Abstract
Circoviruses are small, nonenveloped icosahedral animal viruses characterized by circular single-stranded DNA genomes. Their genomes are the smallest possessed by animal viruses. Infections with circoviruses, which can lead to economically important diseases, frequently result in virus-induced damage to lymphoid tissue and immunosuppression. Within the family Circoviridae, different genera are distinguished by differences in genomic organization. Thus, Chicken anemia virus is in the genus Gyrovirus, while porcine circoviruses and Beak and feather disease virus belong to the genus Circovirus. Little is known about the structures of circoviruses. Accordingly, we investigated the structures of these three viruses with a view to determining whether they are related. Three-dimensional maps computed from electron micrographs showed that all three viruses have a T=1 organization with capsids formed from 60 subunits. Porcine circovirus type 2 and beak and feather disease virus show similar capsid structures with flat pentameric morphological units, whereas chicken anemia virus has stikingly different protruding pentagonal trumpet-shaped units. It thus appears that the structures of viruses in the same genus are related but that those of viruses in different genera are unrelated.
A number of small isometric animal viruses contain a covalently closed circular single-stranded DNA genome. The circular nature of their genomes, which are the smallest possessed by animal viruses, has led to the family being termed Circoviridae. Within the family, different viruses have differences in genomic organization, leading to their classification in different genera (15). Thus, Porcine circovirus (PCV) and Beak and feather disease virus (BFDV) are classified in the genus Circovirus, while Chicken anemia virus (CAV) is the only member of the genus Gyrovirus.
CAV causes disease in young chickens which is characterized by anemia, lymphoid depletion, and hemorrhaging, with associated increased mortality. The capsid of CAV consists of a single type of protein VP1, which encapsidates a negative-strand genome of about 2,300 bases (19). VP1, which has a molecular mass of 50 kDa, has an extremely basic N-terminal region of about 50 amino acids that is likely to interact with the packaged DNA. The C-terminal region of the protein carries motifs associated with rolling circle replication (RCR) of DNA (10), suggesting that VP1 has both structural and functional roles.
The first identified PCV, now known as PCV type 1 (PCV-1), was isolated as a contaminant of cultured pig kidney cells (17) but was later found to be widespread, although it apparently caused no symptoms in pigs. More recently, a clear disease association was demonstrated between a second PCV (PCV-2) and an economically important disease of pigs known as postweaning multisystemic wasting syndrome (4). The capsid of PCV-2 consists of a single type of protein, encoded by open reading frame 2 (ORF2), which encapsidates an ambisense genome of approximately 1,767 bases. The ORF2-encoded capsid proteins of PCV-1 and PCV-2 are 66% identical (12). Like VP1 of CAV, the ORF2-encoded capsid protein has a very basic N-terminal region expected to interact with the packaged DNA; however, with a molecular mass of only 28 kDa, it lacks a region containing RCR motifs. Instead, the replication function in PCVs is carried out by a separate protein. BFDV has a genomic organization similar to that of PCVs, and the ORF2-encoded capsid protein of BFDV shows 26% identity to that of PCV-2, as determined by use of published sequence alignments (2, 12).
The present study was undertaken to analyze the detailed structures of these recently characterized viruses and to determine their morphological relationships. An earlier study of negatively stained preparations of the three viruses showed CAV to have a larger diameter (19.1 to 26.5 nm) than PCV-1 (16.8 to 20.7 nm) or BFDV (14 to 20.7 nm) (20). CAV also has a more distinctive surface morphology, which was interpreted visually to indicate a T=3 surface lattice with 32 hexamer-pentamer clustered morphological units (8, 11). Here we calculated three-dimensional maps from unstained cryopreserved specimens of CAV and PCV-2 and from a negatively stained specimen of BFDV. We showed that all three viruses have an icosahedral T=1 structure containing 60 capsid protein molecules arranged in 12 pentamer clustered units. Whereas PCV-2 and BFDV have very similar appearances with relatively flat capsomeres, in CAV each capsomere has a very striking pentagonal trumpet-shaped appearance. The results indicate that viruses in the genera Gyrovirus and Circovirus are not structurally related.
MATERIALS AND METHODS
Virus preparation.
