Abstract
Instead of displaying the wild-type selective export of virions containing mature genomes, human hepatitis B virus (HBV) mutant I97L, changing from an isoleucine to a leucine at amino acid 97 of HBV core antigen (HBcAg), lost the high stringency of selectivity in genome maturity during virion export. To understand the structural basis of this so-called “immature secretion” phenomenon, we compared the stability and morphology of self-assembled capsid particles from the wild-type and mutant I97L HBV, in either full-length (HBcAg1-183) or truncated core protein contexts (HBcAg1-149 and HBcAg1-140). Using negative staining and electron microscopy, full-length particles appear as “thick-walled” spherical particles with little interior space, whereas truncated particles appear as “thin-walled” spherical particles with a much larger inner space. We found no significant differences in capsid stability between wild-type and mutant I97L particles under denaturing pH and temperature in either full-length or truncated core protein contexts. In general, HBV capsid particles (HBcAg1-183, HBcAg1-149, and HBcAg1-140) are very robust but will dissociate at pH 2 or 14, at temperatures higher than 75°C, or in 0.1% sodium dodecyl sulfate (SDS). An unexpected upshift banding pattern of the SDS-treated full-length particles during agarose gel electrophoresis is most likely caused by disulfide bonding of the last cysteine of HBcAg. HBV capsids are known to exist in natural infection as dimorphic T=3 or T=4 icosahedral particles. No difference in the ratio between T=3 (78%) and T=4 particles (20.3%) are found between wild-type HBV and mutant I97L in the context of HBcAg1-140. In addition, we found no difference in capsid stability between T=3 and T=4 particles successfully separated by using a novel agarose gel electrophoresis procedure.
Human hepatitis B virus (HBV) core antigen (HBcAg) is 183 to 185 amino acids long. The arginine-rich C terminus of HBcAg is involved in binding to the HBV RNA pregenome and DNA genome (Fig. 1A) but is dispensable for HBV capsid assembly in Escherichia coli (2, 7, 9, 17, 31, 37) and insect cells (1). A truncated version of HBV core antigen, HBcAg1-140, lacking 43 amino acids of the carboxy terminus, is known to be sufficient for self-assembly into capsid particles (Fig. 1A) (see references cited above). The four-helix bundle structure of HBV capsid particles has been studied at a high degree of resolution by cryoelectron microscopy and X-ray diffraction analysis (3, 6, 32).
FIG. 1.
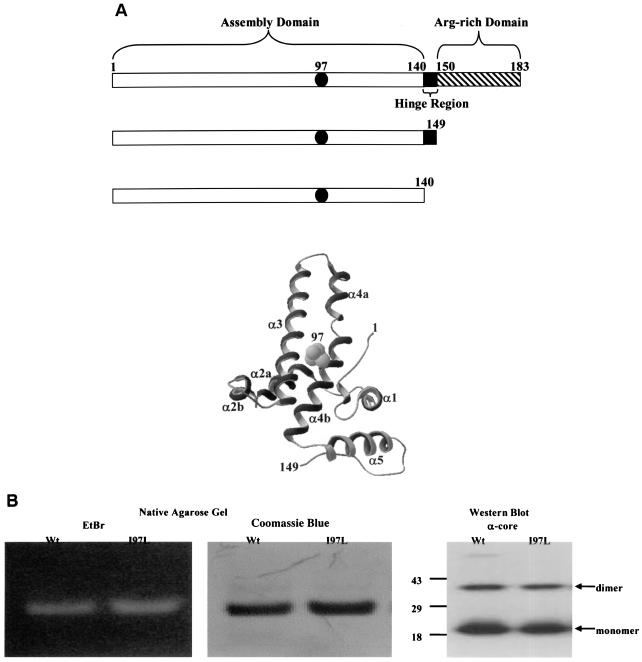
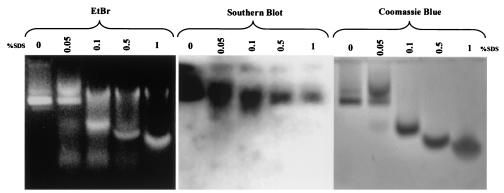
(A) Functional domains and three-dimensional structure of HBV core protein. Filled circles represent a wild-type isoleucine or a mutant leucine at amino acid 97. Truncated wild-type and mutant I97L clones consist of either amino acids 1 to 140 or amino acids 1 to 149 lacking the arginine-rich carboxy termini (▧). A hinge region, which connects the capsid assembly and arginine-rich domains, is exposed on the capsid surface and is protease sensitive (24). The ribbon diagram depicts the three-dimensional structure of the core monomer (3, 6, 32) and the location of amino acid 97. A Swiss PDB Viewer was used to render the diagram from 1QGT obtained from GenBank. (B) Characterizations of the recombinant HBcAg1-149 particles by native agarose gel electrophoresis and immunoblot analysis. Encapsidated nucleic acids were stained with EtBr, and the same gel was subsequently stained with protein-specific Coomassie blue. A rabbit polyclonal anticore antibody was used to detect the HBcAg (27).
The self-assembled icosahedral capsid particles of HBV are somewhat heterogeneous in size. Larger particles with an averaged diameter of 28 to 30 nm have an icosahedral symmetry of T=4 and consist of 240 copies of HBcAg, while smaller particles contain 180 copies with an average diameter of 25 nm and have an icosahedral symmetry of T=3 (3, 6, 7, 32). The T=4 particles are the predominant fraction (>95%) when either full-length HBcAg1-183 or truncated HBcAg1-149 is expressed in E. coli. Interestingly, when the HBcAg1-140 version is expressed in E. coli, the predominant population of self-assembled icosahedral particles are T=3 (29, 38). Both larger and smaller particles can be found not only in the laboratory setting but also in human liver in natural infections (5, 15).
The frequent occurrence of a core I97L mutation in chronic carriers, changing from an isoleucine to a leucine at amino acid 97 of HBcAg, was first recognized by Ehata et al. (8). Despite the prevalence of this mutation reported by many different research groups (see references in Suk et al. [27]), the functional significance of this hotspot mutation remains to be investigated. Previously, we noted two distinct phenotypes associated with this core mutation. First, there is an immature secretion phenotype with nonselective secretion of virions containing immature HBV genomes (single-stranded DNA) (34, 36). This phenotype represents an interesting exception to the dogma of preferential secretion of virions containing mature genomes (partially double-stranded relaxed circle DNA) (11, 21, 28, 30). Second, a replication advantage and host factor-independent phenotype was found to be associated with the mutant I97L in the human hepatoma cell line Huh7, but not in a hepatoblastoma cell line HepG2 (27). To date, the structural basis for either the immature secretion or replication advantage phenotypes has remained unclear. Furthermore, it is also unclear what could be the structural basis for compensatory mutations that can offset the I97L immature secretion (4, 16, 35). Since isoleucine and leucine are isomers with identical molecular weights, it is puzzling that such a subtle structural difference in side chains can cause a significant functional consequence on viral replication and virion secretion.
