Abstract
Prenatal cytomegalovirus infection may cause pregnancy complications such as intrauterine growth restriction and birth defects. How virus from the mother traverses the placenta is unknown. PCR analysis of biopsy specimens of the maternal-fetal interface revealed that DNA sequences from cytomegalovirus were commonly found with those of herpes simplex viruses and pathogenic bacteria. Cytomegalovirus DNA and infected cell proteins were found more often in the decidua than in the placenta, suggesting that the uterus functions as a reservoir for infection. In women with low neutralizing titers, cytomegalovirus replicated in diverse decidual cells and placental trophoblasts and capillaries. In women with intermediate to high neutralizing titers, decidual infection was suppressed and the placenta was spared. Overall, cytomegalovirus virions and maternal immunoglobulin G were detected in syncytiotrophoblasts, villus core macrophages, and dendritic cells. These results suggest that the outcome of cytomegalovirus infection depends on the presence of other pathogens and coordinated immune responses to viral replication at the maternal-fetal interface.
Human cytomegalovirus (CMV) is a ubiquitous virus that causes asymptomatic infections in healthy individuals. Since breast feeding (63), exposure to young children (46), and sexual contact (15) are major risk factors for infection, most adults are seropositive. Diverse organs and specialized cells, including polarized epithelial cells (67) and endothelial cells (13, 34), are susceptible to CMV infection. Latent infection in granulocyte-macrophage progenitors (26) reactivates upon cellular differentiation (18, 60). Although maternal immunity reduces the risk of symptomatic congenital disease in the fetus (1), prenatal infection is estimated to affect 1 to 2% of infants in the United States annually. Productively infected early-gestation human cytotrophoblasts downregulate the expression and functions of stage-specific antigens that are necessary for placental development (14). The routes of CMV infection at the fetal-maternal interface are unknown, as are the types of immune responses elicited. Nevertheless, both phenomena are likely linked to the unusual cellular interactions that give rise to the placenta.
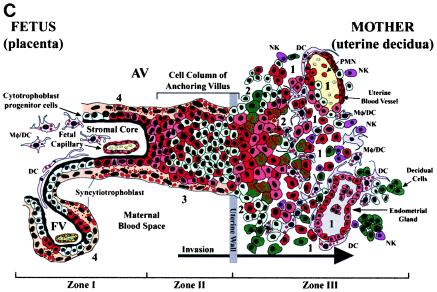
Placentation is a stepwise process that entails differentiation of cytotrophoblast stem cells along two pathways. In the pathway that gives rise to floating villi, cytotrophoblasts differentiate by fusing into multinucleate syncytiotrophoblasts that cover the villus surface, which is in direct contact with maternal blood (Fig. 1, zone I). This trophoblast population is specially adapted for transporting a wide variety of substances to the fetus, including maternal immunoglobulin G (IgG), via the neonatal Fc receptor (59). In the pathway that gives rise to anchoring villi, which attach the placenta to the uterine wall (Fig. 1, zone II), cytotrophoblasts aggregate into cell columns and invade the maternal decidua and the first third of the myometrium (interstitial invasion). During this process they remodel uterine blood vessels, thereby diverting blood flow to the placenta (endovascular invasion) (Fig. 1, zone III). By midgestation, cytotrophoblasts have completely replaced the endothelial lining and partially replaced the muscular wall of uterine blood vessels. Invasive cytotrophoblasts in anchoring villi express adhesion molecules and proteinases that enable attachment to the uterine wall and immune modulating factors that likely elicit maternal tolerance of the hemiallogeneic fetus (10, 27, 31).
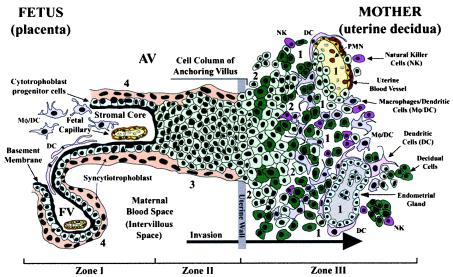
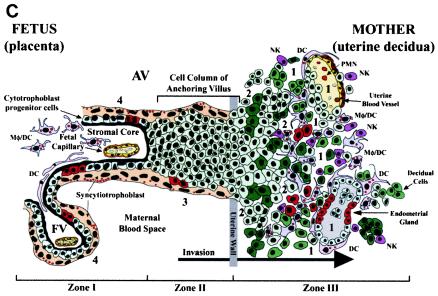
FIG. 1.
Diagram of the placental (fetal)-decidual (uterine) interface near the end of the first trimester of human pregnancy (10 weeks of gestational age). A longitudinal section includes a floating villus and an anchoring chorionic villus. The anchoring villus (AV) functions as a bridge between the fetal and maternal (decidual) compartments. The floating villus (FV), bathed by maternal blood, contains the fetal capillaries. Cytotrophoblasts in AV (zone I) form cell columns that attach to the uterine wall (zone II). Cytotrophoblasts then invade the uterine interstitium, decidua and first third of the myometrium and maternal vasculature (zone III), thereby anchoring the placenta to the uterus and gaining access to the maternal circulation. Colors illustrate different cell types: syncytiotrophoblasts (beige), cytotrophoblast progenitor cells and invasive cells (light green), decidual cells (dark green), endothelial cells (yellow), smooth muscle cells (brown), epithelial cells in endometrial glands (gray); innate immune cells: DC-SIGN-positive macrophage/dendritic cells (Mφ/DC) (purple), another dendritic cell type (DC) (pink), neutrophils (PMN) (red), and natural killer cells (NK) (dark pink). Sites proposed as routes of CMV infection in utero are numbered 1 to 4 (modified from reference 69).
In response to implantation, the uterine lining develops into the decidua, which is maintained by progesterone (44). Interglandular tissues increase in quantity, and the cytoplasm of resident stromal cells is distended with glycogen, lipid, and vimentin-type intermediate filaments (Fig. 1, zone III). Temporal and spatial expression of growth factors and cytokines (e.g., insulin-like growth factor 1 and its binding protein [IGFBP-1]) suggests that these molecules may influence decidualization (9). An unusual population of granular leukocytes is found in the decidua, intermingling with resident maternal cells and invasive fetal cytotrophoblasts (11, 50, 64). These include cells involved in innate pattern recognition, mostly natural killer cells, with some macrophages, dendritic cells, and T lymphocytes. Novel patterns of cytokine/chemokine expression in the decidua as well as specialized adhesion molecules on uterine vessels (28) probably attract this unusual leukocyte population, which functions in immunity and cytotrophoblast differentiation.
The cellular organization of the decidual-placental interface suggests potential routes by which CMV reaches the placenta (14). Virus might disseminate from infected maternal blood cells to the decidua (Fig. 1, site 1), interstitial and endovascular cytotrophoblasts in the uterine wall (site 2), cytotrophoblast columns of anchoring villi (site 3), and/or floating villi (site 4). An endothelial cell-tropic CMV strain replicates in uterine microvascular endothelial cells and spreads to invasive cytotrophoblasts in vitro (34), suggesting that hematogenous transmission occurs in utero.
