Abstract
Alphavirus nsP3 phosphoprotein is essential for virus replication and functions initially within polyprotein P123 or P23 components of the short-lived minus-strand replicase, and upon polyprotein cleavage, mature nsP3 likely functions also in plus-strand synthesis. We report the identification of a second nsP3 mutant from among the A complementation group of Sindbis virus (SIN) heat-resistant strain, ts RNA-negative mutants. The ts138 mutant possessed a change of G4303 to C, predicting an Ala68-to-Gly alteration that altered a conserved His-Ala-Val tripeptide in the ancient (pre-eukaryotic), “X” or histone 2A phosphoesterase-like macrodomain that in SIN encompasses nsP3 residues 1 to 161 and whose role is unknown. We undertook comparative analysis of three nsP3 N-terminal region mutants and observed (i) that nsP3 and nsP2 functioned initially as a single unit as deduced from complementation analysis and in agreement with our previous studies, (ii) that the degree of phosphorylation varied among the nsP3 mutants, and (iii) that reduced phosphorylation of nsP3 correlated with reduced minus-strand synthesis. The most striking phenotype was exhibited by ts4 (Ala268 to Val), which after shift to 40°C made significantly underphosphorylated P23/nsP3 and lost selectively the ability to make minus strands. After shift to 40°C, mutant ts7 (Phe312 to Ser) made phosphorylated P23/nsP3 and minus strands but failed to increase plus-strand synthesis. Macrodomain mutant ts138 was intermediate, making at 40°C partially phosphorylated P23/nsP3 and reduced amounts of minus strands. The mutants were able to assemble their nsPs at 40°C into complexes that were membrane associated. Our analyses argue that P23/P123 phosphorylation is affected by macrodomain and Ala268 region sequences and in turn affects the efficient transcription of the alphavirus genome.
The alphavirus Sindbis virus (SIN) is a plus-strand RNA virus whose 49S genome encodes four nonstructural proteins (nsPs), nsP1, nsP2, nsP3, and nsP4, numbered according to their gene order. These essential components of the viral replicase and transcriptase are synthesized initially as two polyproteins, P1234 and P123, the former by readthrough of an opal termination codon between the nsP3 and nsP4 genes (66). The viral replicase synthesizes genome-length plus- or minus-strand RNA, and the transcriptase synthesizes a subgenomic 26S mRNA that encodes the viral structural proteins. Functions of the four nsPs are beginning to be elucidated (reviewed in references 26, 54, and 66). The nsP1 methyltransferase and guanylyltransferase activities function in capping of the viral 49S and 26S plus-strand RNA (4-6, 9, 45, 59, 72), and nsP1 downstream regions function in the initiation of minus-strand RNA synthesis (23, 58, 74) and interaction with the nsP4 polymerase (15, 16, 60). The N-terminal region of nsP2 expresses RNA 5′-triphosphatase (69), and NTPase and helicase activities (12, 19, 29, 32, 33, 52). The C-terminal domain expresses a thiol protease that is responsible for processing the viral nonstructural polyprotein precursors (11, 13, 18, 23, 24, 28). It also functions in internal initiation for subgenomic mRNA synthesis (22, 27, 56, 63) and encodes a nuclear localization signal (48). The nsP4 polymerase contains the conserved, GDD-containing sequence motif of RNA-dependent RNA polymerases (14, 19, 21, 25, 29) and affects host cell-dependent replication (15, 39). Only some plant virus members of the SIN superfamily have an nsP2-like thiol protease sequence, and only animal virus members have an nsP3-like gene (2, 19, 25, 29, 64).
The 549-amino-acid-long SIN nsP3 provides an essential although unknown function needed for viral RNA synthesis, including minus-strand and subgenomic 26S mRNA syntheses (22, 29, 34, 35, 49, 73). Comparison of the N-terminal sequences of SIN nsP3 with those of rubella virus, hepatitis E virus, and coronaviruses showed a highly conserved region of unknown function, originally called the “X” domain, flanking the papain-like protease domains of each virus (29). This is a homologue of the “macrodomain” identified initially in macrohistone 2 of vertebrates (46) and is also present as a repeated sequence in open reading frames in the human genome that include BAL, a gene implicated in diffuse large B-cell lymphoma migration (1). The macrodomain product of yeast gene YBR022w exhibited ADP-ribose-1"-phosphate (Appr-1"-p)-processing activity, indicating that the macrodomain is a phosphoesterase (41). Only the N-terminal one-half to two-thirds of nsP3 is conserved among alphaviruses, and it is essential for infectivity (35, 66). SIN and Semliki Forest virus (SFV) nsP3 are phosphoproteins. This posttranslational modification occurs on Ser and Thr residues primarily in the C-terminal region and likely involves kinases resembling casein kinase II (40) and protein kinase C (71). SIN nsP3 phosphorylated species range in size from 76 to 106 kDa (23, 40, 49). Sixteen phosphorylation sites were mapped in the smaller SFV nsP3, identifying Ser320, Ser327, Ser332, and Ser350 and ∼10 possible residues within the region containing amino acids G338 to K415, and especially Thr344/345 residues, as phosphorylation targets (71). Several observations support a functional role for phosphorylated nsP3 species. Phosphorylated nsP3 species are enriched in the 15,000 × g pellet (P15) cell fraction that contains viral replicative intermediates and active polymerases (35, 38, 40, 47) and in active replication complexes solubilized from P15 membranes (7). Eliminating phosphorylation of nsP3 greatly reduced pathogenicity in mice (68, 70).
Initially, nsP3 seems to function within polyproteins P123 and P23. Using complementation to assess the ability of SIN heat-resistant (HR) nsPs to dissociate, exchange, and assemble in replication complexes, we found evidence for only three complementation groups (73). These would represent nsP1 (group B), P23 (mutants formerly in group A or G), and nsP4 (group F). We argued that nsP2 and nsP3 would act as a single cistron because they associated immediately after their synthesis and before their cleavage or because polyprotein P23 was their functional form. The only exceptions to this to date are SIN linker insertion mutants carrying an extra six amino acids between residues 58 and 59 or 226 and 227 of nsP3 (35). The low or selective complementation observed may reflect enhanced 2/3 site cleavage or the partial activity of replicases composed of mixtures of both mutant P23 proteins. Biochemical (36, 39, 61, 62) studies also provided evidence that P23 polyproteins are components of the minus-strand replicase. The present model is that cleavage of the nascent P1234 first at the 3/4 site forms P123-nsP4 complexes able to bind genome RNA and synthesize a minus strand. Then, cleavage at the 1/2 site forms nsP1-P23-nsP4 replicases active in 49S minus-strand and plus-strand amplification. Last, cleavage of P23 results in stable nsP1-nsP2-nsP3-nsP4 complexes that continue 49S genome synthesis and can recognize the internal promoter on minus strands for synthesis of 26S mRNA. Cleavage of P23 is a key regulatory event because it inactivates the minus-strand activity of the replicase and forms the transcriptase activity for 26S mRNA synthesis.
We report the identification of a new nsP3 mutant from among those assigned originally to the A complementation group, which includes many nsP2 mutants. This provides additional evidence for an essential role for P23 species. A comparative analysis of this and two other nsP3 mutants showed that the degree of phosphorylation of nsP3/P23 differed in the three mutants and that reduced nsP3/P23 phosphorylation correlated with reduced minus-strand synthesis. It is interesting that the new nsP3 mutant ts138 changed a conserved His-Ala-Ala tripeptide sequence that is predicted to be part of the active site of the putative nsP3 macrodomain enzyme.
MATERIALS AND METHODS
Cells and virus.
Chicken embryo fibroblast (CEF) cells were prepared from 10-day-old embryos from the eggs of leukosis-free (SPF-COFAL/Marek-negative) flocks (Spafas, Roanoke, Ill.) and were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with tryptose phosphate broth (5%, vol/vol) and fetal bovine serum (FBS; 6%, vol/vol). BHK-21 cells, a continuous cell line derived from baby hamster kidney cells, were grown in DMEM containing 6% fetal bovine serum.