Previously, we demonstrated by Western blot analysis that the steady-state levels of HBcAg are very similar between wild-type HBV and mutant I97L (36). Rigorously speaking, although this result provides support that the core “protein” of the wild type and mutant I97L are equally stable, it provides no direct evidence that their core “particles” are equally stable. HBV replication, mediated by reverse transcriptase, is known to be capsid dependent (10, 33). The fact that viral DNA synthesis measured by Southern blot analysis is similar between wild-type HBV and mutant I97L in HepG2 cells (35) suggests that the stabilities of their capsid particles are similar. However, more direct experimental proof on this issue is still needed. Furthermore, despite the fact that wild-type and C-terminally truncated HBV capsid particles have previously been characterized in vitro (2, 9, 31, 37), no characterizations of naturally occurring mutant capsid particles have yet been reported (25).
A “positive selection” hypothesis has been postulated that, at a late stage in HBV genome replication, the nucleocapsid acquires and/or displays a “morphogenetic signal” that allows for the envelopment of the particles leading to egress from the cell by the secretory pathway (28). An alternative “negative selection” hypothesis is that the capsid particles containing the earlier replicative intermediates are less stable particles that fall apart somehow during the envelopement and are thus underrepresented in the population of extracellular virions. Indeed, it has been reported that extracellular core particles are more stable than intracellular particles due to differential formation of disulfide linkages (14). Although the cause-and-effect relationship between capsid stability and virion release remains unclear, it might be interesting to determine whether the immature secretion phenotype of the core mutant I97L simply results from an increased intracellular capsid stability.
To elucidate the structure and function relationship of HBV capsid proteins and particles, we expressed the wild-type and mutant I97L core proteins in E. coli and characterized the purified self-assembled core particles by comparing their stability and morphology under several different denaturing conditions. Taking advantage of a simple procedure recently invented in our laboratory, we also compared the stability of T=3 and T=4 particles in the context of HBcAg1-140. In almost all comparisons, we found no significant difference in the capsid stability between wild-type and mutant HBVs in either the smaller T=3 or larger T=4 icosahedral particles. Unexpectedly, we found that full-length and truncated capsid particles can be clearly differentiated by agarose gel electrophoresis after sodium dodecyl sulfate (SDS) treatment in the absence of reducing agents.
MATERIALS AND METHODS
Southern and Western blot analyses were conducted as described elsewhere (34). Other methods are detailed below.
Construction of expression plasmids.
HBV subtype adr DNA was used for all constructs. For expression in E. coli, truncated versions of wild-type and mutant I97L HBV core proteins, including amino acids 1 to 140 or amino acids 1 to 149, and full-length versions from amino acids 1 to 183 were isolated and amplified by using PCR from pSVC-adr and pSVC-I97L (36). The upstream primer 5′-ATGGACATTGACCCGTATAA-3′ included the AUG start codon and was used for all versions. For HBcAg1-140, the downstream primer (5′-TCCGGAAGTGTTTATAAGATAGGGGCATTT-3′) included a stop codon at amino acid 141. The downstream primer for HBcAg1-149 (5′-CCTCGTCGTCAAACAACAGTA-3′) included a UGA stop codon at amino acid 150. Full-length coding regions were amplified with the downstream primer (5′-TCGAAGGGATACTAACATTGAGATTCCCG-3′), which spans the authentic UGA stop codon for the HBV core. Two full-length mutants which changed codon 183 from a cysteine to either an alanine (C183A) or a serine (C183S) were constructed by using downstream primers (5′-CAAGGGATACTAAGATTGAGATTCCCG-3′) and (5′-CAAGGGATACTAAGCTTGAGATTCCCG-3′), respectively. The amplified fragments were recovered from agarose gel sections with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.). Fragments were cloned into pET-Blue1 by using Novagen's Perfectly Blunt cloning kit and transformed into NovaBlue cells. Colonies were screened for the inserts and correct orientation by using BglII for the truncated versions and TacI for the full-length clones (both Gibco, Rockville, Md.) and confirmed by sequencing.
Expression and purification of core particles.
The plasmid constructs for the truncated versions were moved into Tuner cells (Novagen, Madison, Wis.) for expression in E. coli. Full-length versions contain numerous arginine codons in the carboxy terminus. Since these arginine codons are rarely used in E. coli and their corresponding tRNAs exist at a very low level, a plasmid expressing these rare tRNAs, pGro-ArgUW (13), was transformed into the Tuner cells prior to introducing the full-length expression plasmid to ensure adequate translation.
One-liter cultures with an A600 of 0.7 to 1.0 were induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C for truncated core particles and for 6 h at 15°C for the full-length versions. The cells were harvested by centrifugation (Beckman J2-MC, JLA-10.500 rotor, 5,000 rpm, 15 min, 4°C), resuspended in 10 ml of lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 100 μg of phenylmethylsulfonyl fluoride/ml, and 2 mg of lysozyme/ml), and frozen at −80°C. Preparations were subjected to three freeze-thaw cycles and then treated with 1 ml of DNase cocktail (0.1 M MgCl2, 0.2 M DNase) for 1 to 2 h. The insoluble fractions were removed by centrifugation (Beckman J2-MC, JA-17 rotor, 12,000 rpm, 30 min, 4°C), and the soluble fractions were treated with a one-fourth volume of saturated ammonium sulfate to precipitate the capsids overnight at 4°C. Precipitated capsid preparations were pelleted (Beckman J2-MC, JS-13.3 rotor, 12,000 rpm, 4°C), resuspended in Tris-buffered saline (TBS; 0.1 M NaCl, 2 mM KCl, 25 mM Tris [pH 7.4]), and dialyzed overnight against TBS. Fractions containing capsids from sucrose gradients of 30 to 60% for truncated or 40 to 70% for full-length capsids (Beckman L8-M ultracentrifuge, SW-28 rotor, 26,000 rpm, 18 h, 4°C) were identified on native agarose gels with ethidium bromide (EtBr) and Coomassie blue staining. Pooled fractions were pelleted through a 20% sucrose cushion (Beckman L8-M Ultracentrifuge, SW-28 rotor, 26,000 rpm, 18 h at 4°C). The pellets were resuspended in TBS and protein quantities were determined by the Warburg-Christian concentration method (Beckman/Coulter DU 640 spectrophotometer). The purity of the preparations was examined on Coomassie blue-stained native agarose gels and SDS-polyacrylamide gel electrophoresis. Typical yields ranged from 30 to 40 mg/liter of induced culture for the truncated versions and from 0.5 to 1 mg for the full-length preparations.