The Cuxhaven-1 isolate of CAV was purified following propagation in Marek's disease virus-transformed chicken lymphoblastoid (MDCC-MSB1) cells by a previously described method (19). Briefly, this method involved treatment of sonicated cell lysates with sodium dodecyl sulfate, differential centrifugation, and equilibrium sucrose density gradient centrifugation. Sucrose gradient fractions containing peak amounts of virus, which were identified by an antigen capture enzyme-linked immunosorbent assay (19), were pooled, and virus was sedimented by ultracentifugation. Virus pellets were resuspended in 0.001 M EDTA-0.01 M Tris-HCl (pH 8.7), and suspensions were stored at −80°C prior to electron microscopic examination.
The 1010 isolate of PCV-2 was grown in a continuous porcine kidney cell line (PK/15) (1, 18). The virus was initially purified by lysis of the cells with Triton X-114 and differential centrifugation as described previously (21). Following sucrose density gradient centrifugation, peak virus-containing fractions were identified by an antigen capture enzyme-linked immunosorbent assay and then dialyzed in 0.01 M phosphate-buffered saline (pH 7.2) prior to storage at −80°C until required for electron microscopic studies.
Cryomicroscopy.
The virus samples had a particle concentration of about 109 per ml, which is adequate for conventional negative staining, where the particles are allowed to adsorb to a carbon support. However, cryomicroscopic methods mostly use “in-solution” imaging, where concentrations 1 or 2 orders of magnitude higher are required. Different methods of concentrating the specimens were attempted. High-speed centrifugation resulted in a loss through aggregation in a pellet that could not be resuspended; a 100-kDa-cutoff Microcon concentrator adsorbed the particles even after preblocking with bovine serum albumin (BSA). It was possible to concentrate CAV with a collodion thimble, although contaminating protein was concentrated with the virus.
For CAV, 4-μl samples were applied to air glow-discharged holey carbon films, blotted with Whatman no. 1 filter paper, and frozen in liquid ethane. PCV-2 was applied to holey carbon films with thin carbon across the holes. This method allowed adsorption, as with negative staining, and most of the water was blotted away before freezing. This procedure left virus particles embedded in amorphous ice over thin carbon films supported by the thicker carbon of the holey grid. Low-dose (about 1,000 electrons/nm2) images of CAV were taken with an FEI Tecnai F30 FEG microscope operating at 300 kV in the defocus range of 2.5 to 3.5 μm. Images of PCV-2 were taken with an FEI Tecnai T12 microscope operating at 120 kV in the defocus range of 1 to 4 μm. The grids were kept at liquid nitrogen temperature with side-entry Gatan 626 cold stages. Kodak SO163 film was developed in D19.
Electron microscopic images of negatively stained preparations of mixed CAV and BFDV particles were prepared as described previously (20).
Image processing.
Micrographs (magnification, ×39,000) of CAV were scanned at a sampling of 7 μm on a Z/I SCAI film scanner and then binned to 14 μm, giving a sampling of 0.359 nm/pixel for the specimen. Micrographs (magnification, ×52,000) of PCV-2 were treated similarly, giving a sampling of 0.269 nm/pixel for the specimen. In each case, particles were selected manually by using Ximdisp (6) and then boxed, floated, and scaled to a common mean and standard deviation. Initial orientations and origins were found by self-common lines with icosahedral symmetry (5). Once a preliminary three-dimensional map was available, all further processing was carried out by using cross-common lines against a computed set of projections from the present best three-dimensional map (7). The maps were improved iteratively by these methods.
For CAV images taken with the F30 microscope, there were sufficient particles per micrograph to compute maps uncorrected for defocus to a high resolution for each micrograph. Preliminary estimates of the defocus of each micrograph were made from the positions of the rings of intensity in the incoherent sum of transforms from all of the particles on a particular micrograph. Using these preliminary estimates, maps with different defocus values could be combined to make an initial corrected map (3). Defocus values were then refined by calculating Fourier shell correlations between an uncorrected map from a particular micrograph and the corrected map calculated from all the other micrographs, excluding the particular one under consideration. The resolution of the final map was estimated by computing Fourier shell correlations between defocus-corrected maps computed from two half data sets. The absolute handednessof the CAV structure was determined with previously described computer programs (16) by comparing particle orientations in two images of the same area of specimen, one untilted and the other with the specimen tilted by 10° in a known direction.
RESULTS
Structure of CAV.