The HR strain of SIN (wild type) and the RNA-negative ts mutants ts4 and ts11 of SIN HR were isolated by Burge and Pfefferkorn (8) and obtained from E. Pfefferkorn. The mutant ts138 was isolated following chemical mutagenesis of a large-plaque variant of SIN HR (63) and was a generous gift of E. G. Strauss. Toto:ts7B5 is a recombinant virus (22), obtained by in vitro transcription and transfection of cells with the plasmid Toto:ts7B5, a gift from E. G. Strauss. It contains the SalI (nucleotide [nt] 4845)-SpeI (nt 5262) region of ts7 in Toto1101, an infectious cDNA clone of SIN HR (51). Virus stocks used in this study were obtained by plaque purification and propagated at a low multiplicity of infection (MOI) of 0.1 to 1 PFU/cell. Revertants of ts138 were obtained from plaques at 40°C, followed by two plaque purifications at 40°C. The twice-plaque-purified virus was then grown in CEF cells at an MOI of 0.1 to 1 PFU/cell, and revertant virus stocks that had an efficiency of plaquing (EOP; the ratio of PFU at 40°C to PFU at 30°C) on CEF cells of 0.70 were obtained.
Plasmids.
Toto1101 and the ΠnsP12 and ΠnsP23 shuttle vectors were constructed by Rice and coworkers (51) and were a generous gift from J. Lemm and C. Rice (Rockefeller University, New York, N.Y.). Plasmid DNA preparation, DNA ligations, restriction enzyme digestion and cloning, and transformation were performed as previously described (10, 53, 74).
Infection and RNA labeling.
In all experiments, CEF or BHK monolayers were infected with SIN HR or one of the ts mutants at an MOI of 100 and, where indicated, were shifted to 40°C, as previously described (10). Monolayers were pulse-labeled with 1 ml of 5′-[3H]uridine (50 μCi/ml unless otherwise indicated) in DMEM containing 20 μg of actinomycin D/ml, 5% FBS, and 20 mM HEPES, pH 7.4. At the end of the labeling period, cells were washed twice with ice-cold phosphate-buffered saline and lysed with 5% lithium dodecyl sulfate in LET buffer (0.1 M LiCl, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.4) containing proteinase K (Amresco, Solon, Ohio), as described previously (10). The amount of 49S genome RNA relative to subgenomic 26S mRNA synthesis at permissive and nonpermissive temperatures was determined after electrophoresis of infected-cell extracts on agarose gels (10).
Isolation of viral RNA.
Viral RNA for sequencing and reverse transcription was obtained as described previously (10, 73). The concentration of viral RNA was determined by measuring absorbance at 260 nm in a Beckman DU 70 spectrophotometer (25 A260 = 1 mg of RNA).
Construction of hybrid genomes.
First-strand cDNA was synthesized as described by Dé et al. (10). Second-strand cDNA synthesis and subsequent amplification (20) were performed by PCR in a Perkin-Elmer thermal cycler (Perkin-Elmer Cetus Corp., version 2.0), using thermostable Taq (Thermus aquaticus) DNA polymerase (Perkin-Elmer or Boehringer Mannheim). Toto1101 DNA was used as a control for each of the reactions. Thermal cycling was done for a total of 20 or 25 cycles (10).
Construction of Toto1101 deletion vectors.
Selection of hybrid clones containing mutant cDNA sequences took advantage of the use of vectors of Toto1101 containing deletions within the region to be cloned. Swapping into these vectors the overlapping cDNA fragments derived from mutant genomes restored the full sequence and gave infectious constructs. The vectors TEB, TSH, and TW20 have been described previously (10, 73).
Construction of Toto:ts138 BglI (nt 2288)-BspEI (nt 4318) clones.
First-strand cDNA of regions of the mutant genome RNA was synthesized using a minus-sense primer, DLS11 (nt 4324 to 4342), prepared by Operon Technologies Inc. (Alameda, Calif.). One-twentieth (5 μl) of the reverse transcription reaction mixture was then amplified by PCR using DLS11 and a second-strand primer, ID2 (nt 1366 to 1380), for 25 cycles. The PCR products were separated on an 0.8% agarose gel in Tris-agarose-EDTA buffer. The desired fragment of 2,976 bp was cut out from the gel and extracted using the GeneClean II (Bio 101 Inc.) protocol. About 100 ng of PCR product was digested with BglI (nt 2288) and BspEI (nt 4318) at 37°C. The restriction fragments were separated on an 0.8% agarose gel, and the DNA fragment was cut out and extracted in 10 μl of water. The DNA fragment was then inserted, using T4 DNA ligase (New England Biolabs, Beverly, Mass.), in place of the corresponding region in the shuttle vector ΠnsP23 (37). The ligation products were used to transform competent MC1061p3 bacteria, and the resulting transformed colonies were screened for the presence of a full-sized plasmid. DNA from each clone was screened by restriction enzyme digestion and separation of the resulting fragments on agarose gels to confirm their size and the presence of the inserted fragment. DNA from positive clones was cut with BglII (nt 2288) and SpeI (nt 5262), and this fragment was swapped into Toto1101 in place of the wild-type sequence.
In vitro transcription and transfection of CEF cells.
Hybrid cDNA clones were linearized at the unique XhoI site and transcribed with SP6 polymerase (Megascript; Ambion), and the full-length transcripts were transfected into CEF cells, as previously described (51).
Sequence analysis.
Sequencing of the mutant recombinant cDNA was as described previously (10). The entire region of the mutant cDNAs swapped to form the final recombinant subclones expressing mutant phenotypes was sequenced.
Protein labeling and immunoprecipitation.
CEF cells were infected with SIN HR, ts4, Toto:ts7B5, or ts138 viruses at 30°C for 1 h with an MOI of 100. After virus adsorption, cell monolayers were refed with complete DMEM (DMEM, 5% FBS, 20 mM HEPES [pH 7.4], and 2 μg of actinomycin D/ml) and incubated at 30°C. Cells were refed with DMEM containing 1% of the normal methionine concentration, 5% dialyzed FBS, 2 μg of actinomycin D/ml, and 20 mM HEPES (pH 7.4) and incubated at 30°C for 1 to 2 h prior to labeling at 30°C or to shift to 40°C. Cultures were shifted to 40°C at a time when the rate of RNA synthesis was approximately 10% of the maximum and incubated with prewarmed hypertonic medium (DMEM with 1% normal methionine and containing NaCl at a final concentration of 335 mM) for 40 to 60 min at 40°C (27, 28). For labeling at 30°C, cultures were incubated in the hypertonic medium for 60 min. Following hypertonic treatment, the cells were labeled for 30 min with 1 ml of [35S]methionine (0.2 mCi/ml) in isotonic DMEM minus methionine, after which cells were lysed with 1% lithium dodecyl sulfate-1 mM EDTA to obtain a final cell concentration of 5 × 106 cells/ml. Following the pulse, a second set of cultures was chased for 1 to 2 h in isotonic medium containing a 20-fold excess of methionine at 30 or 40°C and was then lysed as above.
Immunoprecipitation of labeled nsPs was performed with monospecific antibodies to nsP1, nsP2, nsP3, or nsP4, as described previously (7, 10) that were a generous gift from J. H. and E. G. Strauss. The final precipitate after solubilization was analyzed on 5 to 10% linear gradient polyacrylamide gels in Laemmli buffer (31).
Preparation of the P15 fraction and solubilization.
CEF cells in 60-mm-diameter petri dishes were infected with each virus for 1 h at 30°C at an MOI of 100. After virus adsorption the cells were incubated with complete DMEM (DMEM, 2 μg of actinomycin D/ml, 6% FBS, 20 mM HEPES, pH 7.4) at 30°C. One and a half hours before shift to 40°C the cells were refed with medium containing DMEM containing 1% of the normal concentration of methionine, 2 μg of actinomycin D/ml, 20 mM HEPES (pH 7.4), and 6% dialyzed FBS for 1.5 h at 30°C. At 4.5 h postinfection (p.i.) the cultures were shifted to 40°C and incubated with hypertonic medium containing 1% of the normal concentration of methionine and a final concentration of 335 mM NaCl for 40 min at 40°C. After hypertonic treatment, the cells were pulsed with 0.2 mCi of [35S]methionine/ml for 30 min at 40°C in isotonic DMEM-1% methionine followed by a 1-h chase with isotonic DMEM containing a 20-fold excess of methionine at 40°C. At the end of the chase period the cells were harvested and broken by Dounce homogenization in hypotonic medium, and the P15 and S15 fractions were obtained as described previously (7). Aliquots of the P15 fraction were adjusted to 100 mM NaCl and 1% deoxycholate (DOC), vortexed, and recentrifuged at 15,000 × g. Proteins in the resultant supernatant (DOC-S15) fractions were immunoprecipitated as described previously (7), by the addition of nsP1 to nsP4 monospecific antibodies (gift of E. and J. Strauss), and were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)-5 to 10% linear gradient polyacrylamide gels (31).