Stability tests and trypsin digests.
Whenever possible, freshly prepared capsid particles were tested for stability under different conditions. Portions (20 μg) of the preparations were used for each test. For the temperature tests, the samples were incubated in TBS at the indicated temperatures for 15 min; for the pH tests, the capsids were incubated in TBS adjusted to the indicated pH for 30 min at 37°C. Samples were then run on a 1% native agarose gel containing 0.5 μg of EtBr/ml, photographed under UV light, stained with Coomassie blue, and rephotographed under fluorescent light. A similar procedure was used for the stability tests with SDS. Samples were placed into TBS with different percentages of SDS and immediately loaded onto 1% native agarose gels with EtBr and stained with Coomassie blue. Trypsin digests of 20-μg samples of capsid preparations were carried out by using 1 μg of sequencing-grade trypsin (Sigma, St. Louis, Mo.) at 37°C for 1 h. The stability of the full-length capsids after trypsin digestion was tested by adding SDS to the samples prior to running them on 1% agarose gels.
GTG gel and electroelution of capsids.
Portions (20 μg) of truncated capsid particles of HBcAg1-149 and HBcAg1-140 were analyzed in a 2.6% GTG low-melting-point agarose gel (FMC Bioproducts, Rockland, Maine), and electrophoresis was run at 45 V for 4 h in TBE buffer (45 mM Tris-borate, 1 mM EDTA). For electroelution, 50 μg of HBcAg1-140 particles was loaded on a 2.6% GTG agarose gel. After the core particles were separated into two discrete bands, both the upper band (T=4) and the lower band (T=3) were excised from the agarose gel and placed into different dialysis tubes. Electroelution of the core particles from the excised gel slice was conducted in 0.5× TBE buffer at 90 V for 2 to 3 h. Approximately 400 to 500 μl of each sample was collected from the supernatant of the dialysis tubes, and 10 μl of each sample was sufficient for electron microscopy (EM).
EM of core particles.
Standard negative staining procedures were used to prepare capsid particles for EM. Capsids in TBS (0.5 μg/μl) were placed on Formvar-carbon-coated copper grids and stained with 2% aqueous uranyl acetate for 2 min. Electron micrographs were taken in a Philips EM201 electron microscope at 60 kV. Chi-test and Student t test in a Microsoft Excel program were used to analyze the size measurement and comparisons of capsid particles. Tobacco mosaic virus particles at a concentration of 0.25 μg/μl were included as an internal reference (16 to 20 nm in width).
RESULTS
Native agarose gel electrophoresis and Western blot analysis.
As detailed in Materials and Methods, we expressed, purified, and characterized wild-type and mutant I97L capsid particles of HBcAg1-183, HBcAg1-149, and HBcAg1-140 from E. coli. As shown in the left panel of Fig. 1B, these particles encapsidated nucleic acids and thus can be stained by EtBr and later restained with protein-specific Coomassie blue (middle panel, Fig. 1B). To confirm the identity of the expressed and purified HBcAg, we performed immunoblot analysis with a rabbit polyclonal anti-core antibody (27). Both monomeric and dimeric HBcAg1-149 can be detected by immunological cross-reactivity using Western blot analysis (right panel, Fig. 1B). Similar results were obtained when using the HBcAg1-140 and HBcAg1-183 capsid particles (data not shown).
Full-length versus truncated core particles under EM.
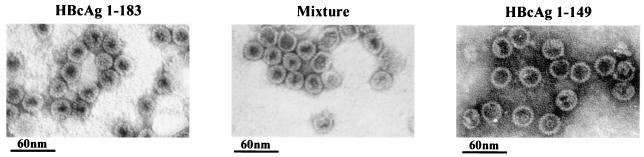
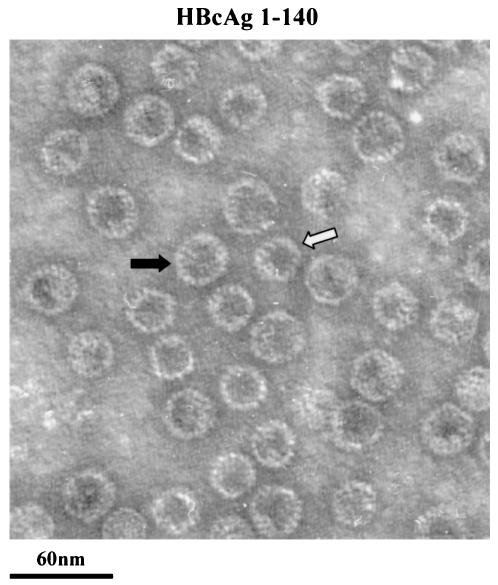
As shown by negative staining and EM in Fig. 2, HBcAg1-149 particles exhibit a “thin-wall” appearance with a much larger intraparticle space, whereas the HBcAg1-183 particles exhibit a “thick-wall” appearance with a relatively smaller intraparticle space. When these two different kinds of purified particles were mixed for the purpose of a closer comparison under EM, HBcAg1-149 particles seem to be very similar or slightly larger in size compared to the HBcAg1-183 particles (Fig. 2). Our results confirmed the previous report that truncated core particles (subtype adw) have a somewhat larger diameter (30 nm) than the full-length particles (28 nm) (9).
FIG. 2.
Electron micrographs of full-length (HBcAg1-183, left panel) and truncated (HBcAg1-149, right panel) HBcAg capsids. Capsids were negatively stained with 2% uranyl acetate and photographed by using a Philips EM201 electron microscope. The middle panel is a 1:1 mixture of full-length and truncated HBcAg capsids. HBcAg1-183 has a thick-walled appearance, whereas the HBcAg1-149 has a thin-walled appearance with more interior space.
Effect of temperature.