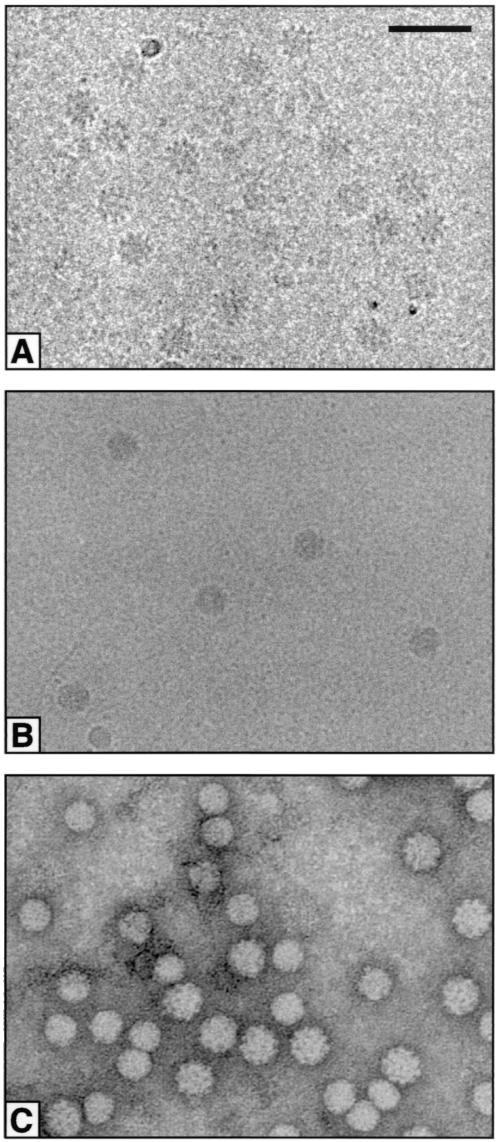
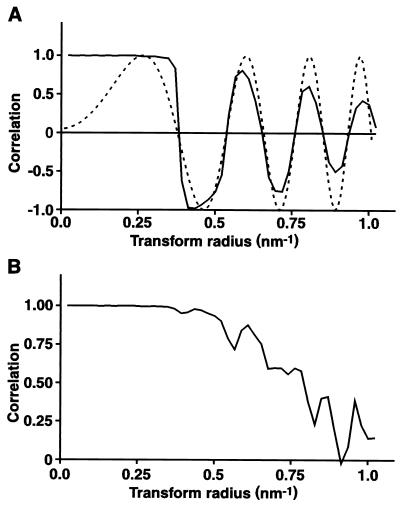
Images of CAV in vitreous ice show a spiky appearance with an overall diameter of about 25 nm (Fig. 1A). The images are of quite low contrast, even when taken at 3 μm under focus, as the accelerating voltage was 300 kV. Images were picked from 11 micrographs, resulting in a total of 1,550 particles, that is, an average of 140 per micrograph. Images were processed on a per-micrograph basis, resulting in a three-dimensional map for each micrograph, uncorrected for defocus. Of the initially picked particles, 85% were included in the final maps, on the criterion of a <55° cross-common line phase residual based on data between spacings of 11.5 and 2.8 nm. An initial map corrected for defocus was then computed by using approximate defocus values obtained from the incoherent sum of transforms of the particles. Fourier shell correlations of the uncorrected map from a particular micrograph versus the corrected map calculated from all of the other micrographs, excluding that particular one, were used to improve the estimates of defocus values (Fig. 2A). The advantage of this approach is that the Fourier shell correlation has sharp zero-crossings, which can be matched easily by the computed phase-contrast transfer function for an appropriate amount of defocus (Fig. 2A). The procedure can be applied iteratively, computing new corrected maps as the improved defocus values are determined, and converges quickly, giving defocus values with a precision of about 10 nm. The resolution of the final map was determined by Fourier shell correlation between corrected maps computed from two half data sets (Fig. 2B). The Fourier shell correlation falls to 0.5 at a spacing of 1.26 nm and to 0.143 at a spacing of 1.11 nm. The latter Fourier shell correlation corresponds to an estimated phase error of 60° in the map computed from all of the data (16).
FIG. 1.
