Abstract
Rhesus monkey rhadinovirus (RRV) is one of the closest phylogenetic relatives to the human pathogen Kaposi's sarcoma-associated herpesvirus (KSHV), yet it has the distinct experimental advantage of entering efficiently into lytic replication and growing to high titers in culture. RRV therefore holds promise as a potentially attractive model with which to study gammaherpesvirus structure and assembly. We have isolated RRV capsids, determined their molecular composition, and identified the genes encoding five of the main capsid structural proteins. Our data indicate that, as with other herpesviruses, lytic infection with RRV leads to the synthesis of three distinct intranuclear capsid species. However, in contrast to the inefficiency of KSHV maturation following reactivation from latently infected B-cell lines (K. Nealon, W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes, J. Virol. 75:2866-2878, 2001), de novo infection of immortalized rhesus fibroblasts with RRV results in the release of high levels of infectious virions with genome-containing C capsids at their center. Together, our findings argue for the use of RRV as a powerful model with which to study the structure and assembly of gammaherpesviruses and, specifically, the human rhadinovirus,KSHV.
Herpesvirus infection persists for the life of the host. In an immunocompromised state, gammaherpesviruses, one of the three subtypes of herpesviruses, can lead to severe disease, including neoplasia (3, 7, 9, 20, 36, 37, 48). The most recently identified human gammaherpesvirus is the rhadinovirus Kaposi's sarcoma-associated herpesvirus (KSHV) (12), the etiologic agent of Kaposi's sarcoma, a human vascular tumor especially frequent in AIDS patients (5, 8, 9, 21, 29, 35). KSHV is also associated with two lymphoproliferative diseases, primary effusion lymphoma and multicentric Castleman's disease (7, 10, 19, 51).
Rhesus monkey rhadinovirus (RRV) is closely related to KSHV (15). The sequences, as well as gene content and overall organization, are well conserved between the two viruses. Furthermore, RRV can lead to the development of a multicentric lymphoproliferative disorder resembling multicentric Castleman's disease in macaques coinfected with simian immunodeficiency virus (34, 58). In contrast to KSHV, however, RRV grows lytically to high titers following de novo infection of immortalized rhesus fibroblasts (15-17, 34, 58).
For all herpesviruses, including both human and rhesus rhadinoviruses, virion formation and release are critical to horizontal spread within the host and transmission within a population. One of the earliest steps in virion formation is assembly of the icosahedral capsid in the nucleus of an infected cell. Release of fully formed virions follows only after such capsids acquire a single copy of the linear viral genome and then an outer proteinaceous tegument layer that is subsequently surrounded by a lipid envelope. The entire process takes approximately 9 h for alphaherpesviruses such as herpes simplex virus type 1 (HSV-1) (6, 14, 27) but up to 3 or more days for gammaherpesviruses such as KSHV (38). The relatively slow kinetics of virion formation and release is characteristic of gammaherpesviruses, including the only other known human member, Epstein-Barr virus, belonging to the Lymphocryptovirus genus (1, 26, 33, 44, 50).
A mature infectious herpesvirus virion contains one copy of the double-stranded DNA viral genome within its capsid. Each capsid is an icosahedron with a triangulation number of 16 and consists of 162 capsomers characteristic of all members of the Herpesviridae family (25). Of the 162 capsomers, 12 are pentons, while the remaining 150 are hexons, composed of five and six subunits of the major capsid protein (MCP), respectively (24, 38, 40, 52). In addition, the capsids have 320 triplex subunits, each consisting of a heterotrimer with α1β2 stoichiometry. During lytic replication of herpesviruses, multiple capsid species arise (22, 24, 25, 38, 52). These include the three major capsid species: (i) A capsids lacking internal structures, (ii) B capsids containing an inner scaffolding protein (SCAF), and (iii) C capsids enclosing the viral genome but lacking SCAF.