Given the importance of understanding how CMV traverses the maternal-fetal interface, we investigated whether the virus was found in isolation or with other pathogens. We determined a role for pathogenic bacteria in viral reactivation in seropositive women and also identified the maternal and fetal cells that are associated with infected foci. Together, the results of this study provide novel information about the cellular mechanisms that allow CMV to reach the placenta, the first step in transmission to the fetus.
MATERIALS AND METHODS
Tissue biopsies.
Approval for this project was obtained from the Institutional Review Board at the University of California, San Francisco. First- and second-trimester placentas were obtained with adjacent specimens of maternal decidua from donors who had normal pregnancies prior to elective termination of pregnancy for nonmedical reasons. First-trimester biopsy specimens were taken from randomly chosen sites. Second-trimester biopsy specimens were taken from both floating villi and the placental bed. These specimens were used for PCR, immunohistochemistry, in situ hybridization, and electron microscopy. DNA for PCR was extracted with the QIAamp DNA kit (Qiagen).
PCR.
Extracted DNA was tested for CMV (33, 43), herpes simplex virus (HSV) (42), Bacteroides (66), Chlamydia (45), Gardnerella (40), Neisseria gonorrhoeae (29), Ureaplasma (32), group B Streptococcus (25), Mycoplasma hominis (32), and Mycoplasma genitalium (CCATGCTGAGAAGTAGAATAGC, TTGACATGCGCTTCCAATAA). An Applied Biosystems 9700HT sequence detection system (Foster City, Calif.) was used for real-time amplification of CMV and HSV sequences according to the manufacturer's instructions. Primers and probes were designed with Primer Express software to amplify fragments of CMV IE1/IE2 (GGAGACCCGCTGTTTCCA, TTGCAATCCTCGGTCACTTG; probe, TTGGCCGAAGAATCCCTCAAAACTTTTG) and UL83 (TGGACCTGCGTACCAACATAGA, TTTCAGGAGAACAAATCTCCGC; probe, CCGGCCCTCGGTTCTCTGCTG) genes, and the HSV DNA polymerase gene (UL30) (TGGATCTGGTGCGCAAAA, CGGATACGGTATCGTCGTAAAAC; probe; CAACCGCACCTCCAGGGCCC). FAM/TAMRA-labeled probes were manufactured by Biosearch Technologies (Novato, Calif.). Analysis of significance (P < 0.05) was determined by Fisher's exact test and McNemar test conducted in Stata (version 7.0) and R (version 1.6.2).
Immunohistochemistry.
Tissues were processed for immunohistochemistry as described (14).
Viral proteins.
Murine monoclonal antibodies to CMV-infected cell proteins and virion gB were used as described (48, 49). Immunoglobulin G (IgG) was affinity-purified from murine ascites with the ImmunoPure IgG purification kit (Pierce). CMV proteins included an antibody pool to gB (49), gH (CH438, UL75), alkaline nuclease (CH19, UL98/UL99), pp65 (CH65, UL83), IE1/IE2 (CH160, UL122/123), and ICP22 (CH41, US22). Guinea pig antiserum to CMV gB was a gift from Chiron Corp. (Emeryville, Calif.).
Cellular proteins.
Purified IgG to cellular proteins was purchased from the following sources: CD45, neutrophils and monocytes (Dako); CD56, natural killer cells (BD PharMingen); DC-SIGN, dendritic cells (BD PharMingen); CD68, macrophage/dendritic cells (Dako); IGFBP-1, decidual cells (goat anti-IGFBP-1, Diagnostic Systems); and vWF, endothelial cells (rabbit anti-human vWF, Novocastra). Anti-human cytokeratin antibody (7D3) was used to stain epithelial cells and cytotrophoblasts (14). Antiserum to neonatal Fc receptor was a gift from Neil Simister (35). Goat anti-human IgG and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated Affini Pure F(ab′) fragment were obtained from Jackson ImmunoResearch. Secondary antibodies (Jackson ImmunoResearch) were goat anti-mouse IgG labeled with fluorescein isothiocyanate (FITC) or TRITC; goat anti-rat IgG labeled with TRITC; goat anti-guinea pig IgG labeled with FITC or TRITC; and goat anti-rabbit IgG labeled with TRITC. Nuclei were counterstained with TO-PRO-3 iodide (Molecular Probes). Laser-scanning confocal images were generated with a Bio-Rad MRC1024 confocal Optiphot II Nikon microscope.
Serological assays.
IgG was purified from conditioned medium that contained biopsy specimens and residual maternal blood with the ImmunoPure IgG purification kit (Pierce). To assess serological status to CMV, IgG was tested by immunofluorescence with strain Toledo-infected fibroblasts and a commercial enzyme-linked immunosorbent assay (OptiCoat CMV [IgG], Biotecx Laboratories). For neutralization titers, strain Toledo was incubated with 100 μg of purified IgG (60 min) and examined in the rapid infectivity assay (41). Titers, calculated as percent neutralization per microgram of IgG compared with positive control IgG, were defined as low (0 to 39%), moderate (40 to 69%), and high (70 to 98%). Positive (neutralizing control CH177) IgG to gB (100 mg/ml) reduced plaque number (86 to 98%) in three sets of duplicate experiments. Nonneutralizing IgG to gB (CH86) was used as a negative control.
Electron microscopy.
Placental biopsy specimens were fixed in 1.5% Karnovsky fixative and postfixed in 1% Palade buffer, dehydrated, and embedded. Sections were stained in uranyl acetate and lead citrate and examined with a JEM-1200EX electron microscope.
Fluorescence in situ hybridization.
Frozen sections were fixed and processed prior to overnight hybridization with a fluorescein oligonucleotide cocktail to a CMV early-gene RNA transcript (Novocastra). A negative control probe was included for hybridization with each biopsy section. Positive control samples were provided by the manufacturer. Sections were incubated with biotinylated anti-fluorescence and then with fluorescein avidin DCS (Vector). TO-PRO-3 above (Molecular Probes) was used for nuclear counterstaining.
RESULTS
Detection of viral and bacterial DNA in placental and decidual specimens.
We used PCR-based strategies to test for viral and bacterial DNA in placental and decidual biopsy specimens collected after the termination of uncomplicated pregnancies. Data from all the placental samples (Table 1) showed that, overall, CMV DNA was detected in 69% of specimens; it was detected with bacteria in 38%. When found in isolation, CMV was detected in 27% of placental samples. Other pathogens were detected as follows: HSV-1 in 3%; HSV-2 in 9%; and more than one bacterial species in 15%. Sixteen percent of placental samples were negative for all of these pathogens. Our findings suggest that early-gestation placentas frequently contain DNA from viral and bacterial pathogens.
TABLE 1.