Micrographs of various circoviruses. (A) Cryomicrograph of CAV. (B) Cryomicrograph of PCV-2. (C) Micrograph of a negatively stained preparation of a mixture of CAV and BFDV. The larger, rough particles are CAV, and the smaller, smoother particles are BFDV. Scale bar, 50 nm.
FIG. 2.
Fourier shell correlations. (A) Fourier shell correlation between a map not corrected for defocus and a map combining data from 10 other micrographs corrected for defocus (solid line). The computed phase-contrast transfer function (broken line) for the appropriate level of defocus of the uncorrected map, in this case, 3,500 nm, was calculated on the assumption of 5% amplitude contrast. Note the good agreement between the zero-crossing positions. (B) Fourier shell correlation between maps computed from two half data sets, showing good agreement out to spacings of about 1.2 nm.
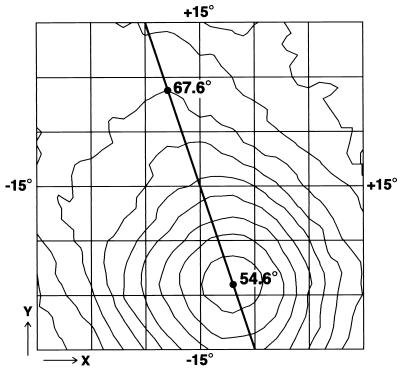
The absolute handedness of the structure was determined from two micrographs of the same area of the specimen, one recorded with zero specimen tilt and the other recorded with a specimen tilt of 10° in a known direction. Pairs of images of 45 particles common to the micrographs of the untilted and tilted specimens were selected. With the three-dimensional map described above, which is of an arbitrary handedness, the program FREALIGN (9) was used to determine, by model-based refinement, the orientations and origins of the untilted particles. Then, using procedures described previously (16), tilt transformations covering all possible directions and tilt angles for the gonioimeter up to ±15° in 1° steps were applied to the orientation of each untilted particle to produce a set of test orientations for the corresponding tilted particle. The phase residual for each of these tilt-transformed orientations was determined for the tilted particle image, and the results were averaged over the 45 selected particles (Fig. 3). With the set of conventions used for this project, if the three-dimensional map were of the correct handedness, the minimum phase residual should occur at −10°, and if the map were of the incorrect handedness, the residual should occur at +10°. The minimum observed phase residual of 54.6° occurs at about a −10° tilt, whereas the residual at a +10° tilt is 67.6° (Fig. 3), indicating that the map (Fig. 4A) is of the correct absolute handedness.
FIG. 3.
Determination of the absolute handedness of CAV. Average phase residuals for 45 particle images recorded at a tilt of 10° were determined by using the three-dimensional model (see Fig. 4A) and tilt-transformed orientation parameters of the corresponding untilted particle images. The contours show phase residuals for tilt transformations up to 15° along the x and y axes. At any point in the plot, the tilt angle (x2+ y2)1/2 is the distance from the origin, and the direction of the tilt axis is arctan(y/x). The direction of the known tilt axis of the goniometer is shown as a diagonal line. The minimum phase residual (54.6°) at a tilt of about −10° around the known tilt axis indicates that the map (see Fig. 4A) is of the correct handedness. The residual (67.6°) at the point corresponding to the opposite handedness is considerably higher, indicating the degree of confidence in the determination.
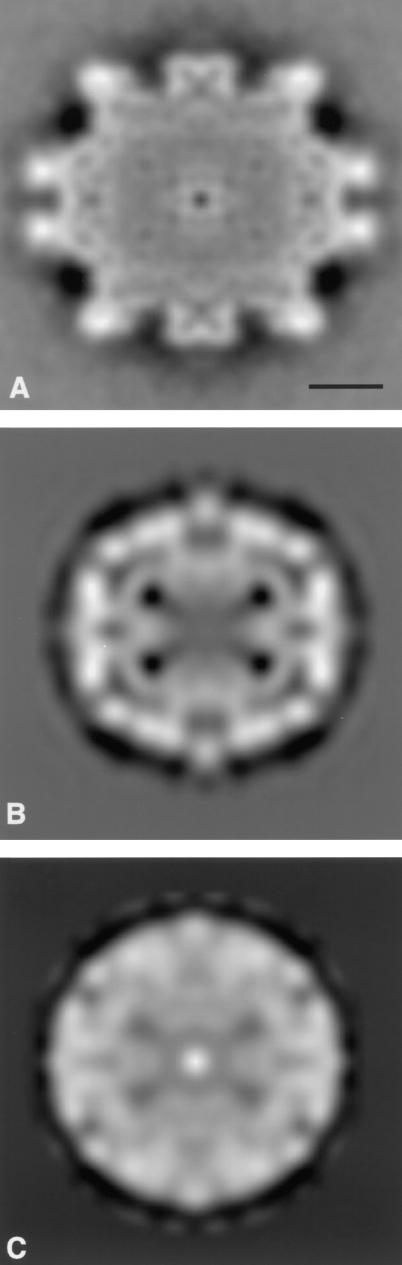
FIG. 4.