Investigation of the structure and assembly of gammaherpesvirus capsids not only will allow a better understanding of specific aspects of the early steps in virion production that may distinguish this subfamily of herpesviruses but also may provide potential insights into future drug targets for human-specific viruses such as KSHV. However, the goal of developing a more detailed picture of human gammaherpesvirus capsid and virion structure and assembly has met with considerable difficulty due to the low yields of virus in in vitro culture systems or the propensity to enter a state of latency after infection (28, 30, 38, 43). In contrast, RRV displays robust lytic-phase growth and high viral titers and thus holds great promise as an attractive model with which to help characterize the formation of its human rhadinovirus counterpart, KSHV, as well as gammaherpesviruses in general.
Capsid and viral isolation.
Herpesvirus capsids assemble in the nuclei of infected cells. To determine if lytic replication of RRV gives rise to different capsid species, we infected telomerase-immortalized rhesus macaque fibroblasts (RhF) for 2 h with crude preparations of RRV as described previously (2, 15, 16) and then monitored lytic replication by both electron microscopy and biochemical analysis. The goal was to generate sufficient numbers of capsids to allow their compositional and structural characterization. We harvested and fixed infected cells for thin-section transmission electron microscopy (TEM) and found that by 4 to 6 days after infection, three capsid species with distinct morphologies arose in the nuclei (Fig. 1A). We reasoned that lysis of infected cells at late stages of lytic replication of RRV would lead to the release of these capsids, as well as viral particles at various stages of maturation, mirroring our earlier findings with KSHV (38).
FIG. 1.
Maturation of RRV particles. (A) TEM of intranuclear RRV capsids 6 days postinfection. Three morphologically distinct capsid species are evident. The inset displays the dashed boxed area at a greater magnification with type A, B, and C capsids indicated. (B) Released RRV particles collected from the supernatant of infected RhF. Virions contain a capsid with an inner density surrounded by a tegument layer (t) and an envelope layer with protrusions consistent with glycoproteins (g). A capsid (c) lacking these two outer layers is also present in this micrograph.
To maximize the yield of viral and subviral particles, we next determined the kinetics of total particle release, collecting the medium at 24-h intervals and subjecting it to centrifugation through a sucrose cushion, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to visualize particle-associated protein bands. Maximum release of particles occurred 6 days postinfection (data not shown). On the basis of these findings, we collected RRV capsids and virions for all subsequent biochemical and imaging studies 6 days postinfection.
We first examined the morphology of released RRV particles to assess the proportion of mature enveloped virions. After partial purification by size exclusion column chromatography, the otherwise untreated population sedimented through a sucrose gradient as a single dominant band. Fractions from the gradient that corresponded to the fastest-sedimenting portion of this visible band contained a fairly homogeneous collection of enveloped virions, although capsids or maturing particles were also occasionally present (Fig. 1B). Of note in three separate experiments, nearly all (239 of 241) of the enveloped particles that we purified in this way and then examined by TEM possessed an inner density suggestive of encapsidated viral DNA (22, 25, 38, 42). The near absence of enveloped particles with empty capsids or no capsid at all (L particles), as can arise during HSV-1 replication (54), suggests that lytic replication of RRV is highly efficient in producing mature virions.
We next focused on characterizing the composition and structure of RRV capsids. Extrapolating from our work on KSHV, as well as studies of other herpesviruses (18, 38, 39, 42), we hypothesized that the three capsid species (A, B, and C) resulting from RRV lytic replication would each have a distinct density, allowing their separation by sedimentation through sucrose gradients (38). We used 2% Triton X-100 detergent to remove tegument and envelope layers from virions and partially wrapped capsids released during the late stages of lytic replication. This approach converts the complex population of particles to a more homogeneous collection of capsid species. It is possible, however, that a subset of tegument proteins, especially those tightly associated with the capsid surface, may remain, at least in part, when this protocol is used. Nevertheless, a similar approach led to moderately pure preparations of KSHV capsids (38, 57), arguing for the efficacy of this approach.
After such treatment, velocity sedimentation through sucrose gradients gave rise to three distinct, light-diffracting bands (Fig. 2) that we tentatively labeled as comprising A, B, and C capsids, respectively, on the basis of earlier studies of herpesvirus capsid purification (38, 42). We found that the C capsid band was consistently the most abundant, followed by B and then A capsid bands (relative intensity of gradient bands in Fig. 2). It is likely that de-enveloped virions were the major contributor to this C capsid predominance.