PCR detection of viruses and pathogenic bacteria in early-gestation biopsy specimensa
| Pathogen | No. (%) of specimens containing viral or bacterial DNA
|
|||
|---|---|---|---|---|
| 1st trimester
|
2nd trimester (placentas) (n = 39) | Total (placentas) (n = 74) | ||
| Decidua (n = 35) | Placentas (n = 35) | |||
| CMV (with or without other pathogens) | 31 (89)* | 22 (63)* | 29 (74) | 51 (69) |
| CMV (with bacteria)b | 9 (26) | 11 (31)† | 17 (44)† | 28 (38) |
| CMV alone | 17 (49) | 9 (26) | 11 (28) | 20 (27) |
| HSV-1 (with or without other pathogens) | 2 (6) | 1 (3) | 1 (3) | 2 (3) |
| HSV-2 (with or without other pathogens) | 5 (14) | 2 (6) | 5 (13) | 7 (9) |
| Bacteria alone | 2 (6) | 4 (11) | 5 (13) | 9 (12) |
| Negative (all pathogens) | 2 (6) | 9 (26)† | 3 (8)† | 12 (16) |
Data based on analysis of one sample per biopsy specimen (n = 282). *, significant association between CMV DNA in the decidua and the placenta in paired biopsy samples (McNemar test; P = 0.038). †, significant difference between first- and second-trimester placentas (Fisher's exact test: negative [all pathogens]: P = 0.039).
Order of occurrence: group B Streptococcus, Gardnerella, Chlamydia, Ureaplasma, M. hominis, M. genitalium, and Bacteroides. Neisseria gonorrhoeae was not detected.
To understand whether the decidua and the placenta from the same pregnancy would contain the same pathogens, we evaluated 35 paired first-trimester biopsy specimens from individual pregnancies (Table 1). We detected CMV DNA in 89% of the decidual samples and 63% of the placentas (P = 0.038). When CMV was found in isolation in the decidua (40% of samples), CMV was also found in the placenta (26%). HSV-1 and HSV-2 were less frequently detected in the decidua (HSV-1, 6%; HSV-2, 14%) and found only half as often in the placenta (HSV-1, 3%; HSV-2, 6%). Bacterial DNA alone, which was often detected in the placenta (11%), was found less frequently in the decidua (6%). Together, these results suggest that CMV can be selectively transferred from a decidual reservoir to the adjacent placenta.
We also examined the effects of gestational age. A high incidence of CMV DNA with or without other pathogens was detected in 63% of first-trimester placentas and increased to 74% in the second trimester (Table 1). Together, samples with both CMV and bacterial DNA increased from 31% in the first trimester to 44% in the second trimester, whereas CMV alone was reduced in the second trimester. Fewer second-trimester placentas were negative for all pathogens (P = 0.039). Our results suggest that CMV is commonly found together with pathogenic bacteria and tends to increase in the second trimester.
Some serologic evidence suggests that reinfection with new CMV strains in seropositive women might be associated with symptomatic fetal infection (2). Therefore, we investigated whether multiple strains colonize the placental-decidual interface. To identify tissues with more than one strain, we sequenced a region of the gB gene with characteristic nucleotide differences (4). Sequence analysis of seven CMV DNA-positive biopsy pairs (Table 1) revealed that the gB genotypes were similar to three variants as classified by Chou and Dennison (4): group 1 (three strains), group 2 (two strains), and group 3 (one strain). Paired samples from one decidua and adjacent placenta from a seropositive donor without detectable neutralizing antibodies contained a mixture of gB genotypes. Together, these results suggest that different CMV strains are present at the maternal-fetal interface and may be found early in infection.
Development of neutralizing antibodies is delayed when primary CMV infection occurs shortly before or during gestation (1, 16, 30, 52, 62), whereas high titers indicate resolution of acute infection and/or reactivation. To evaluate the antibody response to CMV in the group of donors from whom we obtained paired biopsy specimens (Table 1), we assessed the presence of IgG to viral proteins with serological assays. Twenty-three of these paired biopsy specimens were also examined by immunofluorescence confocal microscopy. With one exception, the donors were seropositive with a range of neutralizing activity, as shown by our evaluation of IgG purified from the conditioned medium of biopsy specimens. Ten women had low neutralizing titers (0 to 32%), nine had moderate titers (43 to 67%), and four had high titers (70 to 98%).
Decidual cells and invasive cytotrophoblasts express CMV proteins in specific patterns.
First, we used immunofluorescence confocal microscopy to determine whether the presence of CMV DNA correlated with expression of proteins from infected cells at the decidual-placental interface during the first trimester. Decidual biopsy samples from 23 paired specimens were incubated with a pool of monoclonal antibodies to CMV-infected cell proteins, indicative of viral replication, and with antisera to gB, an abundant virion envelope glycoprotein and neutralization target. Antibodies to cytokeratin (a marker for uterine glandular epithelial cells and invasive placental cytotrophoblasts) and immune cell markers were used for costaining.
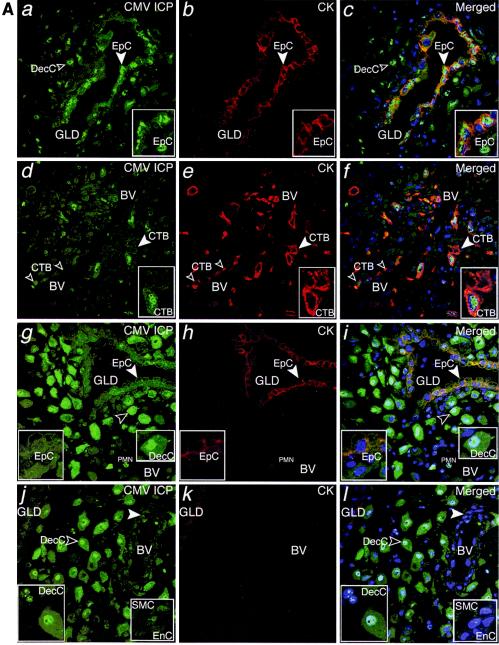
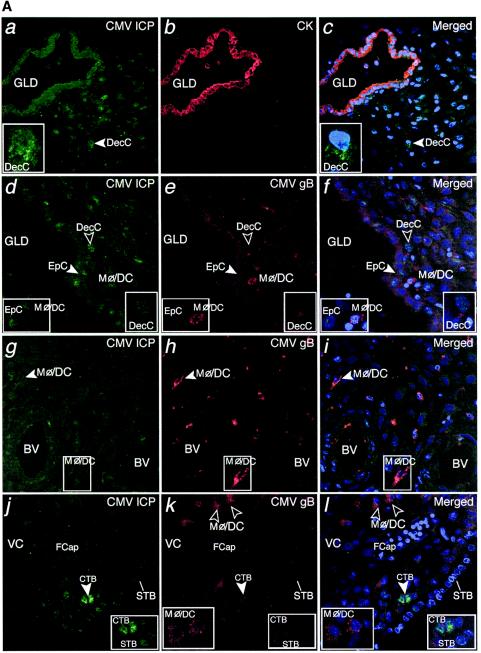
Staining revealed islands of infected cells among much larger uninfected areas. Extensive analysis indicated several common staining patterns. For example, we detected CMV-infected cell proteins in the nuclei and cytoplasm in endometrial glandular epithelial cells (Fig. 2A, a to c), endovascular cytotrophoblasts (Fig. 2A, d to f) and resident decidual cells that stained brightly (Fig. 2A, g to i). Endothelial cells in unmodified uterine blood vessels also stained (Fig. 2A, j to l). These data indicate that CMV infects a diverse population of resident maternal cells within the uterine wall and fetal invasive cytotrophoblasts.