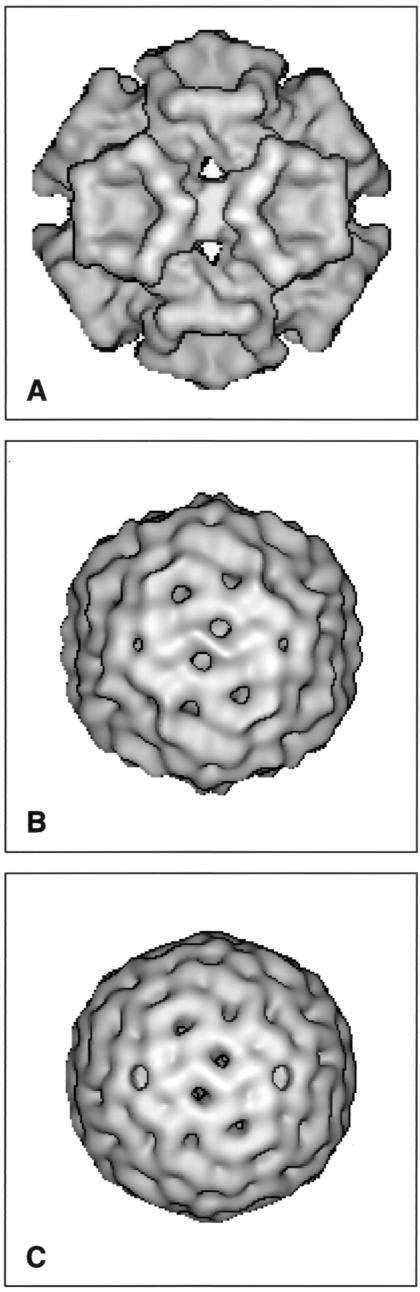
Three-dimensional maps of circoviruses. (A) CAV computed from cryomicrographs. The capsid is formed from 12 pentagonal trumpet-shaped capsomeres, indicating a T=1 surface lattice containing 60 subunits. (B) PCV-2 computed from cryomicrographs. (C) BFDV computed from a micrograph of a negatively stained preparation. The structures in panels B and C are very similar, showing flat pentamer units making contacts across the twofold positions and around the threefold positions and indicating a T=1 surface lattice containing 60 protein subunits in each structure. All maps are viewed along a twofold axis.
The three-dimensional map of CAV shows a capsid structure consisting of 12 pentagonal trumpet-shaped capsomeres (Fig. 4A), indicating a T=1 lattice containing 60 copies of capsid protein VP1. The trumpets extend to an overall radius of about 12.5 nm. The trumpets are joined by bridges across the twofold axes with an outer radius of about 10 nm. The bridges and inner parts of the trumpets give a fairly continuous inner shell, which appears to have small holes at the threefold positions. The outer parts of the trumpets are separated by a gap of about 1.5 nm at their closest approach across the twofold axes. The trumpets display a small degree of handedness, seen chiefly in the ridge of material running down to the twofold bridge. It is also clear that the vertices of neighboring outermost pentagons do not point directly toward each other across the twofold axis but have a small angular displacement (∼5°). The cavities on the fivefold axes extend inward for about 3.5 nm from the plane of the outermost pentagonal surface. Their pentagonal shape means that the wall of the trumpet is about 1.5 to 2.0 nm thick, although modulated into domain-like features. The inner surface of the protein shell is not well defined and appears to merge with disorganized material in the interior of the capsid, as shown in the central section of the map (Fig. 5A).
FIG. 5.