FIG. 2.

Separation of RRV capsids by velocity sedimentation. Purified capsids were separated by velocity sedimentation on a 20 to 50% sucrose gradient (see reference 38). The three light-scattering bands in the gradient are labeled A, B, and C capsids on the basis of a similar banding pattern previously observed for KSHV.
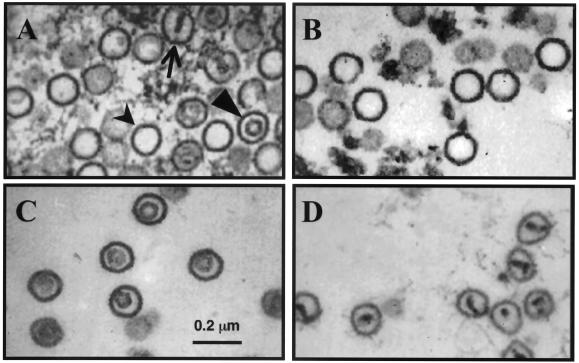
To confirm that the three light-scattering bands in the sucrose gradient represented intact capsid particles, we collected fractions by bottom puncture and subjected each to TEM. Prior to gradient separation, the detergent-treated particles comprised a mixture of capsid species with cross sections characteristic of A, B, and C herpesvirus capsid morphologies (Fig. 3A). In contrast, after velocity sedimentation, the fractions corresponding to the visible bands in the gradient demonstrated relatively homogeneous populations of RRV capsids, each with a distinct morphology. The capsids in band A appeared empty (Fig. 3B), the capsids in band B possessed an inner ring-like structure (Fig. 3C), and the capsids in band C demonstrated a single inner density, sometimes with thin and irregular spoke-like projections extending to the edges of the capsid's interior (Fig. 3D). These morphologies are consistent with the A, B, and C capsids comprising the three bands, respectively, in similarly purified capsids from other herpesviruses, including KSHV and HSV-1 (22, 38).
FIG. 3.
Thin-section TEM of RRV capsids. (A) Mixture of three morphologically distinct capsids prior to separation. A capsids (thick arrow) are empty icosahedral structures, B capsids (arrowhead) contain an inner ring-like structure likely composed of SCAF (see text), and C capsids (thin arrow) contain a dense core consistent with encapsidated viral DNA. Velocity sedimentation separated the mixed capsid population into distinct populations of A, B, and C capsids (B to D, respectively).
Protein characterization of RRV capsids.
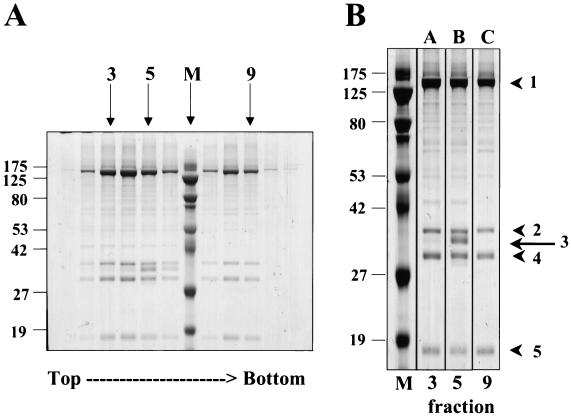
To determine the protein composition of the three capsid species, we next analyzed each fraction of the capsid gradient by SDS-PAGE (Fig. 4) and found protein banding patterns similar to those of KSHV (38) and other herpesviruses (22, 55). The peak intensities of these particle-associated proteins coincided with the fractions containing the A, B, and C capsid particles (Fig. 3), and the most prominent proteins migrated with apparent molecular masses of 134, 41, 34, and 16 kDa (Fig. 4, bands 1, 2, 4, and 5, respectively). Additionally, the fraction that appeared in TEM as an essentially pure population of type B capsids (Fig. 3C) contained a fifth protein migrating with an apparent molecular mass of 37 kDa (Fig. 4, band 3). This protein, unique to B capsids, was therefore a good candidate for the RRV scaffolding homolog (SCAF). Of note, the fractions containing the slowest- and fastest-sedimenting capsids (Fig. 2), respectively, had identical protein profiles (Fig. 4, lanes 3 and 9). This last result is consistent with our initial prediction that these two bands were composed of type A and C capsids, respectively (see above).