FIG.2.
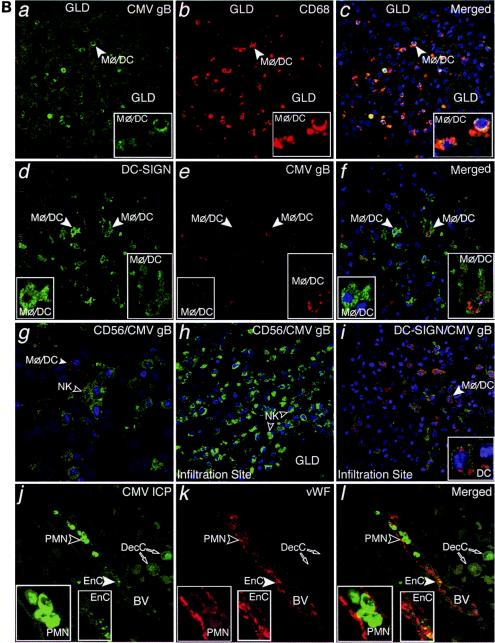
CMV replicates in diverse cell types in maternal uterine decidua. (A) CMV infects endometrial glands (GLD), uterine blood vessels (BV), resident decidual cells (DecC) and cytotrophoblasts (CTB) in the decidua. a to c, Decidual biopsy specimens stained for CMV-infected cell proteins (ICP, green) and cytokeratin (CK, red) in epithelial cells (EpC). d to i, CMV-infected interstitial and endovascular CTB and DecC. j to l, Endothelial cells (EnC) and smooth muscle cells (SMC) of uterine blood vessels (BV) are infected. Merged, colocalized proteins (yellow). Large arrowheads, insets. (B) Abundant innate immune cells infiltrating the decidua contain CMV proteins. a to c, CMV gB (green), macrophages (Mφ/DC, CD68, red). d to f, DC-SIGN-positive (green) macrophage/dendritic cells (Mφ/DC) take up CMV gB (red). g and h, CD56-positive (green) natural killer (NK) cells each target infection sites. i, DC-SIGN-positive cells containing gB. j to l, Neutrophils (PMN) with phagocytosed proteins from virus-infected cells and endothelial cells (EnC) positive for von Willebrand factor (vWF) in blood vessels (BV). Merged, colocalized proteins (yellow). Large arrowheads, insets.
Innate immune cells showed a staining pattern that was distinctly different from that of CMV-infected cells, suggesting phagocytosis of enveloped virions. Macrophages (CD68+) contained cytoplasmic vesicles, of which a subset stained strongly for CMV gB (Fig. 2B, a to c). These abundant macrophage/dendritic cells (Mφ/DC) also stained for dendritic cell ICAM-3-grabbing nonintegrin (DC-SIGN) (24, 61) and contained gB-positive cytoplasmic vesicles (Fig. 2B, d to f). In contrast to CD68 staining, DC-SIGN and gB did not colocalize, suggesting they were in different compartments (Fig. 2B, f). Natural killer (NK) cells (CD56+) were often dispersed among Mφ/DC that were filled with gB-positive vesicles (Fig. 2B, g). Occasionally, striking numbers of NK cells and Mφ/DC were found together (Fig. 2B, h and i). Additionally, neutrophils were associated with uterine blood vessels located near endothelial cells positive for von Willebrand factor (vWF) and decidual cells that expressed CMV-infected cell proteins, suggesting phagocytosis (Fig. 2B, j to l).
Patterns of CMV-infected cell proteins in decidual samples, mirrored in adjacent placentas.
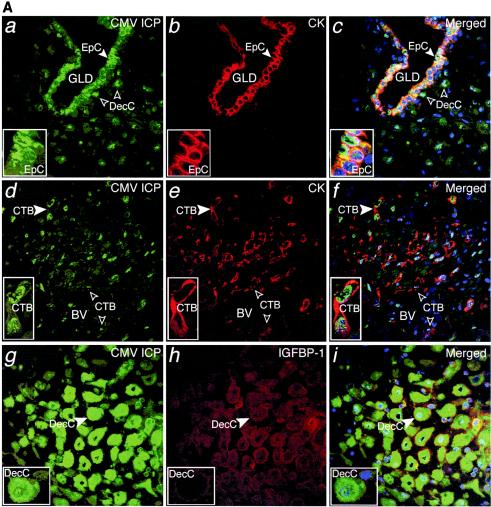
Next, we analyzed CMV proteins in paired decidual and placental biopsy specimens and found three staining patterns. In the first, islands in both decidual and placental compartments stained strongly for expression of CMV-infected cell proteins. This pattern predominated in samples from five donors with low neutralizing titers and one with intermediate neutralizing titer, three of which contained other pathogens. In the decidua, cytokeratin-positive glandular epithelial cells (Fig. 3A, a to c), endovascular cytotrophoblasts in remodeled uterine blood vessels, and interstitial cytotrophoblasts were sometimes positive (Fig. 3A, d to f). Strikingly, IGFBP-1-positive resident decidual cells strongly stained for viral proteins, suggesting that these cells were permissive for viral replication (Fig. 3A, g to i).
FIG. 3.
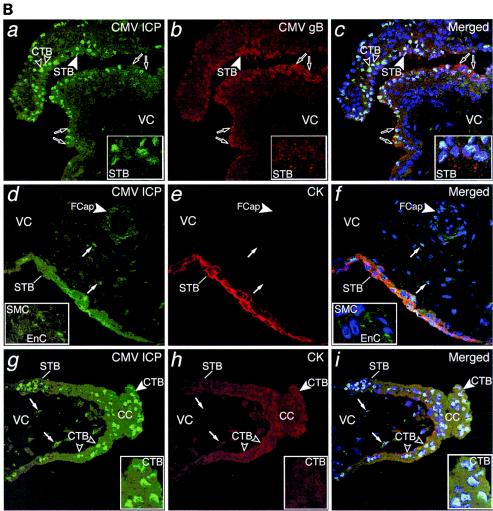
Extensive CMV replication in maternal decidua correlates with transmission of infection to the placenta. (A) a to c, CMV-infected cell proteins (green) expressed in cytokeratin (CK, red)-stained epithelial cells (EpC) in endometrial glands (GLD). d to f, CK-stained endovascular cytotrophoblasts (CTB) in blood vessels (BV) and interstitial CTB infiltrating the decidua (insets). g to i, Decidual cells (DecC) expressing IGFBP-1 (red). Merged, colocalized proteins (yellow). Large arrowheads, insets. (B) a to c, Syncytiotrophoblasts (STB) and cytotrophoblast (CTB) stem cells expressing CMV-infected cell proteins (ICP, green) and abundant gB-containing vesicles (red) on the villus surface. d to f, Infected endothelial cells (EnC) in fetal capillaries (FCap) and fibroblasts in the villus core (VC). g to i, CMV proteins expressed in differentiating/invasive cytotrophoblast cell columns (CC). Macrophages contain cytoplasmic vesicles with infected cell membranes (arrows). Large arrowheads, insets. (C) Schematic that illustrates and summarizes the pattern of CMV protein expression in the decidua that was associated with transmission of infection to the placenta. CMV-infected cells (red) and gB-containing vesicles (red) in Mφ/DC at the placental-decidual interface. Islands of infected cells were present in endometrial glands, uterine blood vessels and invasive cytotrophoblasts, suggesting extensive decidual infection. CMV infection was transmitted to portions of the adjacent placenta, as indicated by widespread expression of replication proteins by trophoblasts and fetal capillaries. Some Mφ/DC contained cytoplasmic vesicles with these proteins, suggesting phagocytosis without productive infection.