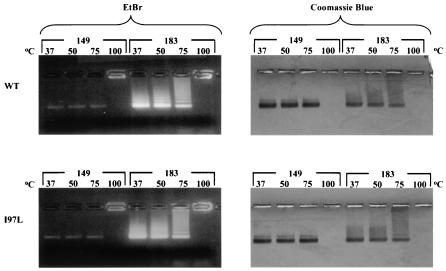
As shown in Fig. 3, these capsid particles were subjected to treatment at various temperatures before native agarose gel electrophoresis. The left panels were stained with EtBr to detect encapsidated nucleic acids, whereas the right panels are the same gels stained subsequently with Coomassie blue. The results shown here indicates that wild-type and mutant capsid particles are more or less equally stable when treated for 15 min at 37, 50, and 75°C. On the other hand, both samples treated at 100°C appeared to have no signals detectable by Coomassie blue staining, whereas EtBr staining revealed that the nucleic acids were stuck in the loading wells and did not enter the gels (Fig. 3). Consistent with earlier reports (2), we noted that the full-length HBcAg1-183 particles exhibited much more intense staining with EtBr than the truncated HBcAg1-149 particles, despite their similar intensities with Coomassie blue staining. We attributed this difference to the arginine-rich domain present in full-length core protein (Fig. 1A), which binds and efficiently packages nucleic acids. These results also suggest that the arginine-rich C terminus of HBcAg1-183, as well as the associated nucleic acids, does not contribute significantly to the heat resistance or sensitivity of capsid particles.
FIG. 3.
Similar stabilities of HBV capsids were observed at various temperatures irrespective of the capsid origins (wild type [top panels] or mutant [lower panels]) or size (full-length, HBcAg1-183 or truncated HBcAg1-149). A total of 20 μg of capsid protein was incubated for 15 min at the indicated temperatures before electrophoresis in a 1% agarose gel containing 0.5 μg of EtBr/ml (left). The same gels were subsequently stained with Coomassie blue (right).
Effect of pH.
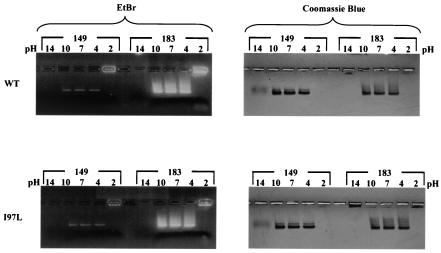
We next tested the capsid stability of wild-type and mutant I97L over a broad pH range. As shown in Fig. 4, both wild-type and mutant I97L capsid particles, in either HBcAg1-149 or HBcAg1-183 context, are equally stable when treated at 37°C for 30 to 60 min at pH 4, 7, and 10, and all particles dissociated at an extreme pH of 2 or 14. Dissociation of capsid particles is operationally defined as the appearance of residual fuzzy banding with decreased intensity or complete loss of staining signals. In other experiments, we observed the dissociation of capsid particles at pH 13 (data not shown). Similar results were obtained when experiments were conducted at room temperature overnight (data not shown).
FIG. 4.
Effects of pH on capsid stability are apparent at pH 2 and pH 14. Portions (20 μg) of E. coli-expressed capsid preparations were incubated in TBS buffers at various pH’s for 30 min at 37°C before electrophoresis in a 1% agarose gel. The upper panels show the results of EtBr (left panel) and Coomassie blue (right panel) staining of the wild-type capsid preparations containing full-length (HBcAg1-183) or truncated (HBcAg1-149) core proteins. The results of the pH challenges with the mutant I97L capsids are shown in the lower panels.
Effect of ionic detergent.
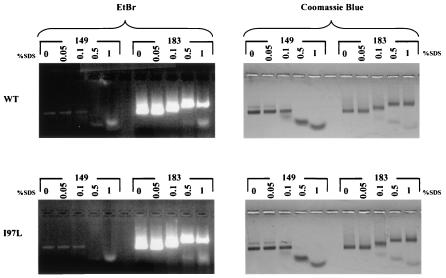
In addition to pH and temperature, we also tested capsid stability in the presence of SDS, an ionic detergent. As shown in Fig. 5, both wild-type and mutant I97L capsid particles, in either an HBcAg1-149 or an HBcAg1-183 context, were equally stable in SDS at concentrations up to 0.1%. In other experiments, we observed capsid stability at a 0.1% SDS concentration (data not shown). Minor variations in SDS resistance seem to be related to differences in capsid sample preparations. Most strikingly, HBcAg1-149 exhibited a downshift pattern when denatured by SDS, whereas the majority of full-length HBcAg1-183 exhibited an unexpected upshift, in addition to a very faint downshift pattern.
FIG. 5.
Stability of E. coli-expressed capsids in the presence of SDS. SDS was added to 20-μg portions of capsid preparations to reach the indicated concentrations. The mixtures were immediately run on a 1% agarose gel containing 0.5 μg of EtBr/ml (left panels) and stained with Coomassie blue (right panels). Wild-type (top panels) and mutant I97L (bottom panels) core proteins, either truncated (HBcAg1-149) or full length (HBcAg1-183), were tested. Note that the truncated capsid particles exhibit a downshift pattern at 0.5 and 1% SDS, whereas the full-length HBcAg1-183 particles exhibit an upshift.
SDS-treated particles under EM.
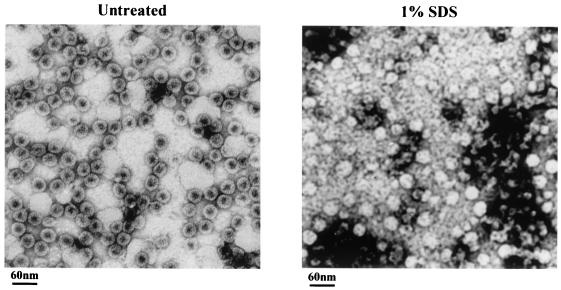
Under an electron microscope, SDS-treated HBcAg1-149 particles exhibited random aggregates rather than any structured entity (data not shown). In contrast, the SDS-treated full-length HBcAg1-183 capsid particles remained largely spherical in shape (Fig. 6), a result consistent with the previous report that full-length particles cannot be dissociated by SDS without reduction (9). A closer examination revealed that these particles were no longer well stained by uranyl acetate, indicating altered intraparticle capsid structure. The upshift gel pattern does not appear to be caused by interparticle cross-linking since we observed no apparent clumping of SDS-treated capsid particles in Fig. 6.
FIG. 6.
Electron micrographs of full-length HBcAg1-183 particles with or without SDS treatment. Capsids consisting of wild-type full-length core proteins were negatively stained with 2% uranyl acetate and photographed by using a Philips EM201 electron microscope. The left panel shows typical full-length HBcAg capsids that were not exposed to SDS, and the right panel is from full-length capsid samples treated with 1% SDS. No structured entity can be found in SDS-treated HBcAg1-149 particles (data not shown).
Effect of trypsin digestion.