FIG. 4.
Protein composition of RRV capsids. (A) SDS-PAGE of capsid fractions from a 20 to 50% sucrose gradient (Fig. 2). Fractions 3, 5, and 9 contain mainly A, B, and C capsids, respectively (see the corresponding micrographs, Fig. 3B to D). Fraction 4 is a mixture of both A and B capsids. (B) Expanded view of fractions 3, 5, and 9. M, molecular mass standards (kilodaltons). The arrow and arrowheads to the right indicate the five capsid-associated proteins migrating with the following apparent molecular masses: 1, 134 kDa; 2, 41 kDa; 3, 37 kDa; 4, 34 kDa; 5, 16 kDa. The values on the left of each panel are molecular sizes in kilodaltons.
To confirm the identity of the five capsid-associated proteins (Fig. 4), we analyzed each by tandem mass spectrometry and then subjected the resultant tryptic peptides to the Sequest search algorithm (National Center for Biotechnology Information). Peptides that were not matched by this algorithm were interpreted individually and searched against the expressed sequence tag databases, also with the Sequest algorithm (38). Figure 5 displays graphically the results of these analyses. Band 1 gave rise to 53 partially overlapping peptides spanning 738 amino acids and the unambiguous assignment of this 150-kDa protein as RRV MCP, encoded by open reading frame 25 (ORF25). Similarly, analyses of the 39-kDa (Fig. 4, band 2) and 30-kDa (Fig. 4, band 4) proteins generated seven peptides spanning 78 amino acids and 12 peptides spanning 146 amino acids, respectively. These results identified bands 2 and 4 as the components of the triplex TRI-1 and TRI-2, encoded by ORF62 and ORF26, respectively. Tryptic digestion of the 17-kDa protein (Fig. 4, band 5) gave rise to five peptides spanning 55 amino acids, and sequence comparisons identified it as RRV SCIP, which is encoded by ORF65. Likewise, the 35-kDa protein (Fig. 4, band 3), present only in the B capsid fraction, generated six peptides spanning 70 amino acids with a sequence identifying it as RRV SCAF, encoded by the predicted ORF17.5 (11; M. Cruise, C. M. O'Connor, and D. H. Kedes, unpublished observations). All five RRV capsid proteins show sequence similarity to their KSHV homologs (Table 1). MCP shows the greatest sequence conservation (84% similarity) between the two viruses, and SCAF shows the least (49% similarity). The remaining capsid-associated proteins show significant homology between the two viruses, with similarities ranging between 56 and 81%.
FIG. 5.
Mass spectrometric analysis of protein bands 1 to 5 in Fig. 4B. Peptides identified by tryptic digestion (shaded) are shown within the sequence of the protein (black) they identified after screening of protein sequence databases. The protein bands are as follows: 1, MCP; 2, TRI-1; 3, SCAF; 4, TRI-2; 5, SCIP. Note that the translational start site and transcript of ORF17.5 (which encodes SCAF) is predicted from its homolog in KSHV and other herpesviruses (see text).
TABLE 1.
Comparison of protein components of RRV and KSHV capsids based on sequence homologya
| Gene | Protein | Predicted mass (kDa)
|
Identity (%) | Similarity (%) | |
|---|---|---|---|---|---|
| RRV | KSHV | ||||
| ORF25 | MCP | 153 | 153 | 71 | 84 |
| ORF62 | TRI-1 | 36 | 36 | 56 | 70 |
| ORF26 | TRI-2 | 34 | 34 | 65 | 81 |
| ORF17.5 | SCAF | 31 | 29 | 38 | 49 |
| ORF65 | SCIP | 17 | 19 | 40 | 56 |
Percent identity and percent similarity were determined by using BLAST2 Sequences (BLAST2.2.1) with a BLOSUM62 matrix (National Center for Biotechnology Information).