In the adjacent portions of the placenta, floating villi contained syncytiotrophoblasts and cytotrophoblast progenitor cells expressing CMV-infected cell proteins that localized to the nuclei and cytoplasm (Fig. 3B, a to c). Abundant vesicles that varied in size amassed close to the plasma membrane of the villus surface and contained gB (and less gH [not shown]). In regions with infected syncytiotrophoblasts, fibroblasts and fetal capillaries in the villus core expressed infected cell proteins (Fig. 3B, d to f). Invasive cytotrophoblasts in developing cell columns that anchor the placenta to the uterine wall also stained (Fig. 3B, g to i). In contrast, Mφ/DC (Hofbauer cells) within the villus stromal cores contained infected cell proteins in cytoplasmic vesicles but not in the nuclei, suggesting phagocytosis. Figure 3C illustrates and summarizes the pattern of CMV protein expression in the decidua that was associated with transmission of infection to the placenta in these cases.
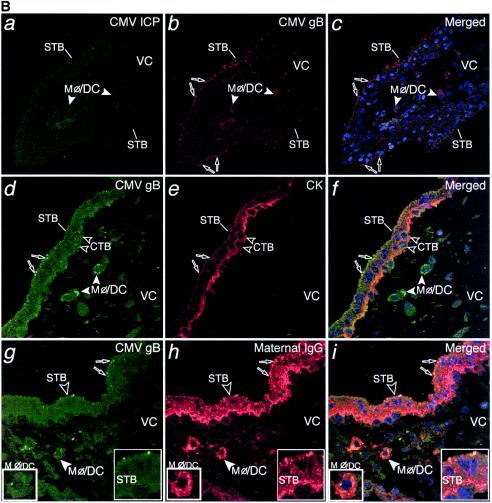
In the second group of paired biopsy specimens, the number of cells that stained for CMV-infected cell proteins was reduced in the decidua, and occasional focal infection was found in the placenta. This pattern predominated in samples from seven donors with low to intermediate neutralizing titers, five of which contained other pathogens. In the decidua, we detected CMV replication in some glandular epithelial cells and decidual cells (Fig. 4A, a to f). In the interstitium, Mφ/DC were abundant throughout, especially near infected glands and blood vessels, and contained gB-positive cytoplasmic vesicles but were not infected (Fig. 4A, d to i). Three of the adjacent placentas contained small clusters of cytotrophoblast progenitor cells underlying syncytiotrophoblasts that expressed CMV-infected cell proteins (Fig. 4A, j to l). Isolated gB-containing vesicles were present in syncytiotrophoblasts (Fig. 4A, j to l). In the villus core, Mφ/DC containing CMV gB-positive vesicles were often observed (Fig. 4A, j to l). In other placental biopsies, only gB-containing vesicles were detected in syncytiotrophoblasts and villus core Mφ/DC without infection.
FIG.4.
Moderate CMV infection in the decidua is mirrored by the adjacent placenta and often associated with the presence of bacteria in women with moderate neutralizing titers. (A) a to c, CMV-infected cell proteins expressed in decidual cells. d to f, Selected glandular epithelial cells are infected, and Mφ/DC internalize CMV gB. g to i, Uninfected Mφ/DC accumulate CMV gB-positive vesicles. j to l, Placental specimen containing a focus of cytotrophoblast (CTB) progenitor cells expressing infected cell proteins. Uninfected Mφ/DC with phagocytosed virion protein are present in the villus core adjacent to a large, uninfected fetal capillary (FCap). Large white and black arrowheads, insets. B, a to c, Placenta that contains many gB-staining vesicles in syncytiotrophoblasts but does not contain cells that express virus-infected cell proteins. g to i, CMV gB-staining vesicles at the apical membrane of STB overlying CK-positive CTB progenitor cells. Villus core Mφ/DC contain gB-positive vesicles. d to f, CMV gB in vesicles that costain with maternal IgG. Selected villus core Mφ/DC take up IgG and gB in some costaining vesicles. (C) Schematic that illustrates and summarizes moderate infection at the placental-decidual interface: CMV-infected cells (red) and gB-containing vesicles (red) in Mφ/DC.
In the last group of paired biopsy specimens, few cells stained for CMV-infected cell proteins in the decidua and none were found in the placenta. This pattern predominated in samples from ten donors with intermediate to high neutralizing titers, seven of which contained other pathogens. In the decidua, neutrophils with viral proteins were found in uterine blood vessels near infected cells (see Fig. 2B, j to l). In the adjacent portions of the placenta, syncytiotrophoblasts contained numerous CMV gB-positive vesicles but were not infected (Fig. 4B, a to c). Cytokeratin staining confirmed that the vesicles were beneath the apical membrane in contact with maternal blood (Fig. 4B, d to f). In villus core Mφ/DC, gB accumulated in large cytoplasmic vesicles (Fig. 4B, b and c). When placentas were stained for IgG, many positive vesicles were found in syncytiotrophoblasts (Fig. 4B, g to i). Close inspection revealed that gB colocalized with a small subset of IgG-positive vesicles (Fig. 4B, g to i, inset). In villus core Mφ/DC, some gB-staining vesicles colocalized with the more abundant IgG-positive vesicles. We also found neonatal Fc receptor-positive vesicles at the apical and basolateral membranes, suggesting IgG transcytosis in first-trimester syncytiotrophoblasts (not shown) (58). Figure 4C illustrates and summarizes the focal pattern of reduced and/or suppressed CMV infection found at the placental-decidual interface.
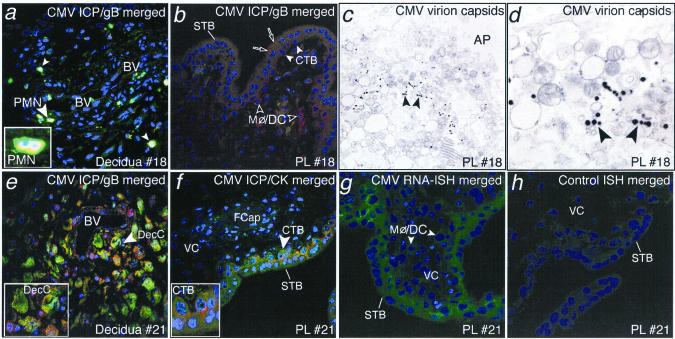
Biopsy specimens with different staining patterns were also examined by with electron microscopy and fluorescence in situ hybridization (Fig. 5). In decidual samples with reduced infection, neutrophils that stained for CMV proteins were present (Fig. 5, a). In the adjacent placenta, vesicular gB staining was found in syncytiotrophoblasts and uninfected villus core Mφ/DC (Fig. 5, b). In five placental biopsy specimens with this pattern, viral nucleocapsids were found in syncytiotrophoblasts (Fig. 5, c and d). In three decidual biopsies with islands expressing CMV-infected cell proteins (Fig. 5, e) and the adjacent placenta (Fig. 5, f), in situ hybridization confirmed the presence of early CMV transcripts in cytotrophoblast progenitor cells and villus core fibroblasts and Mφ/DC (Fig. 5, g). A positive control showed similar hybridization (not shown), whereas a negative control probe failed to react (Fig. 5, h). These results suggest that placentas with gB-staining vesicles in syncytiotrophoblasts also contain nucleocapsids and that viral transcripts are present in placentas that express infected cell proteins.