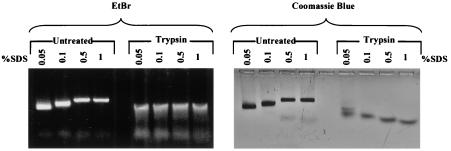
Intrigued by the upshift pattern of the full-length capsid particles with SDS denaturation, we sought to determine whether the carboxy terminus could be responsible for this phenomenon. Previously, Seifer and Standring demonstrated that trypsin can cleave around amino acid 150 of HBcAg, which is within the hinge region connecting the arginine-rich carboxy-terminal domain and the N-terminal assembly domain of HBcAg1-183 (Fig. 1A) (23, 24). We exposed capsid particles of wild-type HBcAg1-183 to trypsin digestion at 37°C for 30 min before the addition of SDS prior to agarose gel electrophoresis. We confirmed a reduction of the molecular weight from a full-length 23-kDa core protein to a 19-kDa protein comigrating with HBcAg1-149 (data not shown). As shown in Fig. 7, the trypsin digestion of the HBcAg1-183 capsid particles abolished the upshift pattern induced by SDS treatment. This result strongly suggests that the C terminus of the full-length HBcAg1-183 is indeed responsible for the upshift pattern. Mutant I97L and wild-type HBcAg1-183 particles are equally sensitive to trypsin cleavage (data not shown).
FIG. 7.
Trypsin predigestion of HBV full-length capsids eliminated the upshift pattern induced by SDS treatment. Portions (20 μg) of capsids consisting of full-length (HBcAg1-183) core proteins were incubated with 25 U of trypsin at 37°C for 30 min. SDS was then added to full-length capsid preparations, with or without trypsin predigestion, to reach the desired concentrations as indicated. They were then run on a 1% agarose gel containing 0.5 μg of EtBr/ml (left panel) and stained with Coomassie blue (right panel). Note that in the left panel, trypsin-digested samples exhibited a slight upshift banding pattern. Further characterizations of this slight upshift banding are shown in Fig. 9.
Upshift versus downshift patterns with SDS treatment.
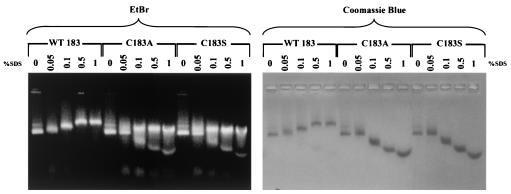
Although the trypsin digestion experiment (Fig. 7) confirmed the role played by the C-terminal polypeptide (amino acids 150 to 183), it was still unclear why the SDS treatment would induce a downshift pattern of the HBcAg1-149 but an upshift pattern for HBcAg1-183 (Fig. 5). Since the C-terminal domain of HBcAg is known to be rich in arginines, a basic amino acid (Fig. 1A), it is possible that SDS treatment significantly altered the charge, mass, and size ratio, which in turn contributed to an altered electrophoretic mobility on the agarose gel. We speculated that the carboxy-terminal cysteine could play a role by forming a disulfide bridge with another cysteine residue from neighboring dimers (2, 9, 37). Indeed, when the SDS treatment was performed in the presence of a reducing agent (β-mercaptoethanol), the upshift pattern was abolished when visualized by Coomassie blue staining (data not shown). To further test this hypothesis, we created C183A and C183S mutant capsid particles, changing the carboxy-terminal cysteine to alanine or serine, respectively. As shown in the right panel of Fig. 8, instead of exhibiting an upshift pattern, both mutant C183A and C183S exhibited the same downshift pattern as the truncated HBcAg1-149 particles upon SDS treatment and Coomassie blue staining.
FIG. 8.
Loss of the SDS-induced upshift pattern of E. coli-expressed C183A and C183S mutant capsids. Portions (20 μg) of capsid preparations of wild-type and mutant C183A or C183S were incubated with the indicated concentrations of SDS. The mixtures were immediately run on a 1% agarose gel containing 0.5 μg of EtBr/ml (left panels) and stained with Coomassie blue (right panels). Note that in the right panel the mutant capsid particles exhibit a downshift pattern at 0.1, 0.5, and 1% SDS, whereas wild-type 183 (WT 183) exhibits an upshift. In the left panel, in addition to the downshift banding pattern, there are strong EtBr signals slightly upshifted after 0.1% SDS in samples C183A and C183S. Unlike the continuous upshift pattern in wild-type HBV, the upshift banding pattern in mutants C183A and C183S did not continue to upshift from 0.1% to 1% SDS. The very low-molecular-weight faint signals in the left panel, stained by EtBr in samples treated with 0.05 to 1% SDS, are likely to be small RNA species of E. coli origin.
A mysterious band induced by SDS treatment can be stained by EtBr but not by protein stains.
As shown in the left panels of Fig. 7 and 8, the EtBr staining pattern of full-length particles after SDS treatment is more complicated than with Coomassie blue. In addition to the upshift or downshift banding, as revealed by Coomassie blue staining, there is another band that does not downshift and only upshifts slightly when SDS concentrations are >0.1%. It appears that this band does not contain any protein since neither Coomassie blue nor the more sensitive SYPRO Ruby (data not shown) can detect this mysterious band. Most likely, the band represents nucleic acids packaged within capsid particles that were released upon SDS treatment. Indeed, when Southern blot analysis was performed after the SDS treatment, HBV specific signals that coincide with this mysterious band were detected (Fig. 9). Based on RNase sensitivity (data not shown), we concluded that the packaged nucleic acid is largely HBV-specific RNA, which is consistent with the existing literature (2). Of note, some lower-molecular-weight signals, which can be detected by EtBr staining but not by Coomassie blue staining, were also observed in samples treated with 0.05 and 0.1% SDS in Fig. 7 to 9. Since these low-molecular-weight signals are not HBV-specific in Fig. 9, they are likely to be of E. coli origin, such as tRNAs, which were copackaged during the process of capsid self-assembly (20).
FIG. 9.
Identification of HBV-specific nucleic acids released from mutant C183S capsid particles after SDS treatment. SDS was added to 20 μg of HBcAg C183S capsid preparations to reach the indicated concentrations. The mixtures were immediately run on a 1% agarose gel containing 0.5 μg of EtBr/ml (left panel) and blotted onto nitrocellulose, and the gel was subsequently stained with Coomassie blue (right panel). The center panel is a Southern blot with an HBV adr probe which demonstrated that the packaged nucleic acids are of HBV origin. The faint lower-molecular-weight signals in the left panel, stained by EtBr in samples treated with 0.05, 0.1, and 0.5% SDS, are likely to be small RNA species of E. coli origin.
Proportions of T=3 versus T=4 capsid particles.