Distinguishing between A and C capsids.
Although TEM demonstrated morphologically distinct capsid species that migrated at different rates through sucrose gradients, corroborative biochemical identification of the three species requires analyses of not only their protein but also their viral DNA content. In contrast to B capsids that uniquely contain SCAF, aiding in their identification (Fig. 4, lane 5), A and C capsids demonstrate effectively identical protein profiles on SDS-PAGE (Fig. 4, lanes 3 and 9). To distinguish between the latter two species, we analyzed sequential fractions from the capsid gradients for relative amounts of encapsidated (DNase-resistant) RRV DNA with a fluorescently labeled DNA probe complementary to RRV ORF65 by Southern dot blot analyses. Results of this analyses are shown in Fig. 6A. The majority of the encapsidated DNA cosedimented with fractions containing the rapidly sedimenting capsid population, labeled C in Fig. 2. In contrast, gradient fractions containing the A and B capsids showed little RRV DNA signal over the background. Coupled with the protein analyses, these data confirm that the slowest-sedimenting, empty capsids (Fig. 3B) are A capsids and the fastest-sedimenting, dense-cored capsids (Fig. 3D) are C capsids.
FIG. 6.
RRV C capsids cosediment with encapsidated DNA and, in contrast to KSHV, represent the most abundant species that arise during lytic replication. (A) RRV capsids were separated by velocity sedimentation and relative amounts of encapsidated RRV-specific DNA (solid squares) in each fraction were measured by Southern dot blot analysis. The RRV DNA peaks in the fractions correlating with the most rapidly sedimenting core-filled capsid species (Fig. 3D). For direct comparison, the MCP profile (open diamonds) determined by densitometry (Fig. 4, band 1) from a second aliquot of each fraction is also shown. A, B, and C indicate the fractions containing the purest populations of A, B, and C capsids (determined by TEM and SDS-PAGE), respectively. (B) In similar experiments (38), KSHV-specific DNA (solid squares) also cosediments with the KSHV C capsids. However, with the human virus, C capsids are the least-abundant species. The peak in KSHV MCP (open diamonds) correlates, instead, with the A and B capsids.
The C capsids are the most abundant of the three RRV capsid species in our preparations (Fig. 2 and 6), representing approximately 60% of the total capsids produced by our isolation method 6 days after infection. This contrasts with our earlier findings with KSHV based on similar capsid preparations from 12-O-tetradecanoylphorbol-13-acetate-induced primary effusion lymphoma cells. During KSHV lytic replication, C capsids represent the least abundant (10 to 15%) of the three capsid species (Fig. 6) (38). Since only those virions containing intact viral genomes are potentially infectious, the number of C capsid-containing particles correlates with the relative infectivity of particles released from lytically infected cells. Thus, this finding may help explain the marked differences in the efficiency of lytic replication between KSHV and RRV.
To address this issue directly with RRV, for which a plaque assay on RhF exists (17), we measured the infectivity of gradient-purified RRV virions and then compared this titer (in PFU and PFU per milliliter) with the number of viral particles or encapsidated genomes per milliliter. We found that a typical 500-ml preparation gave a titer of 6.6 × 106 PFU/ml in unconcentrated medium 6 days after infection. (This represents a calculated titer after viral purification by size exclusion chromatography and velocity sedimentation through a sucrose gradient.) We then determined the concentration of RRV particles in two ways. In the first, we estimated the amount of MCP in each viral sample after its separation by SDS-PAGE and subsequent staining with SYPRO Orange (Bio-Rad). The intensity of staining with SYPRO Orange correlates with protein mass and is affected little by other protein properties. We compared the amount of MCP in each sample with known concentrations of bovine serum albumin electrophoresed and stained on the same gel by quantitative densitometry. Since each particle contains 960 copies of MCP, we were then able to approximate the number of virions in each sample. In the second method, we determined the concentration of encapsidated (DNase-resistant) RRV genomes within the purified virions with a plasmid containing RRV ORF65 as our standard and a dot blot Southern analysis with a probe to ORF65 as described above. These two methods gave surprisingly close estimates of 2.0 × 109 particles and 3.0 × 109 genomes per ml of unconcentrated medium, respectively, indicating that nearly all of the virions that we purified from medium 6 days after infection contained RRV DNA. Nevertheless, these data also suggest that the particle-to-infectivity ratio in the plaque assay on RhF is 300:1 to 450:1.