FIG. 5.
CMV virion uptake and replication in the placenta. a, Decidua expressing CMV-infected cell proteins (ICP). b, Adjoining placenta with CMV gB-stained vesicles in syncytiotrophoblasts (STB) and Mφ/DC in the villus core. c and d, Electron micrographs of placenta showing CMV virion capsids clustered near the apical (AP) surface of the STB membrane. e, Decidua expressing CMV proteins in decidual cells and uterine blood vessels (BV). f, Placenta contains infected CTB progenitor cells and infected fetal capillaries (FCap). g and h, Fluorescence in situ hybridization showing CMV-specific probe and negative control in reactions with CMV early RNA transcripts in trophoblast layers of the placenta surface and Mφ/DC in the villus core. White arrowheads, inset.
DISCUSSION
Here we describe a novel experimental system for examining CMV biology in the context of concurrent bacterial and viral infections in human hemiallogeneic fetal tissues that are “transplanted” to the uterus during pregnancy. CMV reactivates from latency in healthy individuals, and this process escalates in pathological situations, allogeneic transplantation and immunosuppression that may occur during the lifetime of an infected person (12, 18, 36, 60). In the context of immunosuppression and bacterial infections, murine CMV reactivates from latently infected Mφ/DC in different organs, suggesting that cells that clear infection could be associated with pathogenesis (7, 20, 21). We showed that CMV and bacterial pathogens are commonly present at the maternal-fetal interface, one possible explanation for why pregnant women shed virus from the cervix (5, 56, 62). Bacteria were often found in donors with intermediate to high neutralizing titers whose uninfected placentas contained virion proteins; this suggests that limited CMV replication in the decidua could create a reservoir.
Reactivation from decidual Mφ/DC might occur as a consequence of inflammatory responses to bacteria and could depend on the number of latently infected Mφ/DC infiltrating the uterus. Placentas from healthy pregnant donors contained isolated areas of infection that were a small part of the whole tissue. Since these pregnancies were normal, placental infection that leads to transmission likely involves the decidual and placental components that stained for infected cell proteins, i.e., an exacerbation of the situation found in those donors with the lowest neutralizing titers and some with intermediate titers and bacterial pathogens at the uterine-placental interface.
Coordinated innate and adaptive immune responses suppressed CMV infection of the placenta in women with intermediate to high neutralizing titers, one explanation for a correlation between high-avidity IgG and protection against vertical transmission (1, 52). The most remarkable result is that healthy women with uncomplicated pregnancies may have infected decidual cells. Virion-IgG complexes may be transported to the placenta without infection, a process that demonstrates the efficacy of innate and adaptive immunity. CMV infection of the decidua is a novel paradigm and further illustrates how this virus utilizes host immunity (39) by exploiting maternal hyporesponsiveness.
Pathogen-associated pattern receptors on innate immune cells could recognize diverse pathogens at the maternal-fetal interface. Mφ/DC attracted to the decidua by the specialized environment created by hormonal changes (11, 24, 50, 64) might carry pathogens from the infected genital and cervical mucosa (8, 47, 68). Like the capture of human immunodeficiency virus by DC-SIGN-positive DC (17), CMV virions are internalized via DC-SIGN interaction with gB (19). The striking concentration of CMV gB in endocytic vesicles and strong DC-SIGN expression by Mφ/DC indicate a central role in virion clearance at the placental-decidual interface.
Recently we discovered that villus core Mφ/DC take up CMV virions within 60 min after infection in vitro (unpublished observation). This extraordinary process could occur in utero and involve sampling of virions and/or IgG-virion complexes across tight junctions of cytotrophoblast progenitor cells, comparable to DC penetration of gut epithelial cell monolayers to sample bacteria (51). Like bacteria, CMV virions may interact with Toll-like receptors that initiate inflammatory responses to invading pathogens via the interferon (IFN)-α/β pathways (6, 22, 23). Moreover, HSV binds the mannose receptor that upregulates IFN-α produced by plasmacytoid dendritic cells (38, 55, 57). Finally, ligation of IgG-virion complexes to the Fc-gamma I receptor on Mφ could remove opsonized virions, suppress proinflammatory responses and reduce the spread of infection (65).
In healthy adults, CMV reactivation is controlled predominantly by T cells (12), which are underrepresented in the decidual leukocyte population in pregnant women (11, 54, 64). The cytokine milieu also diminishes adaptive responses to pathogens, presenting an opportunity for viral infection and/or bacterial colonization (53). In the present study, Mφ/DC contained virion proteins, suggesting that they were in an immature state characterized by antigen uptake (3)-one reason why CMV replication that occurs in mature Mφ/DC was not detected (18, 60). Regardless, CMV can infect decidual cells of immune women, suggesting that interruption of IFN-α signal transduction pathways can occur under certain conditions (37). Understanding the molecular mechanisms that repress infection in the microenvironment at the maternal-fetal interface could generate novel antiviral therapies for pregnancy and organ transplantation.
FIG. 2.FIG. 2—Continued.
Acknowledgments
We are grateful to members of the Pereira and Fisher laboratories, Ed Mocarski, Ann Arvin, and Tony DeFranco for thoughtful discussions; Eduardo Caceres, Mirhan Kapidzic, and Hsin-Ti Chang for excellent technical assistance and Vibeke Petersen for electron microscopy; Paul Lepp for bacterial primer design; Stephen Shiboski for statistical analyses; and Mary McKenney for editing the manuscript.
This work was supported by Public Health Service grants AI46657, AI53782 (L.P. and S.F.), EY13683 (L.P.), and HD30367 (S.F.) from the National Institutes of Health, grants from the March of Dimes Birth Defects Foundation and the University of California Academic Senate (L.P. and S.F.), and a gift from GlaxoSmithKline.