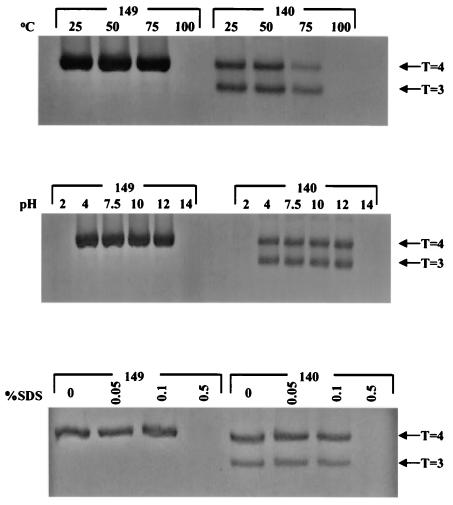
We examined the HBcAg1-140 particles under EM and observed two different-sized particles (Fig. 10) (7, 31). The relative proportion between the smaller capsids with a triangulation number of T=3 and the larger capsids with T=4 is known to be influenced by a linker peptide of HBcAg at amino acids 141 to 149 (29, 38). To date, it remains unknown whether other parts of the HBcAg molecule, such as amino acid 97, might influence the relative proportions between the two. To determine whether the mutation I97L could alter the ratio between T=3 and T=4 particles, we compared wild-type HBV and mutant I97L in the context of HBcAg1-140. We used HBcAg1-140 because the T=3 population constitutes only a very minor fraction (<5%) of HBcAg1-183 and HBcAg1-149 particles. The proportion of T=3 particles increased to ca. 78% and T=4 particles decreased to ca. 20.3%, when the morphogenic linker peptide from amino acids 141 to 149 is deleted (data not shown) (29, 38). Our results indicated that the ratio between putative T=3 particles (average diameter, 25.5 nm) and T=4 particles (average diameter, 28.7 nm) is not significantly different between wild-type HBV and mutant I97L in the context of HBcAg1-140 (Fig. 10 and data not shown).
FIG. 10.
Electron micrographs of mutant I97L HBcAg1-140 capsid particles. The proportion of T=3 particles increased to ca. 78% and T=4 particles decreased to ca. 20.3%, when the morphogenic linker peptide 141-149 was deleted (data not shown). Capsids were negatively stained with 2% uranyl acetate and photographed by using a Philips EM201 electron microscope. White arrow, T=3; black arrow, T=4. Similar results were obtained with wild-type HBcAg1-140 particles (data not shown).
Comparison of stability of T=3 versus T=4 capsid particles separated by using a novel method.
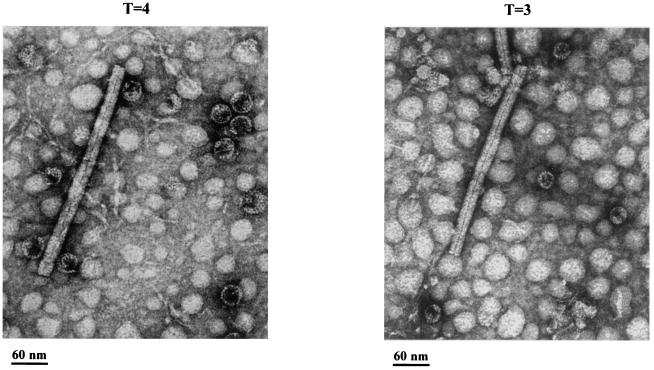
It remains unknown if the two different-sized particles have any difference in biological activity or structural stability (7, 31, 38). To compare the capsid stability between T=3 and T=4 particles, we challenged HBcAg1-140 particles with various pHs, temperatures, and SDS concentrations before subjecting them to 2.6% GTG low-melting-point agarose gel electrophoresis. As shown in Fig. 11, putative T=3 and T=4 particles of HBcAg1-140 are well separated and are almost equally stable in all conditions. The upper and lower bands in Fig. 11 were excised and electroeluted into separate dialysis tubes. The recovered particles were then stained with 2% uranyl acetate, and the size identity of the upper and lower bands were confirmed by EM (Fig. 12). We also noted that HBcAg1-140 particles may be less stable than HBcAg1-149 particles at 75°C, suggesting that the morphogenic linker peptide (HBcAg amino acids 141 to 149) could contribute to increased thermal stability of HBcAg1-149 capsids. We also compared the stability of T=3 HBcAg1-140 capsid particles from wild-type and mutant I97L origins and found no difference in stability between the two (data not shown).
FIG. 11.
No significant difference in stabilities between T=3 and T=4 wild-type HBcAg1-140 capsid particles separated by GTG low-melting-point agarose gel electrophoresis. Portions (20 μg) of truncated wild-type HBcAg1-149 or HBcAg1-140 particles were treated with different SDS concentrations, pHs, and temperatures. Similar results were obtained with mutant I97L HBcAg1-140 particles (data not shown). Note that, at 75°C, HBcAg1-149 particles seemed to be more stable than HBcAg1-140.
FIG. 12.
Electron micrographs of HBcAg1-140 particles prepared by electroelution from the upper band (A) and the lower band (B) on the GTG gel in Fig. 11. The average diameter of the capsid particles in panel A is ∼27 nm (T=4), whereas in panel B it is ∼25 nm (T=3) (38). The rod-like particles are tobacco mosaic virions and were included as an internal standard. The high background in these micrographs is in part due to the impurities coeluted from the GTG gel.
DISCUSSION
To elucidate the structural basis of a so-called immature secretion phenomenon, we have expressed, purified, and characterized both truncated and full-length HBV capsid particles with or without an I97L mutation (Fig. 1 and 2). Consistent with previous findings (31), HBV capsid particles exhibited a rather robust resistance to treatments with temperature (Fig. 3), pH (Fig. 4), and nonionic detergents (data not shown). Although treatment with an ionic detergent, such as SDS, can differentiate between full-length and truncated particles, it cannot distinguish between wild-type and mutant I97L capsid particles (Fig. 5). The SDS-induced upshift gel pattern of full-length particles is mainly caused by the interdimer disulfide bridge mediated by the carboxy-terminal cysteines (Fig. 7 and 8 and data not shown). The encapsidated HBV specific RNA is dissociated from the protein moiety of full-length particles after SDS treatment (Fig. 9 and data not shown). Interestingly, despite the differences in size and architecture of T=3 and T=4 icosahedral particles (Fig. 10) (7, 31, 38), we found no apparent difference in their capsid stabilities (Fig. 11 and 12). To our knowledge, this is probably the first report that compares directly the stability between T=3 and T=4 icosahedral particles of human HBV. Further discussions on these results are detailed below.
Capsid stability and pH treatment.