Isolation of SIN RF RNA and quantitation of minus-strand RNA.
The rate of minus-strand RNA synthesis was determined as described by Dé et al. (10). Duplicate cultures at 30°C were pulse-labeled for 1 h or for 20 min at the time of shiftup and for 1 h or for 20 min before shiftup to determine the amount of minus strands synthesized at 30°C prior to shift as well as after shift to 40°C over the same period of time. In experiments where incorporation was measured at 5-min intervals for 20 min at 40°C, cells also were labeled at 30°C, starting at the time of shiftup, for two 20-min intervals to determine the trend of minus-strand RNA synthesis at 30°C. Deproteinized cell lysates were digested with RNase A and chromatographed on CF-11 cellulose (Whatman, Clifton, N.J.) as described previously (17) for the isolation of the replicative-form RNA (RF RNA). Minus-strand RNA was measured by nuclease protection assays that determine the amount of [3H]uridine-labeled RF RNA that after denaturation will hybridize to an excess (about 100-fold) of unlabeled 49S plus-strand RNA (53).
RESULTS
Identification of a new nsP3 mutant.
Reversion frequencies of A complementation group mutant ts138 ranged between 10−4 and 10−5 (data not shown), suggesting that the phenotype of ts138 was the result of a single point mutation. The A complementation group is complex and comprised of mutants whose causal lesions can reside in the gene for either nsP2 or nsP3. Thus, it is likely that the A complementation group represents the functional P23 cistron. For this reason, sequences coding for both nsP2 (nt 1601 to nt 4101) and the N-terminal part of nsP3, which encompasses nt 4102 to nt 5750, were cloned initially and exchanged in place of the corresponding wild-type sequence in the SIN cDNA clone pToto1101 (Fig. 1). In vitro RNA transcripts were synthesized and used to transfect CEF cells. The recombinant viruses that resulted were screened for growth at 30 and 40°C. Results are summarized in Table 1.
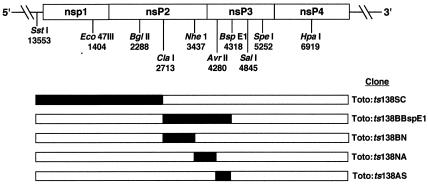
FIG. 1.
Strategy for mapping the responsible mutation in ts138. Infectious cDNA clones that contained parts of each mutant nsP coding region in place of the corresponding regions of parental pToto1101 cDNA were made as described in Materials and Methods. Unique restriction endonucleases and their cleavage sites are marked under the schematic of the viral nsP1 to nsP4 regions. The solid regions represent sequences from the mutant; the open regions are those from pToto1101.
TABLE 1.
Analysis of recombinant viruses indicates that ts138 is an nsP3 mutant
| Virus | Fragment replaced (nt) | EOP (PFU at 40°C/PFU at 30°C) | Growth |
|---|---|---|---|
| ts138 | 2 × 10−4 | ts | |
| 138R | 7 × 10−1 | wta | |
| Toto:ts138SC | 1-2713 | 3 × 10−1 | wt |
| Toto:ts138BBs | 2288-4318 | 0.1 × 10−5-3 × 10−5 | ts |
| Toto:ts138BN | 2288-3734 | 3 × 10−1-9 × 10−1 | wt |
| Toto:ts138NA | 3734-4280 | 4 × 10−1-6 × 10−1 | wt |
| Toto:ts138ABs | 4280-4318 | 2 × 10−4 | ts |
| Toto:ts138BN+NA | 2288-4280 | 1 × 10−1-5 × 10−1 | wt |
wt, wild type.
Only Toto:ts138BBs (BglII [nt 2288]-BspEI [nt 4318]) viruses were ts for growth and had an EOP that ranged between 0.1 × 10−5 and 3 × 10−5. This region was subcloned as three separate pieces, and the resulting viruses were analyzed. Toto:ts138BN (BglII [nt 2288]-NheI [nt 3734]) and Toto:ts138NA (NheI [nt 3734]-AvrII [nt 4280]) viruses were wild type, but Toto:ts138ABs (AvrII-BspEI) viruses that contained the region from nt 4280 to nt 4318 were ts for growth. This latter virus gave an EOP similar to the original ts138 mutant EOP, confirming that the causal lesion(s) was in the region between nt 4280 and nt 4318. Sequencing identified two changes from the published SIN nsP3 sequence (65) at nt 4227 (A to G, predicting a change of Thr43 to Ala) and at nt 4303 (C to G, predicting a change of Ala68 to Gly). We found that Toto:ts138ABs recombinants with only the nt 4303 change expressed fully the ts phenotype, while Toto:ts138NA recombinants with only the nt 4227 change were wild type. Therefore, the change at nt 4303 was necessary and sufficient to confer the ts138 phenotype.
In addition to the causal mutation, the nsP2 sequence of ts138 and ts138 revertant viruses differed from the published SIN HR small-plaque virus sequence (65) in having U at nt 2992 instead of C, which predicts a change of Pro438 to Leu, and in having U at nt 3035 instead of C, which would not change the Asp452 codon. The substitution at nt 2992 is present also in our laboratory strain of SIN HR and mutants derived from SIN HR (10), in Toto1101 (S. Barnhart and D. Sawicki, unpublished results), and in several strains of SIN including AR339 (43). The change at nt 3035 was unique to ts138 and its revertants, as it was not present in the genomes of Toto1101, SIN HR, or other SIN HR mutants such as ts16 and ts19 (data not shown). We tested whether the nt 2992 and nt 3035 base changes together might confer an additional conditionally lethal phenotype by combining subclones Toto:ts138BN and Toto:ts138NA into one genome (Toto:ts138BN+NA). Neither of these mutations made virus growth ts (Table 1) or significantly affected the temperature sensitivity of viruses containing the Gly68 change also (Toto:ts138BBs, Table 1).
Finding that A group mutants ts4 (73) and ts138 map to nsP3, while eight other A complementation group mutants map to nsP2 (10, 22, 57), supports our earlier finding that nsP2 and nsP3 behave as a single cistron and the interpretation that this reflects early, essential functions of the P23 polyprotein intermediate (73). To date, there are three conditionally lethal, single-nucleotide alterations known to affect nsP3 function, a change of Ala268 to Val in ts4 (73), the Gly68 change in ts138 (this study), and a change of Phe312 to Ser in ts7 that in the recombinant Toto:ts7B5 is expressed in the absence of an nsP2 mutation also found in ts7 (22). Some of the multiple-base insertion mutations constructed by LaStarza et al. (35) are also in this N-terminal region (Fig. 2). Mutants CR3.36, with an insertion between residues 58 and 59, and CR3.39, with an insertion between residues 226 and 227, were ts for growth and defective in minus-strand synthesis at 40°C (35). Mutant CR3.34, with an insertion between amino acids 252 and 253, was defective in 26S mRNA synthesis independent of temperature.
FIG. 2.
Change of Ala68 to Gly alters an invariant residue in the nsP3 N-terminal macrodomain sequence. The symbol ! denotes invariant residues among nsP3-like proteins encoded in the genomes of hepatitis E virus, rubella virus, and at least the alphaviruses SIN, SFV, O'nyong-nyong virus, Ross River virus, Middelburg virus, and Venezuelan equine encephalitis virus. Computer analysis and sequences were reported by Koonin et al. (29), who found that the macrodomain represented residues 1 to 161 of SIN nsP3, residues 785 to 942 of the hepatitis E virus protein, and residues 816 to 984 of the rubella virus protein. Additional insertion mutations and deletions were reported by LaStarza et al. (35), and five of these mutants mapping within the N-terminal 325 amino acids of nsP3 are also indicated (designated CR3, followed by a number).
