. III. Genetic analysis and properties of mutants resitant to ultraviolet-induced forward mutation. Mutat. Res., 43, 179–204. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence C. (1994) The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do and how does it do it? Bioessays, 16, 253–258. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence C.W. and Hinkle,D.C. (1996) DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv., 28, 21–31. [PubMed] [Google Scholar]
- 7.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 9.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- 10.Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg E.C., Feaver,W.J. and Gerlach,V.L. (2000) The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc. Natl Acad. Sci. USA, 97, 5681–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson R.E., Prakash,S. and Prakash,L. (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 14.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 16.Cleaver J.E. and Kraemer,K.H. (1989) Xeroderma pigmentosum. In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D. (eds), The Metabolic Basis of Inherited Disease, 6th Edn. McGraw-Hill, New York, NY, pp. 2949–2971.
- 17.McGregor W.G., Wei,D., Maher,V.M. and McCormick,J.J. (1999) Abnormal, error-prone bypass of photoproducts by xeroderma pigmentosum variant cell extracts results in extreme strand bias for the kinds of mutations induced by UV light. Mol. Cell. Biol., 19, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaisman A., Masutani,C., Hanaoka,F. and Chaney,S.G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry, 39, 4575–4580. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Yuan,F., Wu,X., Talor,J.-S. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 21.Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haracska L., Yu,S.L., Johnson,R.E., Prakash,L. and Prakash,S. (2000) Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet., 25, 458–461. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L., Prakash,S. and Prakash,L. (2000) Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol., 20, 8001–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Yuan,F., Wu,X. and Wang,Z. (2000) Preferential incorporation of G opposite template T by the low fidelity human DNA polymerase ι. Mol. Cell. Biol., 20, 7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tissier A., McDonald,J.P., Frank,E.G. and Woodgate,R. (2000) polι, a remarkably error-prone human DNA polymerase. Genes Dev., 14, 1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Yuan,F., Xin,H., Wu,X., Rajpal,D., Yang,D. and Wang,Z. (2000) Human DNA polymerase κ synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res., 28, 4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Yuan,F., Wu,X., Wang,M., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Error-free and error-prone lesion bypass by human DNA polymerase κin vitro. Nucleic Acids Res., 28, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C.A. and Taylor,J.S. (1993) Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6–4) and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J. Biol. Chem., 268, 11143–11151. [PubMed] [Google Scholar]
- 30.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 31.Belguise-Valladier P. and Fuchs,R.P. (1995) N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J. Mol. Biol., 249, 903–913. [DOI] [PubMed] [Google Scholar]
- 32.Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 33.Tissier A., Frank,E.G., McDonald,J.P., Iwai,S., Hanaoka,F. and Woodgate,R. (2000) Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase ι. EMBO J., 19, 5259–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klinedinst D.K. and Drinkwater,N.R. (1992) Mutagenesis by apurinic sites in normal and ataxia telangiectasia human lymphoblastoid cells. Mol. Carcinog., 6, 32–42. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Low fidelity DNA synthesis by human DNA polymerase η. Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- 36.Ohashi E., Bebenek,K., Matsuda,T., Feaver,W.J., Gerlach,V.L., Friedberg,E.C., Ohmori,H. and Kunkel,T.A. (2000) Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem., 275, 39678–39684. [DOI] [PubMed] [Google Scholar]
- 37.Ohashi E., Ogi,T., Kusumoto,R., Iwai,S., Masutani,C., Hanaoka,F. and Ohmori,H. (2000) Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev., 14, 1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Castano,I.B., De Las Penas,A., Adams,C. and Christman,M.F. (2000) Pol κ: a DNA polymerase required for sister chromatid cohesion. Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- 39.Lin W., Wu,X. and Wang,Z. (1999) A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- 40.Murakumo Y., Roth,T., Ishii,H., Rasio,D., Numata,S., Croce,C.M. and Fishel,R. (2000) A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem., 275, 4391–4397. [DOI] [PubMed] [Google Scholar]