Central sections of the three maps shown in Fig. 4. (A) CAV. (B) PCV-2. (C) BFDV. The sections are normal to a twofold axis. Protein or nucleic acid appears white. Scale bar, 5 nm.
Structure of PCV-2.
Images of PCV-2 (Fig. 1B) have a quite different appearance from those of CAV (Fig. 1A). PCV-2 particles appear smaller and much smoother than CAV particles, but many of the images show a distinctly polygon outline. PCV-2 particles show more contrast, partly because these images were taken at an accelerating voltage of 120 kV. The micrographs also show a fine granular background, because the virus particles had to be captured on thin carbon film over the normal holey carbon support. There were too few particles per micrograph to be able to make high-resolution maps, so no attempt was made to correct for defocusing, the calculations being limited to data within the first zero of the contrast transfer function at 2-nm spacings. A total of 236 particles from eight micrographs was included.
As would be expected from the raw images, the computed three-dimensional map of PCV-2 (Fig. 4B) shows a much less strongly modulated surface than does that of CAV. The morphology can still be described in terms of 12 pentamer units, again indicating a T=1 lattice containing 60 subunits. However, in this case, the pentamer units protrude only slightly, giving an overall diameter of about 20.5 nm. The pentamers protrude sufficiently for projected views of the particle to have a polygonal outline, as noted in the raw images. Material extends from the pentamers to form bridges at an outer radius across the twofold axes and to produce Y-shaped features at the threefold axes. There appear to be small holes in the capsid between the twofold bridges and the threefold Y-shaped features. There also may be small holes at the fivefold axes at the centers sections of the pentamers. The inner surface of the protein shell at a radius of about 7.5 to 8 nm is more clearly delineated in PCV-2 than in CAV, and the capsid has a thickness of about 2.5 nm. There is an inner shell of material, likely to represent DNA, at a radius of about 6.5 nm, and the capsid appears to contact this inner shell under the threefold Y-shaped features (Fig. 5B).
Structure of BFDV.
Negatively stained particles of BFDV (Fig. 1C) look rather smooth and featureless, with an approximately circular outline. Initial orientations and origin positions were found for a few particles by cross-common line refinement against appropriately scaled projections of the PCV-2 map. A map of BFDV was then computed, and the parameters of additional particles were determined by cross-common line refinement against this map. After iteration of this procedure, the final map (Fig. 4C) was made from 72 of the 100 particles initially selected from the micrograph, with data included to 2-nm spacings. The map of BFDV has an appearance very similar to that of PCV-2, with 12 flattened pentamer units making bridges at the twofold positions and Y-shaped contacts around the threefold positions. There appear to be holes in the capsid shell between the twofold bridges and the Y-shaped features and also on the fivefold positions. A small difference between the PCV-2 map and the BFDV map is that the material around the fivefold positions does not project from BFDV quite so much as it does from PCV-2. This finding was clear from the nature of the images, where the PCV-2 particles show a polygonal outline and the BFDV particles look circular, a point emphasized by comparison of the central sections of the two maps (Fig. 5B and C). However, whether this is a genuine difference between the particles or arises from staining of the BFDV preparation remains to be determined.
DISCUSSION
We have presented here the structures of three circoviruses: PCV-2 and BFDV from the genus Circovirus and CAV from the genus Gyrovirus. PCV-2 and BFDV share a very similar genomic organization, with an ambisense genome and with capsid and replication proteins encoded by separate genes. The capsid proteins of PCV-2 and BFDV (molecular mass, 28 kDa) show 26% sequence identity, suggesting that they have similar folding. CAV has a negative-strand genome, and the capsid protein (molecular mass, 50 kDa) is considerably larger than and shows no sequence homology to those of PCV-2 and BFDV. The capsid proteins of all three viruses have a very basic N-terminal region, which is expected to interact with the packaged DNA.
The three-dimensional maps show PCV-2 and BFDV to have a diameter of about 20.5 nm, while the diameter of CAV is about 25 nm. In PCV-2 and BFDV, the capsids show very similar structures, consisting of 12 flat pentameric morphological units, whereas the capsid of CAV consists of 12 pentagonal trumpet-shaped units. Thus, all three viruses have a T = 1 structure containing 60 protein subunits. However, the capsids of the two Circovirus members (PCV-2 and BFDV) are very different from the capsid of the Gyrovirus member (CAV). Previous studies based on visual inspection of micrographs had interpreted images of negatively stained CAV in terms of a T=3 hexamer-pentamer structure (8, 11). The results presented here show that this interpretation was incorrect. Earlier images of BFDV and PCV-1 showed too little detail for interpretation by simple visual inspection to be attempted, but the more sophisticated analysis undertaken here has allowed details of the similar capsid structures of BFDV and PCV-2 to be established.