Lesions in nsP3 are within the essential N-terminal domain and one overlaps the “X” or macrodomain.
Of the 549 amino acid residues in SIN nsP3, only the N-terminal 325 residues are conserved and essential in cultured cells (34, 35). Substituted residues Val268 and Ser312 are nearer the C-terminal end of this conserved domain (Fig. 2). The N-terminal part of nsP3 contains residues identified as invariant in the genomes of at least four alphaviruses, rubella virus, and hepatitis E virus and are within the ancient macrodomain (29). In SIN HR, the macrodomain comprises amino acid residues 1 to 161. Of interest, the change of Ala68 to Gly in ts138 alters one of these invariant residues and the middle residue in an His-Ala-Val tripeptide sequence (the sequence is His-Ala-Ala in newly described fish alphaviruses, sleeping disease of trout and salmon pancreas disease viruses (50, 77). The His-Ala-Val (or His-Ala-Ala) sequence is predicted by structural analysis to be part of the active site of an enzyme residing in nsP3 (A. E. Gorbalenya, personal communication). We undertook a comparative analysis of the phenotypes of the nsP3 Gly68, Val268, and Ser312 mutants.
Polyprotein processing by the nsP3 mutants.
The presence of nsP3 Gly68 (ts138), Val268 (ts4), or Ser312 (Toto:ts7B5) proteins in nascent or intermediate polyproteins did not affect cleavage and release of nsP2 (10, 56, 73) or of nsP3 and nsP4 (Fig. 3). Infected cells shifted to 40°C at 3.5 h p.i., a time when viral replication complexes are being formed and nsP production is maximal, contained cleaved nsP3, nsP4, and P34 polypeptides (Fig. 3). The formation of nsP3 in large amounts indicated that pulse-labeled P123 and P23 were cleaved efficiently during the chase. Newly synthesized nsP3 migrated as 76-kDa proteins, similar to the fast-migrating, nonphosphorylated forms of nsP3a proteins detected immediately after a short pulse (40). Phosphorylation is responsible for the reduced mobility on gels of nsP3 proteins and polyproteins (40). Consistent with their becoming phosphorylated during the chase period, nsP3 proteins were found as several higher-molecular-massspecies, two of which have been called nsP3b (78 kDa) and nsP3c (106 kDa) by Li et al. (40). In addition to these two major phosphorylated species, a significant amount of a third species with intermediate mobility, here labeled nsP3*, was present. A protein of similar mobility was detected in C7/10 mosquito cells (40).
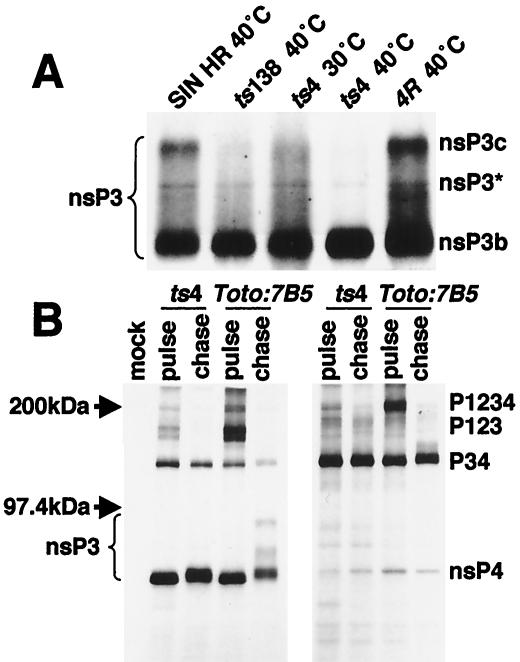
FIG. 3.
Altered migration of nsP3 proteins made at 40°C indicated that certain mutant nsP3 proteins were temperature sensitive for phosphorylation. (A) CEF cells were infected with either an nsP3 mutant, the revertant of ts4, or parental SIN HR and were maintained at 30°C or shifted to 40°C late in infection. Viral proteins were pulse-labeled with [35S]methionine for 30 min as described above (Materials and Methods) followed by a chase period of 1 h before the cells were lysed. Infected-cell lysates were immunoprecipitated with nsP3 antibodies, and the precipitates were analyzed by gel electrophoresis. (B) Lysates from ts4-, Toto:ts7B5-, or mock-infected cells labeled and chased at 40°C were immunoprecipitated with antibodies specific for nsP3 (left panel) or for nsP4 (right panel), and the immunoprecipitates were analyzed on 5 to 10% Laemmli linear gradient polyacrylamide gels (31).
Two of the nsP3 mutants showed a ts defect in phosphorylation based on the higher electrophoretic mobility of their nsP3 species. This modification occurred normally at 40°C in virus-infected CEF cells producing Ser312 (Toto:ts7B5) nsP3 and P34 proteins (Fig. 3B). However, Val268 (ts4) nsP3 produced at 40°C included little or none of the higher-molecular-weight nsP3* or nsP3c species (Fig. 3). Gly68 (ts138) nsP3 proteins accumulated as mostly nsP3b and nsP3*, with little of the nsP3c species (Fig. 3A). There was a similar failure to fully modify their P34 proteins at 40°C. The ts4 Val268 nsP3 proteins were found as nsP3a species immediately after the pulse and were converted to slightly higher molecular weight forms equivalent to parental nsP3b after a chase. This indicated that a level of phosphorylation typical of nsP3b was likely occurring in ts4-infected cells at 40°C (Fig. 3B) but that little or no nsP3* or nsP3c species were formed. Revertants of ts4 regained the ability to fully phosphorylate Val268 nsP3 at 40°C (Fig. 3A).
Also noted was an increased formation of P34 relative to nsP4 by nsP3 mutant-infected cultures (Fig. 3B) compared to wild type or non-nsP3 mutants (data not shown) (10). Accumulation of P34 argued that P1234 polyproteins were cleaved efficiently at the 2/3 site but less well at the 3/4 site. Failure to efficiently release large amounts of nsP4 would be rate limiting for the formation of minus-strand replicases and in turn would affect levels of plus-strand replicases and transcriptases.
Transport of mutant nsPs to P15 membrane-associated replication complexes at 40°C.
SIN HR replication complexes are composed of all four nsPs, are enriched selectively in the 15,000 × g mitochondrial membrane pellet (P15) fraction, and can be released from membranes with DOC (7). We found evidence that mutant nsP3 proteins made at 40°C were transported to P15 membranes and became complexed with other nsPs at 40°C. Cultures of CEF cells in 60-mm-diameter petri dishes that were infected at an MOI of 100 at 30°C were monitored for the composition of replication complexes early in infection by shifting these cultures to 40°C when each virus's rate of RNA synthesis was ca. 25% of its eventual maximal 30°C rate (monitored in a separate experiment). Cultures were incubated for 1 h with 40°C hypertonic medium to clear translating ribosomes, allowed to initiate translation in the presence of [35S]methionine in isotonic medium for 30 min, and chased for 1 h to allow nsPs to be processed and assembled into complexes that normally accumulate in the P15 fraction. When made and processed at 40°C, mutant nsP3 was found in the P15 fraction with other nsPs and was released into the soluble phase after DOC treatment (Fig. 4A), similar to parental nsPs. At 40°C, Toto- and nsP3 mutant-infected cells had greater amounts of nsP2 in the soluble fraction than the P15 fraction even without DOC treatment, confirming results with SFV nsP2 (67). Only at 40°C and only in Toto:ts7B5-infected cells were several high-molecular-weight species present of the size expected for P12 or P23 intermediate polyproteins.