A previous report indicated that truncated core particles (HBcAg1-144 and HBcAg1-149) are less stable than full-length particles (HBcAg1-183) at pH 10 and 12 by using sucrose gradient centrifugation analysis (2). In contrast, we found truncated particles to be stable at pH 10 (Fig. 3). The discrepancy could be caused by a number of possibilities. (i) The ayw subtype used previously compared to the adr subtype used here differs at eight amino acids in HBcAg (36). (ii) Differences in the assay procedures, such as durations and temperatures of pH treatments, or the use of a two-step sucrose gradient centrifugation (2) versus the native agarose gel electrophoresis used here could also have caused the discrepancy. It is well known that HBcAg, upon denaturation, can be converted into HBeAg (19). Previously, the presence or absence of HBc- or HBe-specific epitopes was used as an assay for particle integrity (2, 9). It is quite possible that measuring the changes of immunological reactivity from HBcAg to HBeAg or hydrodynamic properties by gradient centrifugation is a more sensitive assay for conformational changes, whereas measuring the electrophoretic mobility of particles by agarose gel electrophoresis and morphological examinations under EM could be a more direct way to measure particle integrity.
Cys-183 and particle stability.
HBcAg contains four evolutionarily conserved cysteine residues at amino acids 48, 61, 107, and 183. Previous reports proposed that Cys-183 is important in forming interdimer disulfide bridges (2, 9, 37). Presumably, the increased stability caused by the Cys-183/Cys-183 bonding is responsible for the unexpected upshift pattern in the presence of SDS. As shown in Fig. 6, in the absence of reducing agents or trypsin digestion, the full-length SDS-treated particles retained a spherical shape, despite the fact that the uranyl acetate staining showed little interior space. In contrast, there is no structured entity in SDS-treated truncated particle preparations under EM (data not shown). The carboxy-terminal cysteine appears to be responsible for both the resistance to dissociation by SDS treatment under EM (Fig. 6 and data not shown) and the upshifted gel pattern (Fig. 5). Characterizations of mutant C183A and C183S particles provided direct proof of the importance of the carboxy-terminal cysteine residue and its involvement in the interdimer disulfide bridges important for particle integrity (Fig. 8) (2, 9, 37).
Encapsidated HBV RNA of full-length particles is released after SDS treatment.
The carboxy terminus of full-length HBcAg1-183 is rich in arginine residues that can bind to DNA and RNA (Fig. 1) (1, 2, 9, 12). Indeed, truncated particles (HBcAg1-149) without this domain encapsidated much less nucleic acid than did the full-length particles, as shown by their significant differences in EtBr staining (Fig. 5). This would explain why the mysterious band detected with EtBr staining was only found when the full-length particles were treated with SDS (Fig. 7 to 9) but not when truncated particles were treated in a similar manner. Apparently, the released nucleic acids include HBV-specific RNA (Fig. 9 and data not shown) which was dissociated from the capsid protein upon SDS treatment. Using spectrophotometric measurement, Zlotnick et al. estimated the stoichiometry of encapsidated RNA and E. coli-derived capsid particles to be near a total of 3,000 ribonucleotides per full-length capsid particle (95% T=4) (39). It would be interesting to quantitate the stoichiometry between the capsid protein and encapsidated RNA in our E. coli system in the near future.
T=3 versus T=4 particles.
In capsids isolated from human liver, the T=4 form was found to be in excess to the T=3 form by about 13 to 1 (15). In laboratory settings, it is also known that HBcAg expressed in E. coli can assemble into two different sizes of particles (7, 31, 38). The relative proportions between T=3 and T=4 particles is known to be influenced by a cysteine-to-alanine mutation at amino acid 61 or by the length of the linker peptide (amino acids 141 to 149) of HBcAg (29, 38). To date, it remains unclear whether other parts of the HBcAg molecule, such as amino acid 97, could influence the relative proportions between the two. Our studies using EM and GTG gel electrophoresis demonstrated that the mutation I97L has no apparent effect on the T=4/T=3 ratio (data not shown; Fig. 10 and Fig. 11). Our study also demonstrated that T=3 and T=4 particles have similar capsid stabilities (Fig. 11). Furthermore, we noted that the linker peptide (amino acids 141 to 149) appeared to contribute to capsid stability at 75°C (Fig. 11, upper panel). Whether these two different-sized particles have the same kinetics or genome maturation during virion secretion remains to be investigated in the future.
Structural basis of immature virion secretion.
Although Ile-97 or Leu-97 is on the α4b helix (Fig. 1A), it is not part of the conserved hydrophobic core that is supposed to be important for the stability of the monomer fold (32). The disulfide bridge formation of the capsid dimer is known to be facilitated by Cys-61 resident on the α3 helix (18, 37). However, amino acid 97 is located on a different helix (α4b), and it remains unclear whether amino acid 97 could affect the stability of the dimer subunit. In addition to the stability of the monomer fold and the dimer subunit, stability of capsid particles could depend on the strength of the interdimer contact (amino acids 120 to 143). The proline-rich loop of HBcAg128-136 is known to be involved in the interdimer interactions. Since amino acid 97 is at least 20 to 30 Å away from this proline-rich loop, it seems unlikely that amino acid 97 can affect capsid stability (32). In summary, on a theoretical ground, immature secretion of mutant I97L does not appear to be caused by either capsid stability or instability. The results from our experiments here lend strong support to that prediction. More likely, immature secretion could be caused by aberrant core-envelope interactions, which lead to the incorrect display of a “genome maturation signal” of mutant I97L capsids prior to envelopment. In this regard, however, it is worth mentioning that the time course study of virion secretion revealed that immature and mature virions of mutant I97L were secreted by similar kinetics (16).
Virion secretion of wild-type hepadnaviruses is a tightly regulated event (11, 21, 28, 30). A genome maturation signal from the capsids must somehow be recognized by the envelopment machinery. A previous study has suggested that dephosphorylation of HBcAg may play a role in signaling genome maturation (22). Since amino acid 97 of HBcAg is never a serine or threonine, and its position on the three-dimensional structure of the monomer fold is far away from the known phosphorylation sites near the C terminus, there is currently no evidence to support the possibility that amino acid 97 could affect dephosphorylation of HBcAg. Further investigation will be needed to elucidate this issue.
One limitation in our current assay system, based on the self-assembled HBcAg in E. coli, is the lack of involvement of HBV polymerase, putative host factors, and a bona fide viral genome. Such a deficiency precludes certain experiments, such as studying the “breathing” or dynamics of nucleocapsids during viral DNA synthesis and intracellular trafficking. Nevertheless, this E. coli-based system may still be useful for future studies, such as comparing the kinetics of capsid assembly between wild-type and mutant viruses (26). To compare the stability and size of capsid particles isolated from mammalian cells, we are in the process of circumventing the problem of detection sensitivity by first establishing stable cell lines that can produce a large quantity of the mutant 97 capsid particles in tissue culture.