Therefore, although it appears that nearly all of the released virions contain genomes, the fraction that remains infectious after our purification is small. This may reflect an inherent inefficiency in the plaque assay, as well as potential damage to the viral particles that we may introduce during the purification process. Even so, the concentration of particles released into the medium (according to either calculation) by RRV-infected RhF exceeds that of induced KSHV-infected BCBL-1 cells by approximately 30-fold (data not shown). Thus, RRV lytic replication, at least in culture, is both qualitatively and quantitatively distinct from that of KSHV, producing a higher proportion of released particles that contain or are C capsids (compare the relative amounts of A, B, and C capsids in Fig. 6A and B) as well as a greater total number of particles. However, comparisons of the infectious titers of the two viruses remain as estimates, since, in contrast to RRV, there is no currently available plaque assay for KSHV.
The differences in capsid and viral production between KSHV and RRV in culture may also reflect, at least in part, the distinctly different culture systems for growing the two viruses rather than fundamental differences in their biology. With KSHV, the capsid and viral preparations depend on reactivation of the viral genome from latently infected B-cell lines. In contrast, the RRV system involves de novo infection of immortalized fibroblasts and is marked by efficient entry into lytic replication. Currently, no equivalent KSHV lytic culture system exists for direct comparison. Even in recently developed methods allowing de novo infection with KSHV, latency remains the dominant mode of infection (4, 13, 32, 56). Significant lytic reactivation arises only through exogenous initiation of the lytic pathway either by chemical induction (e.g., addition of 12-O-tetradecanoylphorbol-13-acetate or sodium butyrate to the cells) (41, 45, 47, 60) or by introduction or activation of an immediate-early gene (23, 32, 46, 53, 56). This notion that the process of reactivation may limit the efficiency of KSHV production in latently infected cells is supported by our preliminary results indicating poor reactivation in rhesus macaque B cells latently infected with RRV (B. Damania, A. Smith, and D. H. Kedes, unpublished observations).
RRV as a model for KSHV.
The present study indicates that RRV is a potentially powerful model to help us understand the intricacies of the structure and assembly of gammaherpesviruses, including the closely related human rhadinovirus, KSHV. The inability to grow KSHV to high titers in cell culture has hampered studies addressing its structure, assembly, and maturation (38, 57, 59). RRV, in contrast, demonstrates robust lytic growth (2, 15-17, 31, 49). Nevertheless, the similarities between RRV and KSHV are more remarkable than their differences. In the present study, we have demonstrated that (i) RRV also produces three capsid species (the A, B, and C capsids) during lytic replication, (ii) the three RRV capsid species (as shown by TEM) are similar in structure to those of KSHV, (iii) RRV and KSHV possess a highly conserved protein composition, and (iv) both viruses assemble with similarly slow kinetics.
The efficient de novo infection and lytic replication of RRV, furthermore, lends itself more readily not only to refined structural analyses (58a) but also to a potentially more tractable means of introducing genetic manipulation into the genes relevant to assembly. These advantages can also lead to finer compositional analysis, including the detection of low-abundance capsid and virion-associated proteins (C. M. O'Connor and D. H. Kedes, unpublished observations). Although work with alpha- and betaherpesviruses continues to lead to further insights into herpesvirus structure and assembly, extrapolation of these findings to gammaherpesviruses runs the risk of missing subtle yet potentially important differences among the subfamilies. The RRV system allows direct study of a gammaherpesvirus that is evolutionarily related to the important human pathogen KSHV. Exploration of both the similarities and differences between these two primate rhadinoviruses should yield further clues to their biology and pathogenesis.