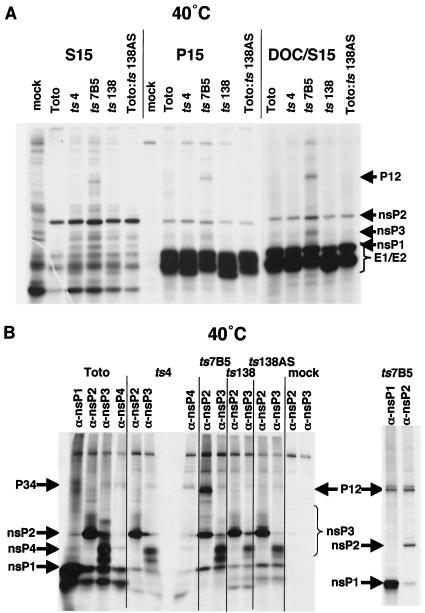
FIG. 4.
nsP3 mutants form replication complexes that associate with P15 membranes and are released with detergent treatment. CEF cells were infected at 30°C, shifted to 40°C between 1.5 and 2 h p.i., and labeled with [35S]methionine as described in Materials and Methods. (A) Lysates from cultures shifted to 40°C. The 15,000 × g pellet (P15) and supernatant (S15) fractions were obtained and analyzed directly on 5 to 10% Laemmli polyacrylamide gels. The P15 fraction was resuspended in 100 mM NaCl-1% DOC, vortexed, and repelleted at 15,000 × g to obtained the detergent-washed pellet fraction (DOC-P15) and the detergent supernatant (DOC-S15). (B) Immunoprecipitation of 40°C nsP3 mutant DOC-S15 fractions. The DOC-S15 fractions, obtained from each infected-cell P15 fraction as described above, were incubated with antibodies specific for each of the viral nsPs, as indicated. Immunoprecipitates were collected and analyzed on 5 to 10% Laemmli polyacrylamide gels. Right panel: DOC-S15 fractions were denatured in 1% SDS at 100°C before immunoprecipitation.
Immunoprecipitation of the solubilized DOC-S15 fractions with antibodies monospecific for each nsP and analysis of the immunoprecipitates after electrophoresis on SDS-polyacrylamide gels found that the four nsPs made at 40°C were complexed together (Fig. 4B). While each nsP antiserum predominantly precipitated its homologous nsP, a consistent finding when this fraction is analyzed by immunoprecipitation (7), each nsP antiserum also coprecipitated detectable amounts of the other nsPs. Intermediate polyproteins present in the 40°C Toto:ts7B5-infected DOC-S15 fraction were P12, and not P23, proteins from their presence in immunoprecipitates formed by both nsP1 and nsP2 antisera (Fig. 4B, right panel). This is of interest because, in vitro, P12 proteases poorly cleave the 2/3 site of P23 proteins that would be components of minus-strand replicases (11, 24). One prediction would be that, at 40°C, its minus-strand synthesis would continue longer than in infected cells where P23 was cleaved more quickly. In addition to nsP1 and nsP2 proteins, nsP3 was present in the DOC-S15 fractions and had similar electrophoretic mobilities (phosphorylated species) as the total infected-cell nsP3 population (Fig. 3). Also, some nsP4, and more P34, was detected in the detergent-solubilized fractions (Fig. 4B). In Toto:ts7B5 DOC-S15 fractions, very little P12 coprecipitated with nsP3 antibodies and may not have been assembled into nsP complexes.
RNA synthesis and plaquing efficiency at 40 and 37°C relative to 30°C.
We determined the replication abilities of the nsP3 mutants and compared them to representative nsP1 and nsP2 mutants. When assayed for the ability to form plaques, all mutant stocks produced less than 10−3 to 10−7 fewer plaques at 40°C compared to 30°C (Table 2). However, only nsP3 mutant ts4, three mutants mapping to the C-terminal domain of nsP2, and nsP1 mutant ts11 were also defective at a temperature of 37°C. The causal lesions in nsP3 mutants Toto:ts7B5, ts138, Toto:ts138BBs, and Toto:ts138ABs and four N-terminal domain nsP2 mutants were more leaky, requiring temperatures higher than 37°C to confer full temperature sensitivity.
TABLE 2.
Plaque formation at 37 and 40°C relative to 30°C
| Virus | Mutant nsPa | EOP
|
|
|---|---|---|---|
| 40/30°C | 37/30°C | ||
| SIN HR | 0.6-1.2 | 0.8-1.2 | |
| Toto1101 | 1.0 | 1.5 | |
| 24R1 | 0.4 | 0.4-0.9 | |
| 24R2 | 0.3 | 0.5-1.0 | |
| 133R | 0.3 | 0.4 | |
| 17R | 0.7 | 0.6 | |
| ts138 | nsP3 | 0.7 × 10−4-5 × 10−4 | 0.2-0.3 |
| Toto:ts138BBs | nsP3 | 0.2 × 10−4 | 0.2 |
| Toto:ts138ABs | nsP3 | 0.3 × 10−4-0.7 × 10−4 | 0.2-0.6 |
| Toto:ts7B5 | nsP3 | 0.9 × 10−4-14 × 10−4 | 0.5-0.6 |
| ts4 | nsP3 | 0.3 × 10−3-0.6 × 10−3 | 0.4 × 10−3-1.3 × 10−3 |
| ts133 | C-nsP2 | <10−7 | <10−7 |
| ts17 | C-nsP2 | 0.4 × 10−4 | 0.8 × 10−4 |
| ts24 | C-nsP2 | <10−7 | <10−7 |
| ts14 | N-nsP2 | 0.1 × 10−4 | 0.3 |
| ts16 | N-nsP2 | 0.2 × 10−4 | 0.2 |
| ts19 | N-nsP2 | 0.2 × 10−4 | 0.5 |
| ts21 | N-nsP2 | 0.1 × 10−5 | 0.4 |
| Toto:ts11A1 | nsP1 | 0.6 × 10−4-0.9 × 10−4 | 0.2 × 10−4-0.3 × 10−4 |
C-nsP2, C-terminal nsP2; N-nsP2, N-terminal nsP2.
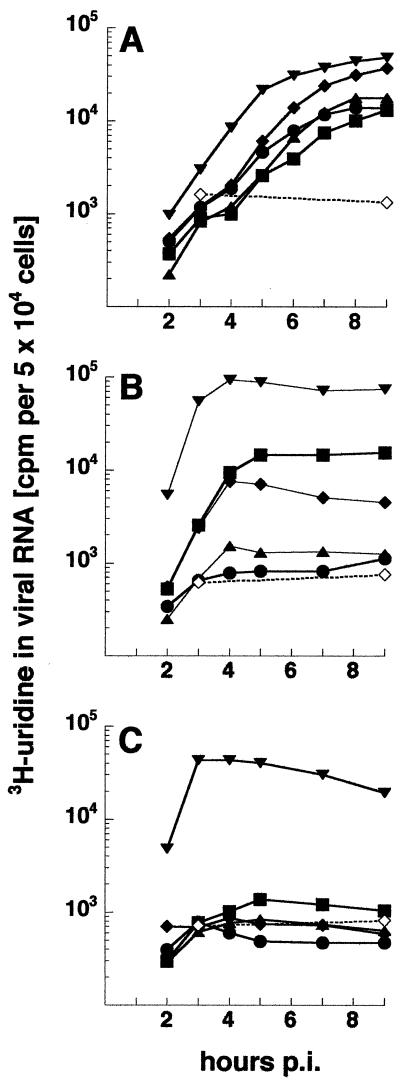
As shown in Fig. 5, levels of RNA synthesis generally reflected the EOP profile. The nsP3 mutant ts4 was inhibited over 97% at 40°C and more than 95% at 37°C, coinciding with EOP values of 10−3 or less at both temperatures. Mutants ts138, Toto:ts138ABs, and Toto:ts7B5 synthesized RNA at 37°C to 15, ca. 7.5, and 100% of the maximum rate observed at 30°C, respectively (Fig. 5), yet PFU yields at 37°C were within parental levels (Table 2). At 40°C, however, RNA synthesis by each virus was 5% or less of that at 30°C, paralleling the low PFU yields at 40°C (Table 2). We interpret the results to indicate that low but stable rates of transcription of at least 7.5% of the 30°C maximum are sufficient to enable formation of plaques, albeit ones generally with smaller diameters, over the 2- to 3-day incubation period of the assay. Recombinant Toto:ts138BBs resembled ts138 fully (data not shown). Differences in the ability to synthesize RNA at 30 and 37°C of ts138 and Toto:ts138BBs compared to Toto:ts138ABs, which has only the nsP3 ts Gly68 lesion common to each virus, suggest that the other, non-ts substitutions in the nt 2288-to-nt 4318 region and expressed in the nsP2 and nsP3 proteins of ts138 and Toto:ts138BBs partially rescue the ts lesion. Finally, ts4, Toto:ts7B5, and Toto:ts138ABs viruses were partially defective even at 30°C and had rates of overall transcription of about one-third of parental values (Fig. 5A).