Acknowledgments
This study was funded by NIH grants R01 CA 70336 and CA 84217 to C.S. F.-M.S. was supported in part by Taipei Medical University Hospital and Juei-Low Sung's Research Foundation of Taiwan.
We thank colleagues in C.S.'s laboratory for careful reading of the manuscript. We also thank Stan Watowich for advice on using the Swiss PDB Viewer Program, Vsevolod Popov for advice on EM, and Philip Sewer for advice on the use of GTG agarose gels. The arginine tRNA plasmid was a kind gift from Hiroshi Matsuzawa. Tobacco mosaic virus particles were generously provided by Gerald Stubbs.
REFERENCES
- 1.Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology 194:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 64:3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottcher, B., S. A. Wynne, and R. A. Crowther. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88-91. [DOI] [PubMed] [Google Scholar]
- 4.Chua, P. K., Y. M. Wen, and C. Shih. 2003. Coexistence of two distinct secretion mutations (P5T and I97L) in hepatitis B virus core produces a wild-type pattern of secretion. J. Virol. 77:7673-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, B. J., and J. E. Richmond. 1982. Electron microscopy of hepatitis B core antigen synthesized in Escherichia coli. Nature 296:677-679. [DOI] [PubMed] [Google Scholar]
- 6.Conway, J. F., N. Cheng, A. Zlotnick, P. T. Wingfield, S. J. Stahl, and A. C. Steven. 1997. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386:91-94. [DOI] [PubMed] [Google Scholar]
- 7.Crowther, R. A., N. A. Kiselev, B. Bottcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 8.Ehata, T., M. Omata, O. Yokosuka, K. Hosoda, and M. Ohto. 1992. Variations in codons 84-101 in the core nucleotide sequence correlate with hepatocellular injury in chronic hepatitis B virus infection. J. Clin. Investig. 89:332-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallina, A., F. Bonelli, L. Zentilin, G. Rindi, M. Muttini, and G. Milanesi. 1989. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol. 63:4645-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2970. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 11.Gerelsaikhan, T., J. E. Tavis, and V. Bruss. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 70:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 66:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamura, H., B. Jeon, T. Wakagi, and H. Matsuzawa. 1999. High level expression of Thermococcus litoralis 4-alpha-glucanotransferase in a soluble form in Escherichia coli with a novel expression system involving minor arginine tRNAs and GroELS. FEBS Lett. 457:393-396. [DOI] [PubMed] [Google Scholar]
- 14.Jeng, K. S., C. P. Hu, and C. M. Chang. 1991. Differential formation of disulfide linkages in the core antigen of extracellular and intracellular hepatitis B virus core particles. J. Virol. 65:3924-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 16.Le Pogam, S., and C. Shih. 2002. Influence of a putative intermolecular interaction between core and the pre-S1 domain of the large envelope protein on hepatitis B virus secretion. J. Virol. 76:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mimms, L., J. Staller, I. K. Mushahwar, K. S. Spiezia, A. Kapsalis, and P. Anderson. 1988. Production, purification, and immunological characterization of a recombinant DNA-derived hepatitis B e antigen, p. 248-251. In A. J. Zuckerman (ed.), Viral hepatitis and liver diseases. Alan R. Liss, Inc., New York, N.Y.
- 18.Nassal, M. 1992. Conserved cysteines of the hepatitis B virus core protein are not required for assembly of replication-competent core particles nor for their envelopment. Virology 190:499-505. [DOI] [PubMed] [Google Scholar]
- 19.Ou, J. H. 1997. Molecular biology of hepatitis B virus e antigen. J. Gastroenterol. Hepatol. 12:S178-S187. [DOI] [PubMed] [Google Scholar]
- 20.Pasek, M., T. Goto, W. Gilbert, B. Zink, H. Schaller, P. MacKay, G. Leadbetter, and K. Murray. 1979. Hepatitis B virus genes and their expression in Escherichia coli. Nature 282:575-579. [DOI] [PubMed] [Google Scholar]
- 21.Perlman, D., and J. Hu. 2003. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J. Virol. 77:2287-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugh, J., A. Zweidler, and J. Summers. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seifer, M., and D. N. Standring. 1995. Assembly and antigenicity of hepatitis B virus core particles. Intervirology 38:47-62. [DOI] [PubMed]
- 24.Seifer, M., and D. N. Standring. 1994. A protease-sensitive hinge linking the two domains of the hepatitis B virus core protein is exposed on the viral capsid surface. J. Virol. 68:5548-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih, C. 2003. Functional significance of naturally occurring hepatitis B virus variants, p. 23-41. In S. Locarnini and C. L. Lai (ed.), Human virus guides-human hepatitis B viruses. International Medical Press, London, United Kingdom.
- 26.Singh, S., and A. Zlotnick. 2003. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 278:18249-18255. [DOI] [PubMed] [Google Scholar]
- 27.Suk, F. M., M. H. Lin, M. Newman, S. Pan, S. H. Chen, J. D. Liu, and C. Shih. 2002. Replication advantage and host factor-independent phenotypes attributable to a common naturally occurring capsid mutation (I97L) in human hepatitis B virus. J. Virol. 76:12069-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 29.Watts, N. R., J. F. Conway, N. Cheng, S. J. Stahl, D. M. Belnap, A. C. Steven, and P. T. Wingfield. 2002. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 21:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei, Y., J. E. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wingfield, P. T., S. J. Stahl, R. W. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 32.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 33.Yuan, T. T., M. H. Lin, S. M. Qiu, and C. Shih. 1998. Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation. J. Virol. 72:2168-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan, T. T., G. K. Sahu, W. E. Whitehead, R. Greenberg, and C. Shih. 1999. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J. Virol. 73:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan, T. T., and C. Shih. 2000. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L). J. Virol. 74:4929-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan, T. T., P. C. Tai, and C. Shih. 1999. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J. Virol. 73:10122-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng, J., F. Schodel, and D. L. Peterson. 1992. The structure of hepadnaviral core antigens: identification of free thiols and determination of the disulfide bonding pattern. J. Biol. Chem. 267:9422-9429. [PubMed] [Google Scholar]
- 38.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]
- 39.Zlotnick, A., N. Cheng, S. J. Stahl, J. F. Conway, A. C. Steven,, and P. T. Wingfield. 1997. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. USA 94:9556-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]