Acknowledgments
We thank N. Sherman at the W. M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia Biomolecular Research Facility, which is supported by a grant from the University of Virginia Pratt Committee. We also thank Jan Redick and Bonnie Sheppard at the Central Electron Microscopy Facility, University of Virginia. In addition, we thank Jay C. Brown and William W. Newcomb for invaluable discussion, as well as help with TEM.
This work was supported by National Institute of General Medical Sciences grant T32GM008136 (C.M.O.), National Institutes of Health-National Cancer Institute grant R-01 CA88768-01 (D.H.K.), Pew Memorial Trust award 97003260-000 (D.H.K.), Doris Duke Charitable Foundation award 20000355 (D.H.K.), and National Institutes of Health-National Cancer Institute grant CA096500 (B.D.).
REFERENCES
- 1.Ablashi, D. V., G. R. Armstrong, U. Heine, and R. A. Manaker. 1971. Propagation of Herpesvirus saimiri in human cells. J. Natl. Cancer Inst. 47:241-244. [PubMed] [Google Scholar]
- 2.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder, R. F. 2001. Epstein-Barr virus associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1209-1216. [DOI] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., Y. Liang, J. Hvidding, and D. Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beral, V. 1991. Epidemiology of Kaposi's sarcoma. Cancer Surv. 10:5-22. [PubMed] [Google Scholar]
- 6.Blomberg, J., E. Bjorck, S. Olofsson, G. Berg, and E. Lycke. 1976. Purification of virions and nucleocapsids of herpes simplex virus by means of metrizamide and sodium metrizoate gradients. Arch. Virol. 50:271-278. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff, C., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52:453-470. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff, C., and R. Weiss. 2002. AIDS-related malignancies. Nat. Rev. Cancer 2:373-382. [DOI] [PubMed] [Google Scholar]
- 9.Boshoff, C., and R. A. Weiss. 1998. Kaposi's sarcoma-associated herpesvirus. Adv. Cancer Res. 75:57-86. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 11.Chang, J., and D. Ganem. 2000. On the control of late gene expression in Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 81:2039-2047. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 13.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeWire, S. M., M. A. McVoy, and B. Damania. 2002. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes. J. Virol. 76:9819-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeWire, S. M., E. S. Money, S. P. Krall, and B. Damania. 2003. Rhesus monkey rhadinovirus (RRV): construction of an RRV-GFP recombinant virus and the development of assays to assess viral replication. Virology 312:122-134. [DOI] [PubMed] [Google Scholar]
- 18.Dolyniuk, M., E. Wolff, and E. Kieff. 1976. Proteins of Epstein-Barr virus. II. Electrophoretic analysis of the polypeptides of the nucleocapsid and the glucosamine- and polysaccharide-containing components of enveloped virus. J. Virol. 18:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupin, N., I. Gorin, J. Deleuze, H. Agut, J. M. Huraux, and J. P. Escande. 1995. Herpes-like DNA sequences, AIDS-related tumors, and Castleman's disease. N. Engl. J. Med. 333:798-799. [PubMed] [Google Scholar]
- 20.Fickenscher, H., and B. Fleckenstein. 2001. Herpesvirus saimiri. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:545-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 22.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haarr, L., and S. Skulstad. 1994. The herpes simplex virus type 1 particle: structure and molecular functions. APMIS 102:321-346. [DOI] [PubMed] [Google Scholar]
- 25.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 26.Horoszewicz, J. S., V. C. Dunkel, and J. T. Grace, Jr. 1968. Biological properties of a herpes-like virus (HLV) from Burkitt lymphoma cell line. Fed. Proc. 27:262. [Google Scholar]
- 27.Jensen, H. L., and B. Norrild. 2002. Temporal morphogenesis of herpes simplex virus type 1-infected and brefeldin A-treated human fibroblasts. Mol. Med. 8:210-224. [PMC free article] [PubMed] [Google Scholar]