FIG. 5.
Comparison of the kinetics of RNA synthesis by nsP3 mutant viruses and parental viruses. CEF cells were infected at 30°C (A) with ts4 (•), ts7B5 (▪), ts138 (♦), Toto:ts138AS recombinant (▴), or parental SIN HR (▾) at an MOI of 100 or were mock infected (⋄). Duplicate cultures were shifted to 37°C (B) or 40°C (C) beginning at 1 h p.i. and were maintained at the respective temperatures until the end of the experiment. Cultures were labeled with [3H]uridine (50 μCi/ml) in the presence of 20 μg of actinomycin D/ml for 1-h periods and were immediately harvested at the end of the pulse. The acid-insoluble incorporation in 50,000 cells is shown.
Similar to ts138, replication complexes formed at 30°C by ts138 recombinants also were stable and continued RNA synthesis when cultures were shifted to 40°C, but few or no new ones formed at 40°C (data not shown). None of the ts138 recombinants were defective in 26S mRNA synthesis at 40°C (Table 3) and thus differed from most C-terminal domain nsP2 mutants (10, 27, 66). The ts138 recombinant fully reproduced the ts RNA-negative phenotype of the original ts138 mutant.
TABLE 3.
Toto:ts138ABs recombinants resemble ts138 and are not ts for 26S mRNA synthesisa
| Virus | Molar ratio of 49S RNA:26S mRNA
|
|||
|---|---|---|---|---|
| At 30°C | At 40°C when shifted up at:
|
|||
| 1 h p.i. | 4 h p.i. | 8 h p.i. | ||
| ts138 | 0.12 | 0.19 | 0.16 | 0.19 |
| Toto:ts138ABs | ||||
| 1 | 0.19 | 0.28 | 0.21 | 0.27 |
| 4 | 0.16 | 0.29 | 0.21 | 0.26 |
CEF cells were infected with each virus at an MOI of 100 PFU per cell. At 4 and at 9 h p.i., one set of cultures was labeled for 1 h at 30°C with [3H]uridine (50 μCi/ml). At the times indicated, cultures were shifted to 40°C and incubated for 1 h before being labeled for 1 h at 40°C. At the end of the labeling period, the monolayers were harvested and the lysates were electrophoresed on agarose gels as described in Materials and Methods. The molar ratio of the two RNAs was calculated from the incorporation present in each RNA band excised from the dried gel after correcting for their respective molecular weights.
Minus-strand RNA synthesis at 40°C.
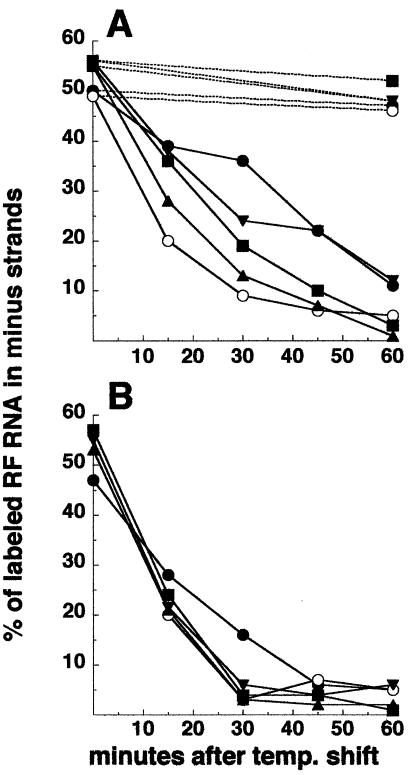
The nsP3 mutants were tested for their ability to produce minus strands after shift to 40°C. Infected cultures were allowed to initiate replication at 30°C for 2 to 3 h. To compare the different mutants at the same relative time in the infectious cycle, each set of infected cultures was shifted to 40°C early and at the time p.i. when only ca. 15% of the maximum rate of plus-strand RNA synthesis had been attained. Cultures were pulse-labeled at 40°C for successive 15-min periods and harvested at the end of each pulse period. As shown in Fig. 6, ts4 nsP3 rapidly inhibited minus-strand synthesis at 40°C and at a rate similar to that following addition of cycloheximide. An intermediate inhibition profile was observed for ts138 and Toto:ts138ABs, while Toto:ts7B5 resembled parental Toto virus and continued minus-strand synthesis longer. Addition of cycloheximide at the time of shift to 40°C prevented continued minus-strand synthesis in all cultures (Fig. 6B), indicating that nsPs synthesized at 40°C were responsible for this replicase activity. Since ts138, Toto:ts138ABs, and Toto:ts7B5 nsP3 function at 37°C similarly to parental nsP3 but do not function at 40°C (Table 2 and Fig. 5), synthesis observed during the first 15 min postshift could be due to production of active nsP3 proteins at temperatures lower than 40°C. However, the observed continuation of minus-strand synthesis beyond this time argues that ts138 nsP3 proteins produced at 40°C are only partially compromised and Toto:ts7B5 nsP3 proteins are in fact functional (Fig. 6). Loss of phosphorylation and loss of minus-strand synthesis were also exhibited by nsP3 insertion mutants CR3.36 and CR3.39 (Fig. 2) (35) that are located near ts138 or ts4, respectively, supporting a role of the two nsP3 subregions in both functions.
FIG. 6.
Minus-strand synthesis by the nsP3 mutants. CEF cells, infected with an MOI of 100 of each virus at 30°C, were maintained at 30°C (A) (dashed lines) or were shifted up to 40°C when about 15% of the maximal rate of RNA synthesis had been achieved at 30°C or between 2 and 3 h p.i. depending on the virus. Duplicate cultures shifted to 40°C were incubated in the presence (B) or absence (A) (solid lines) of cycloheximide beginning at the time of shift. Minus-strand synthesis was determined by pulse-labeling cells with 200 μCi of [3H]uridine/ml for 15-min periods over the first 1 h of shift and analysis of the purified viral RF RNA for radiolabeled minus-strand RNA, as described in Materials and Methods. Incorporation into newly made minus strands is expressed as the percentage of the total labeled RF RNA that was in minus-strand RNA. Symbols: •, SIN HR; ○, ts4; ▴, ts138; ▪, Toto:ts138AS recombinant; ▾, ts7B5.
DISCUSSION
The results of our studies identified a new nsP3 mutant, ts138 of the A complementation group of SIN HR RNA-negative mutants. Its lesion mapped to nt 4303 in the nsP3 gene and to a predicted change of Ala68 to Gly in the N-terminal region of the nsP3 protein. The nsP3 sequence first functions in infected cells as nascent P1234 polyproteins, then as intermediate P123 and P23 polyproteins, and later as mature, fully cleaved forms. It was possible that a phenotype observed for individual nsP3 mutants would be expressed by only one of these forms. Our observations were (i) that nsP3 and nsP2 seem to function initially as a single unit (P23/P123) as deduced from complementation experiments and as predicted earlier, (ii) that the degree of phosphorylation changed with different nsP3 mutants, and (iii) that reduced phosphorylation of nsP3 (as P23/P123) correlated with reduced minus-strand synthesis. The ability to block the majority of nsP3 phosphorylation (nsP3c) with single changes in the macrodomain or at residue 268 suggests that these sites are important for recruiting or targeting kinases and that the higher phosphorylated nsP3 species are functionally important. LaStarza et al. (34, 35) reported similar effects on nsP3 phosphorylation in some but not all nsP3 insertion or deletion mutants. Mutant CR3.39 (insertion at residue 226-227) and mutant CR3.36 (insertion at residue 58-59) exhibit phenotypes similar to ts4 and ts138, including reduced plus-strand synthesis (20 to 30% of wild type) at 30°C.