REFERENCES
- 1.Boppana, S. B., and W. J. Britt. 1995. Antiviral antibody responses and intrauterine transmission after primary maternal cytomegalovirus infection. J. Infect. Dis. 171:1115-1121. [DOI] [PubMed] [Google Scholar]
- 2.Boppana, S. B., K. B. Fowler, W. J. Britt, S. Stagno, and R. F. Pass. 1999. Symptomatic congenital cytomegalovirus infection in infants born to mothers with preexisting immunity to cytomegalovirus. Pediatrics 104:55-60. [DOI] [PubMed] [Google Scholar]
- 3.Cella, M., A. Engering, V. Pinet, J. Pieters, and A. Lanzavecchia. 1997. Inflammatory stimuli induce accumulation of MHC Class II complexes on dendritic cells. Nature 388:782-787. [DOI] [PubMed] [Google Scholar]
- 4.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 5.Collier, A. C., H. H. Handsfield, R. Ashley, P. L. Roberts, T. DeRouen, J. D. Meyers, and L. Corey. 1995. Cervical but not urinary excretion of cytomegalovirus is related to sexual activity and contraceptive practices in sexually active women. J. Infect. Dis. 171:33-38. [DOI] [PubMed] [Google Scholar]
- 6.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, C. H., Y. Zhang, B. J. McGuinness, M. C. Lahm, D. D. Sedmak, and R. M. Ferguson. 2002. Intra-abdominal bacterial infection reactivates latent pulmonary cytomegalovirus in immunocompetent mice. J. Infect. Dis. 185:1395-1400. [DOI] [PubMed] [Google Scholar]
- 8.Coonrod, D., A. C. Collier, R. Ashley, T. DeRouen, and L. Corey. 1998. Association between cytomegalovirus seroconversion and upper genital tract infection among women attending a sexually transmitted disease clinic: a prospective study. J. Infect. Dis. 177:1188-1193. [DOI] [PubMed] [Google Scholar]
- 9.Crossey, P. A., C. C. Pillai, and J. P. Miell. 2002. Altered placental development and intrauterine growth restriction in IGF binding protein-1 transgenic mice. J. Clin. Investig. 110:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damsky, C. H., C. Librach, K. H. Lim, M. L. Fitzgerald, M. T. McMaster, M. Janatpour, Y. Zhou, S. K. Logan, and S. J. Fisher. 1994. Integrin switching regulates normal trophoblast invasion. Development 120:3657-3666. [DOI] [PubMed] [Google Scholar]
- 11.Drake, P. M., M. D. Gunn, I. F. Charo, C. L. Tsou, Y. Zhou, L. Huang, and S. J. Fisher. 2001. Human placental cytotrophoblasts attract monocytes and CD56(bright) natural killer cells via the actions of monocyte inflammatory protein 1alpha. J. Exp. Med. 193:1199-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, H. S., D. J. Haney, S. A. Ghanekar, P. Stepick-Biek, D. B. Lewis, and H. T. Maecker. 2002. Dynamics of CD4 and CD8 T cell responses to cytomegalovirus in healthy human donors. J. Infect. Dis. 186:15-22. [DOI] [PubMed] [Google Scholar]
- 13.Fish, K. N., C. Soderberg-Naucler, L. K. Mills, S. Stenglein, and J. A. Nelson. 1998. Hum. cytomegalovirus persistently infects aortic endothelial cells. J. Virol. 72:5661-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, S., O. Genbacev, E. Maidji, and L. Pereira. 2000. Hum. cytomegalovirus infection of placental cytotrophoblasts in vitro and in utero: implications for transmission and pathogenesis. J. Virol. 74:6808-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowler, K. B., and R. F. Pass. 1991. Sexually transmitted diseases in mothers of neonates with congenital cytomegalovirus infection. J. Infect. Dis. 164:259-264. [DOI] [PubMed] [Google Scholar]
- 16.Fowler, K. B., S. Stagno, R. F. Pass, W. J. Britt, T. J. Boll, and C. A. Alford. 1992. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N. Engl. J. Med. 326:663-667. [DOI] [PubMed] [Google Scholar]
- 17.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houles, F. Fieschi, F. Arenzana-Seisdedos, J. F. Moreau, and J. Dechanet-Merville. 2002. Hum. cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 20.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. t. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummel, M., Z. Zhang, S. Yan, I. DePlaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 23.Jarrossay, D., G. Napolitani, M. Colonna, F. Sallusto, and A. Lanzavecchia. 2001. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol. 31:3388-3393. [DOI] [PubMed] [Google Scholar]
- 24.Kammerer, U., A. O. Eggert, M. Kapp, A. D. McLellan, T. B. Geijtenbeek, J. Dietl, Y. Van Kooyk, and E. Kampgen. 2003. Unique appearance of proliferating antigen-presenting cells expressing DC-SIGN (CD209) in the decidua of early human pregnancy. Am. J. Pathol. 162:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke, D., C. Menard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 26.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Hum. cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovats, S., E. K. Main, C. Librach, M. Stubblebine, S. J. Fisher, and R. DeMars. 1990. A class I antigen, HLA-G, expressed in human trophoblasts. Science 248:220-223. [DOI] [PubMed] [Google Scholar]
- 28.Kruse, A., R. Hallmann, and E. C. Butcher. 1999. Specialized patterns of vascular differentiation antigens in the pregnant mouse uterus and the placenta. Biol. Reprod. 61:1393-1401. [DOI] [PubMed] [Google Scholar]
- 29.Lanham, S., A. Herbert, A. Basarab, and P. Watt. 2001. Detection of cervical infections in colposcopy clinic patients. J. Clin. Microbiol. 39:2946-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazzarotto, T., P. Spezzacatena, P. Pradelli, D. A. Abate, S. Varani, and M. P. Landini. 1997. Avidity of immunoglobulin G directed against human cytomegalovirus during primary and secondary infections in immunocompetent and immunocompromised subjects. Clin. Diagn. Lab. Immunol. 4:469-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Librach, C. L., Z. Werb, M. L. Fitzgerald, K. Chiu, N. M. Corwin, R. A. Esteves, D. Grobelny, R. Galardy, C. H. Damsky, and S. J. Fisher. 1991. 92-kD type IV collagenase mediates invasion of human cytotrophoblasts. J. Cell Biol. 113:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luki, N., P. Lebel, M. Boucher, B. Doray, J. Turgeon, and R. Brousseau. 1998. Comparison of polymerase chain reaction assay with culture for detection of genital mycoplasmas in perinatal infections. Eur. J. Clin. Microbiol. Infect. Dis. 17:255-263. [DOI] [PubMed] [Google Scholar]
- 33.Lurain, N. S., K. S. Kapell, D. D. Huang, J. A. Short, J. Paintsil, E. Winkfield, C. A. Benedict, C. F. Ware, and J. W. Bremer. 1999. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J. Virol. 73:10040-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maidji, E., E. Percivalle, G. Gerna, S. Fisher, and L. Pereira. 2002. Transmission of human cytomegalovirus from infected uterine microvascular endothelial cells to differentiating/invasive placental cytotrophoblasts. Virology 304:53-69. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy, K. M., Y. Yoong, and N. E. Simister. 2000. Bidirectional transcytosis of IgG by the rat neonatal Fc receptor expressed in a rat kidney cell line: a system to study protein transport across epithelia. J. Cell Sci. 113:1277-1285. [DOI] [PubMed] [Google Scholar]