- 28.Keating, S., S. Prince, M. Jones, and M. Rowe. 2002. The lytic cycle of Epstein-Barr virus is associated with decreased expression of cell surface major histocompatibility complex class I and class II molecules. J. Virol. 76:8179-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 30.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In P. M. Howley, D. M. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 31.Kirchoff, V., S. Wong, S. St. Jeor, and G. S. Pari. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321-333. [DOI] [PubMed] [Google Scholar]
- 32.Lagunoff, M., J. Bechtel, E. Venetsanakos, A. M. Roy, N. Abbey, B. Herndier, M. McMahon, and D. Ganem. 2002. De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 76:2440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leibold, W., T. D. Flanagan, J. Menezes, and G. Klein. 1975. Induction of Epstein-Barr virus-associated nuclear antigen during in vitro transformation of human lymphoid cells. J. Natl. Cancer Inst. 54:65-68. [DOI] [PubMed] [Google Scholar]
- 34.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, J. N., D. E. Ganem, D. H. Osmond, K. A. Page-Shafer, D. Macrae, and D. H. Kedes. 1998. Sexual transmission and the natural history of human herpesvirus 8 infection. N. Engl. J. Med. 338:948-954. [DOI] [PubMed] [Google Scholar]
- 36.Mistrikova, J., H. Raslova, M. Mrmusova, and M. Kudelova. 2000. A murine gammaherpesvirus. Acta Virol. 44:211-226. [PubMed] [Google Scholar]
- 37.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 41.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perdue, M. L., J. C. Cohen, M. C. Kemp, C. C. Randall, and D. J. O'Callaghan. 1975. Characterization of three species of nucleocapsids of equine herpesvirus type-1 (EHV-1). Virology 64:187-204. [DOI] [PubMed] [Google Scholar]
- 43.Prince, S., S. Keating, C. Fielding, P. Brennan, E. Floettmann, and M. Rowe. 2003. Latent membrane protein 1 inhibits Epstein-Barr virus lytic cycle induction and progress via different mechanisms. J. Virol. 77:5000-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabin, H., G. Pearson, H. C. Chopra, T. Orr, D. V. Ablashi, and G. R. Armstrong. 1973. Characteristics of Herpesvirus saimiri-induced lymphoma cells in tissue culture. In Vitro 9:65-72. [DOI] [PubMed] [Google Scholar]
- 45.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 46.Sakurada, S., H. Katano, T. Sata, H. Ohkuni, T. Watanabe, and S. Mori. 2001. Effective human herpesvirus 8 infection of human umbilical vein endothelial cells by cell-mediated transmission. J. Virol. 75:7717-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schalling, M., M. Ekman, E. E. Kaaya, A. Linde, and P. Biberfeld. 1995. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat. Med. 1:707-708. [DOI] [PubMed] [Google Scholar]
- 49.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonds, J. A., W. G. Robey, B. J. Graham, H. Oie, and G. F. Vande Woude. 1975. Purification of herpesvirus saimiri and properties of the viral DNA. Arch. Virol. 49:249-259. [DOI] [PubMed] [Google Scholar]
- 51.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 52.Steven, A. C., B. L. Trus, F. P. Booy, N. Cheng, A. Zlotnick, J. R. Caston, and J. F. Conway. 1997. The making and breaking of symmetry in virus capsid assembly: glimpses of capsid biology from cryoelectron microscopy. FASEB J. 11:733-742. [DOI] [PubMed] [Google Scholar]
- 53.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szilagyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661-668. [DOI] [PubMed] [Google Scholar]
- 55.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomescu, C., W. K. Law, and D. H. Kedes. 2003. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:9669-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trus, B. L., J. B. Heymann, K. Nealon, N. Cheng, W. W. Newcomb, J. C. Brown, D. H. Kedes, and A. C. Steven. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., P. Lo, X. Yu, J. K. Stoops, B. Forghani, and Z. H. Zhou. 2000. Three-dimensional structure of the human herpesvirus 8 capsid. J. Virol. 74:9646-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Yu, X.-K., C. M. O’Connor, I. Atanasov, B. Damania, D. H. Kedes, and Z. H. Zhou. 2003. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J. Virol. 77:13182-13193. [DOI] [PMC free article] [PubMed]
- 60.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]