As summarized in Table 4, the phenotype of Gly68 (ts138) nsP3 had several properties in common with mutant nsP3 Val268 (ts4) and Ser312 (Toto:ts7B5) proteins. All three single amino acid substitutions conferred ts phenotypes in replication and PFU production (EOP of 10−3 to 10−5) at 40°C. None of the mutations inhibited overall polyprotein processing or the ability to internally initiate subgenomic mRNA synthesis at 40°C; thus, none drastically blocked functions of the nsP2 carboxyl-terminal domain that in nascent and intermediate polyproteins was covalently attached to the N-terminal end of nsP3. However, all three affected the processing of P1234 in more subtle ways by reducing cleavage at the 3/4 site, thereby increasing accumulation of inactive P34 relative to the active nsP4 polymerase core. Finding that the nsP3 mutants successfully complemented defective ts6 nsP4 proteins argues that their cleaved nsP4 levels were sufficient and trans-active at 40°C (56, 73). All three mutant nsP3 proteins also were transported to membrane compartments of the cell and associated with nsP1, nsP2, and nsP4 in detergent-resistant and immunoprecipitation-resistant complexes, similar to parental nsP complexes (7). Thus, their functional defects occur after these initial steps. A recent study of the biogenesis of alphavirus SFV complexes suggested that assembly occurs at the plasma membrane, where nascent P1234 polyproteins are targeted initially by signals within the nsP1 protein sequence; intriguingly, over time, such complexes appear to cycle between endosomal-lysosomal compartments and the plasma membrane (30).
TABLE 4.
Phenotypic analysis of mutant nsP3 proteins at 40°Ca
| nsP3 protein | Polyprotein processing | nsP3 phosphorylation | RNA synthesis | 26S RNA synthesis | 37°C EOP | Forms nsP complex | Minus-strand RNA synthesis |
|---|---|---|---|---|---|---|---|
| Gly68 | Incr. P34 | Partially ts | ts | wt | wt | + | Intermediate |
| Val268 | Incr. P34 | ts | ts | wt | ts | + | ts |
| Ser312 | Incr. P34, P12 | wt | ts | wt | wt | + | wt |
Incr., increased; wt, wild type or parental phenotype; +, replication-transcription nsP complexes were detected.
Table 4 also summarizes the differences observed among the three mutant nsP3 proteins. Of these three, ts4 was the most extreme, and only it failed to complement ts11, which has a mutated nsP1 (73). At 40°C, both P23 and nsP3 of ts4 were underphosphorylated (both nsP3* and nsP3c were undetectable) and minus-strand synthesis stopped immediately. This is in contrast to Toto:ts7B5, which at 40°C produced Ser312 nsP3 that ranged in size from 76 to 106 kDa (Fig. 3B), similarly to parental fully phosphorylated nsP3, and retained the ability to engage in minus-strand synthesis after shift to 40°C, also like parental SIN HR. Unlike parental virus, only Toto:ts7B5-infected cells accumulated P12, suggesting that the mutated nsP3 decreased recognition of the 1/2 cleavage site. The Toto:ts7B5 Ser312 lesion in nsP3 also required temperatures greater than 37°C to inactivate its RNA synthetic functions. The ts138 Gly68 change was intermediate between these two extremes. Its ts lesion was leaky at 37°C, its nsP3 proteins were partially phosphorylated (reduced nsP3* and little nsP3c), and its P23 polyproteins were partially active in minus-strand synthesis at 40°C. Also, because cleavage of the 1/2 site occurred efficiently, the ts138 P123/P23 polyproteins did not appear to be grossly altered in overall conformation. Also, plus-strand synthesis by ts138 was unaffected at 40°C. Whatever the role of the macrodomain, our observations suggest that it exerts its effect at the polyprotein (P23/P123) level and not on mature, cleaved nsP3 proteins. This property is shared by ts4 Val268 nsP3 proteins, whose ts inhibition of minus-strand synthesis was specific for newly made, polyprotein-containing complexes and did not inhibit a resumption of minus-strand synthesis by mature, normally plus-strand-synthesizing complexes whose nsP1, nsP2, and nsP3 components also included a mutant form of the nsP4 polymerase (73). We conclude that the Val268 nsP3 proteins (ts4) form only inactive nsP complexes at 40°C. The Gly68 nsP3 proteins (ts138 and its recombinants) are weakly active, reducing the efficiency with which new complexes form or function; this in turn would limit accumulation of plus-strand RNA-synthesizing complexes and prevent rapid increases in the rate of plus-strand synthesis. Because alphavirus minus-strand synthesis is blocked by the infected cell at about 4 h p.i. (at 37°C), a failure to sufficiently amplify minus-strand templates during this early period could explain its ts RNA-negative phenotype. The Ser312 nsP3 proteins (Toto:ts7B5) are essentially fully active, arguing that the defect with this virus occurs after formation of the minus-strand replicase and may be in a step needed for its conversion to a mature, plus-strand-synthesizing form. Consistent with this interpretation, our earlier studies showed that the Toto:ts7B5 Ser312 nsP3 defect did not block reactivation of minus-strand synthesis by preformed replication complexes containing an altered nsP4 polymerase (73).
The ts138 Gly68 nsP3 mutant provided the first opportunity to probe the role of the histone 2A macrodomain in nsP3. The macrodomain homologue in yeast (gene YBR022w) is a phosphoesterase; it acts to process ADP-ribose-1"-phosphate (Appr-1"-p) generated during processing of tRNA precursors. Removal of a 2′-phosphate from the exon junction after ligation involves its transfer to NAD with formation of Appr>p (1",2"-cyclic phosphate) and nicotinamide, and the subsequent cleavage of Appr>p to Appr-1"-p by the 2′,3′-cyclic phosphoesterase CPD1p (41). Some viruses encode a 2′, 3′-phosphodiesterase activity that is part of a novel 2H phosphoesterase superfamily, and coronaviruses and a few others possess both this and the Appr-1"-p phosphoesterase (42). Their functions have been postulated to include posttranscriptional gene silencing, RNA degradation, capping (rotavirus VP3 protein active in capping contains a 2H phosphodiesterase domain at its C terminus), or even signal transduction (42). While nsP2 is transported and accumulates in the nucleus and nucleolus, P23/nsP3 proteins do not enter the nucleus or nucleolus (reviewed in reference 26), ruling out a role for the viral macrodomain in modifying cellular RNA processing events. Structural analysis supports the likelihood that the alphavirus nsP3 macrodomain retains phosphoesterase activity (M. Lakshminarayan, personal communication). Of particular interest, the His-Ala-Val (His-Ala-Ala) tripeptide is part of the putative active site of this predicted enzyme, with its conservation due, at least partly and possibly fully, to this enzymatic activity (A. Gorbalenya, personal communication).
A role for a putative macrodomain phosphoesterase in alphavirus replication is not obvious at this time. It may play some role affecting the response to infection, not actually replication itself. Its role might be cell type specific. There is no macrodomain sequence in the genomes of plant virus members of the SIN superfamily. Alphavirus replication does not involve cytoplasmic splicing or RNA processing-ligation events such as occur with tRNA precursors. Rather, replication involves an internal initiation process for subgenomic synthesis (66) that may require template remodeling. Also, alphavirus replication utilizes a novel form of capping whereby GTP is first methylated and then transferred to the 5′ end of nascent plus-strand transcripts (3). And, alphavirus replication results in the addition of a nontemplated G residue to the 5′ end of nascent viral minus strands (75, 76). In addition to possible roles in the above functions, a putative phosphoesterase or other activity might contribute to antihost defenses if it inactivates small, cyclic, signaling molecules (55) or might recruit activated protein kinases to the initial viral replication complexes (42). Finally, the histone 2A variant H2A.Z protein protects euchromatin from transcriptional silencing (44), a function that if shared by the histone 2A-like macrodomain in nsP3 could contribute to the transcriptional efficiency or active state of alphavirus replicative and transcriptive intermediates.
In summary, for the macrodomain His-Ala-Val peptide and residue 268, the extent of nsP3 phosphorylation and the ability of each mutant to engage in minus-strand synthesis varied in parallel at nonpermissive temperature. Thus, although the present evidence argues that the nsP3 sequence is not an essential accessory component of group 3 RNA-dependent RNA core polymerases from its presence in only the animal members of the SIN superfamily (19), the N-terminal nsP3 sequence and the enzyme predicted to reside within the nsP3 macrodomain appear to play a role in overall viral replication in vertebrate cells that affect principally minus-strand synthesis.