- 36.Meyers, J. D., N. Flournoy, and E. D. Thomas. 1986. Risk factors for cytomegalovirus infection after human marrow transplantation. J. Infect. Dis. 153:478-488. [DOI] [PubMed] [Google Scholar]
- 37.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Hum. cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 38.Milone, M. C., and P. Fitzgerald-Bocarsly. 1998. The mannose receptor mediates induction of IFN-alpha in peripheral blood dendritic cells by enveloped RNA and DNA viruses. J. Immunol. 161:2391-2399. [PubMed] [Google Scholar]
- 39.Mocarski, E. S. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10:332-339. [DOI] [PubMed] [Google Scholar]
- 40.Nath, K., J. W. Sarosy, and S. P. Stylianou. 2000. Suitability of a unique 16s Rrna gene PCR product as an indicator of gardnerella vaginalis. BioTechniques 28:222-226. [DOI] [PubMed] [Google Scholar]
- 41.Navarro, D., S. Tugizov, J. La Vail, and L. Pereira. 1993. Glycoprotein B of human cytomegalovirus promotes virion penetration, transmission of infection from cell to cell, and fusion of infected cells. Virology 197:143-158. [DOI] [PubMed]
- 42.Nicoll, S., A. Brass, and H. A. Cubie. 2001. Detection of herpes viruses in clinical samples with real-time PCR. J. Virol. Methods 96:25-31. [DOI] [PubMed] [Google Scholar]
- 43.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, and W. Siegert. 1999. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem. 45:1932-1937. [PubMed] [Google Scholar]
- 44.Norwitz, E. R., D. J. Schust, and S. J. Fisher. 2001. Implantation and the survival of early pregnancy. N. Engl. J. Med. 345:1400-1408. [DOI] [PubMed] [Google Scholar]
- 45.Ossewaarde, J. M., and A. Meijer. 1999. Molecular evidence for the existence of additional members of the order chlamydiales. Microbiolog. 145:411-417. [DOI] [PubMed] [Google Scholar]
- 46.Pass, R. F., E. A. Little, S. Stagno, W. J. Britt, and C. A. Alford. 1987. Young children as a probable source of maternal and congenital cytomegalovirus infection. N. Engl. J. Med. 316:1366-1370. [DOI] [PubMed] [Google Scholar]
- 47.Patterson, S., A. Rae, N. Hockey, J. Gilmour, and F. Gotch. 2001. Plasmacytoid dendritic cells are highly susceptible to human immunodeficiency virus type 1 infection and release infectious virus. J. Virol. 75:6710-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira, L., M. Hoffman, D. Gallo, and N. Cremer. 1982. Monoclonal antibodies to human cytomegalovirus. I. Three cell surface proteins with unique immunologic and electrophoretic properties specify cross-reactive determinants. Infect. Immun. 36:924-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qadri, I., D. Navarro, P. Paz, and L. Pereira. 1992. Assembly of conformation-dependent neutralizing domains on human cytomegalovirus glycoprotein B. J. Gen. Virol. 73:2913-2921. [DOI] [PubMed] [Google Scholar]
- 50.Red-Horse, K., P. M. Drake, M. D. Gunn, and S. J. Fisher. 2001. Chemokine ligand and receptor expression in the pregnant uterus: reciprocal patterns in complementary cell subsets suggest functional roles. Am. J. Pathol. 159:2199-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rescigno, M., M. Urbano, B. Valzasina, M. Francolini, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361-367. [DOI] [PubMed] [Google Scholar]
- 52.Revello, M. G., M. Zavattoni, M. Furione, D. Lilleri, G. Gorini, and G. Gerna. 2002. Diagnosis and outcome of preconceptional and periconceptional primary human cytomegalovirus infections. J. Infect. Dis. 186:553-557. [DOI] [PubMed] [Google Scholar]
- 53.Roth, I., D. B. Corry, R. M. Locksley, J. S. Abrams, M. J. Litton, and S. J. Fisher. 1996. Hum. placental cytotrophoblasts produce the immunosuppressive cytokine interleukin 10. J. Exp. Med. 184:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sacks, G., I. Sargent, and C. Redman. 1999. An innate view of human pregnancy. Immunol. Today 20:114-118. [DOI] [PubMed] [Google Scholar]
- 55.Sallusto, F., M. Cella, C. Danieli, and A. Lanzavecchia. 1995. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 182:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen, C. Y., S. F. Chang, M. S. Yen, H. T. Ng, E. S. Huang, and C. W. Wu. 1993. Cytomegalovirus excretion in pregnant and nonpregnant women. J. Clin. Microbiol. 31:1635-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 284:1835-1837. [DOI] [PubMed] [Google Scholar]
- 58.Simister, N. E., and C. M. Story. 1997. Human placental Fc receptors and the transmission of antibodies from mother to fetus. J. Reprod. Immunol. 37:1-23. [DOI] [PubMed] [Google Scholar]
- 59.Simister, N. E., C. M. Story, H. L. Chen, and J. S. Hunt. 1996. An IgG-transporting Fc receptor expressed in the syncytiotrophoblast of human placenta. Eur. J. Immunol. 26:1527-1531. [DOI] [PubMed] [Google Scholar]
- 60.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 61.Soilleux, E. J., L. S. Morris, B. Lee, S. Pohlmann, J. Trowsdale, R. W. Doms, and N. Coleman. 2001. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J. Pathol. 195:586-592. [DOI] [PubMed] [Google Scholar]
- 62.Stagno, S., D. Reynolds, A. Tsiantos, D. A. Fuccillo, R. Smith, M. Tiller, and C. A. Alford, Jr. 1975. Cervical cytomegalovirus excretion in pregnant and nonpregnant women: suppression in early gestation. J. Infect. Dis. 131:522-527. [DOI] [PubMed] [Google Scholar]
- 63.Stagno, S., D. W. Reynolds, R. F. Pass, and C. A. Alford. 1980. Breast milk and the risk of cytomegalovirus infection. N. Engl. J. Med. 302:1073-1076. [DOI] [PubMed] [Google Scholar]
- 64.Starkey, P. M., I. L. Sargent, and C. W. Redman. 1988. Cell populations in human early pregnancy decidua: characterization and isolation of large granular lymphocytes by flow cytometry. Immunology 65:129-134. [PMC free article] [PubMed] [Google Scholar]
- 65.Sutterwala, F. S., G. J. Noel, P. Salgame, and D. M. Mosser. 1998. Reversal of proinflammatory responses by ligating the macrophage Fcgamma receptor type I. J. Exp. Med. 188:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tran, T., M. J. Flynn, C. Chen, and J. Slots. 1997. Absence of porphyromonas asaccharolytica, bacteroides fragilis and chlamydia pneumoniae in human subgingival plaque. Oral Microbiol. Immunol. 12:377-378. [DOI] [PubMed] [Google Scholar]
- 67.Tugizov, S., E. Maidji, and L. Pereira. 1996. Role of apical and basolateral membranes in replication of human cytomegalovirus in polarized retinal pigment epithelial cells. J. Gen. Virol. 77:61-74. [DOI] [PubMed] [Google Scholar]
- 68.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pohlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975-983. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, Y., S. J. Fisher, M. Janatpour, O. Genbacev, E. Dejana, M. Wheelock, and C. H. Damsky. 1997. Hum. cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J. Clin. Investig. 99:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]