Acknowledgments
We gladly acknowledge the generous gift of Toto1101 cDNA from C. M. Rice and of the monospecific nsP antisera from Ellen and Jim Strauss. We thank E. Koonin, A. Gorbalenya, and M. Iyer Lakshminarayan for stimulating discussions.
Support for this study was derived from Public Health Service grant AI-15123 from the National Institutes of Health.
REFERENCES
- 1.Aguiar, R. C., Y. Yakushijin, S. Kharbanda, R. Salgia, J. A. Fletcher, and M. A. Shipp. 2000. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96:4328-4334. [PubMed] [Google Scholar]
- 2.Ahlquist, P., E. G. Strauss, C. M. Rice, J. H. Strauss, J. Haseloff, and D. Zimmern. 1985. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J. Virol. 53:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola, T., and L. Kaariainen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahola, T., P. Kujala, M. Tuittila, T. Blom, P. Laakkonen, A. Hinkkanen, and P. Auvinen. 2000. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J. Virol. 74:6725-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahola, T., P. Laakkonen, H. Vihinen, and L. Kaariainen. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71:392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burge, B. W., and E. R. Pfefferkorn. 1966. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology 30:204-213. [DOI] [PubMed] [Google Scholar]
- 9.Cross, R. K. 1983. Identification of a unique guanine-7-methyltransferase in Semliki Forest virus (SFV) infected cell extracts. Virology 130:452-463. [DOI] [PubMed] [Google Scholar]
- 10.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot, R. J., W. R. Hardy, Y. Shirako, and J. H. Strauss. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 9:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinant, S., M. Janda, P. A. Kroner, and P. Ahlquist. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 67:7181-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez, G., C. Y. Wang, and T. K. Frey. 1990. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology 177:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fata, C. L., S. G. Sawicki, and D. L. Sawicki. 2002. Alphavirus minus-strand RNA synthesis: identification of a role for Arg183 of the nsP4 polymerase. J. Virol. 76:8632-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fata, C. L., S. G. Sawicki, and D. L. Sawicki. 2002. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 76:8641-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin, R. M. 1966. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc. Natl. Acad. Sci. USA 55:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya, A. E., and E. V. Koonin. 1993. Comparative analysis of amino acid sequences of key enzymes of replication and expression of positive-strand RNA viruses: validity of approach and evolutionary implications. Soc. Sci. Rev. D Physiochem. Biol. 11:1-84. [Google Scholar]
- 20.Grady, L. J., and W. P. Campbell. 1989. Amplification of large RNAs (greater than 1.5 kb) by polymerase chain reaction. BioTechniques 7:798-800. [PubMed] [Google Scholar]
- 21.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA-temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 63:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA-temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy, W. R., Y. S. Hahn, R. J. de Groot, E. G. Strauss, and J. H. Strauss. 1990. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology 177:199-208. [DOI] [PubMed] [Google Scholar]
- 24.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haseloff, J., P. Goelet, D. Zimmern, P. Ahlquist, R. Dasgupta, and P. Kaesberg. 1984. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. USA 81:4358-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaariainen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71:187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keranen, S., and L. Kaariainen. 1979. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 32:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keranen, S., and L. Ruohonen. 1983. Nonstructural proteins of Semliki Forest virus: synthesis, processing, and stability in infected cells. J. Virol. 47:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin, E. V., A. E. Gorbalenya, M. A. Purdy, M. N. Rozanov, G. R. Reyes, and D. W. Bradley. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 89:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lain, S., M. T. Martin, J. L. Riechmann, and J. A. Garcia. 1991. Novel catalytic activity associated with positive-strand RNA virus infection: nucleic acid-stimulated ATPase activity of the plum pox potyvirus helicaselike protein. J. Virol. 65:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lain, S., J. L. Riechmann, and J. A. Garcia. 1990. RNA helicase: a novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acids Res. 18:7003-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lastarza, M. W., A. Grakoui, and C. M. Rice. 1994. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202:224-232. [DOI] [PubMed] [Google Scholar]
- 35.LaStarza, M. W., J. A. Lemm, and C. M. Rice. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 68:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 41.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153-1155. [DOI] [PubMed] [Google Scholar]
- 42.Mazumder, R., L. M. Iyer, S. Vasudevan, and L. Aravind. 2002. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 30:5229-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKnight, K. L., D. A. Simpson, S. C. Lin, T. A. Knott, J. M. Polo, D. F. Pence, D. B. Johannsen, H. W. Heidner, N. L. Davis, and R. E. Johnston. 1996. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J. Virol. 70:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 45.Mi, S., and V. Stollar. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184:423-427. [DOI] [PubMed] [Google Scholar]
- 46.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 26:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peranen, J. 1991. Localization and phosphorylation of Semliki Forest virus non-structural protein nsP3 expressed in COS cells from a cloned cDNA. J. Gen. Virol. 72:195-199. [DOI] [PubMed] [Google Scholar]
- 48.Peranen, J., M. Rikkonen, P. Liljestrom, and L. Kaariainen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 64:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peranen, J., K. Takkinen, N. Kalkkinen, and L. Kaariainen. 1988. Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J. Gen. Virol. 69:2165-2178. [DOI] [PubMed] [Google Scholar]
- 50.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawicki, D., D. B. Barkhimer, S. G. Sawicki, C. M. Rice, and S. Schlesinger. 1990. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology 174:43-52. [DOI] [PubMed] [Google Scholar]
- 54.Sawicki, D., and S. Sawicki. 1994. Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch. Virol. Suppl. 9:393-405. [DOI] [PubMed] [Google Scholar]
- 55.Sawicki, D. L., R. H. Silverman, B. R. Williams, and S. G. Sawicki. 2003. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J. Virol. 77:1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 144:20-34. [DOI] [PubMed] [Google Scholar]
- 57.Sawicki, D. L., and S. G. Sawicki. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J. Virol. 67:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawicki, D. L., S. G. Sawicki, S. Keranen, and L. Kaariainen. 1981. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J. Virol. 39:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheidel, L. M., and V. Stollar. 1991. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology 181:490-499. [DOI] [PubMed] [Google Scholar]
- 60.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148-160. [DOI] [PubMed] [Google Scholar]
- 61.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shirako, Y., and J. H. Strauss. 1998. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J. Virol. 72:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 1976. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology 74:154-168. [DOI] [PubMed] [Google Scholar]
- 64.Strauss, E. G., R. Levinson, C. M. Rice, J. Dalrymple, and J. H. Strauss. 1988. Nonstructural proteins nsP3 and nsP4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology 164:265-274. [DOI] [PubMed] [Google Scholar]
- 65.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. USA 80:5271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 58:491-562. (Erratum, 58: 806.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kaariainen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79:309-319. [DOI] [PubMed] [Google Scholar]
- 68.Tuittila, M., and A. E. Hinkkanen. 2003. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J. Gen. Virol. 84:1525-1533. [DOI] [PubMed] [Google Scholar]
- 69.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 70.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kaariainen. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 276:5745-5752. [DOI] [PubMed] [Google Scholar]
- 71.Vihinen, H., and J. Saarinen. 2000. Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J. Biol. Chem. 275:27775-27783. [DOI] [PubMed] [Google Scholar]
- 72.Wang, H. L., J. O'Rear, and V. Stollar. 1996. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology 217:527-531. [DOI] [PubMed] [Google Scholar]
- 73.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wengler, G., and H. J. Gross. 1982. Terminal sequences of Sindbis virus-specific nucleic acids: identity in molecules synthesized in vertebrate and insect cells and characteristic properties of the replicative form RNA. Virology 123:273-283. [DOI] [PubMed] [Google Scholar]
- 76.Wengler, G., and H. S. Gross. 1979. Replicative form of Semliki Forest virus RNA contains an unpaired guanosine. Nature 282:754-756. [DOI] [PubMed] [Google Scholar]
- 77.Weston, J., S. Villoing, M. Bremont, J. Castric, M. Pfeffer, V. Jewhurst, M. McLoughlin, O. M. Rodset, K. E. Christie, J. Koumans, and D. Todd. 2002. Comparison of two aquatic alphaviruses, salmon pancrease disease virus and sleeping disease virus, by using genome sequence analysis, monoclonal reactivity, and cross-infection. J. Virol. 76:6155-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]