The capsid protein of CAV contains motifs for RCR in its C-terminal half, suggesting that it has a functional role in replication as well as a structural role in encapsidation. Since the basic N-terminal region of the capsid protein is likely to interact with the packaged DNA, this part of the protein will be inside the capsid. The next part of the polypeptide chain therefore is likely to form the inner shell of the capsid, where contacts between pentamers take place, and the C-terminal half of the polypeptide, containing replication motifs, is likely to form the outer part of the pentagonal trumpets. It would be interesting to know whether the replication function is carried out by polymeric forms of the capsid/replication protein or whether polymeric forms occur only during virion assembly.
The human TT viruses (TTV) also have been shown to possess circular single-stranded DNA genomes (13, 14). The genomic organization of TTV appears to be similar to that of CAV, although very little sequence homology is detectable. As in CAV, the capsid and replication functions of TTV appear to be combined in a single polypeptide which is considerably larger than the corresponding CAV protein but still has a very basic N-terminal region. TTV sizes have been estimated to be in the range of 30 to 50 nm (14). It is thus possible that the capsid of TTV is organized in the same way as that of CAV but with the larger capsid/replication protein yielding a virion with a larger diameter.
Acknowledgments
We thank Peter Rosenthal for advice on determining the absolute handedness of CAV.
REFERENCES
- 1.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clark, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like virus from pigs with a wasting disease in the United States of America and Europe. J. Vet. Diagn. Investig. 10:3-10. [DOI] [PubMed] [Google Scholar]
- 2.Bassami, M. R., D. Berryman, G. E. Wilcox, and S. R. Raidal. 1998. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anemia virus. J. Virol. 249:453-459. [DOI] [PubMed] [Google Scholar]
- 3.Böttcher, B., and R. A. Crowther. 1996. Difference imaging reveals ordered regions of RNA in turnip yellow mosaic virus. Structure 4:387-394. [DOI] [PubMed] [Google Scholar]
- 4.Clark, E. G. 1997. Post weaning multisystemic wasting syndrome, p. 499-501. In Proceedings of the 28th Annual Meeting of the American Association of Swine Practitioners, Quebec City, Quebec, Canada.
- 5.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos. Trans. R. Soc. Lond. B 261:221-230. [DOI] [PubMed] [Google Scholar]
- 6.Crowther, R. A., R. Henderson, and J. S. Smith. 1996. MRC image processing programs. J. Struct. Biol. 166:9-16. [DOI] [PubMed] [Google Scholar]
- 7.Crowther, R. A., N. A. Kiselev, B. Böttcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 8.Gelderblom, H., S. Kling, R. Lurz, I. Tischer, and V. von Bülow. 1989. Morphological characterization of chicken anaemia agent (CAA). Arch. Virol. 109:115-120. [DOI] [PubMed] [Google Scholar]
- 9.Grigorieff, N. 1998. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22Å in ice. J. Mol. Biol. 277:1033-1046. [DOI] [PubMed] [Google Scholar]
- 10.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNulty, M. S., W. L. Curran, D. Todd, and D. P. Mackie. 1990. Chicken anemia agent: an electron microscopic study. Avian Dis. 34:736-743. [PubMed] [Google Scholar]
- 12.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171-2179. [DOI] [PubMed] [Google Scholar]
- 13.Miyata, H., H. Tsunoda, A. Kazi, A. Yamada, M. A. Kahn, J. Murakami, T. Kamahora, S. Shiraki, and S. Hino. 1999. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J. Virol. 73:3582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mushahwar, I. K., J. C. Erker, A. S. Muerhoff, T. P. Leary, J. N. Simons, L. G. Birkenmeyer, M. L. Chalmers, T. J. Pilot-Matias, and S. M. Dexai. 1999. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc. Natl. Acad. Sci. USA 96:3177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 16.Rosenthal, P. B., and R. Henderson. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single particle electron cryomicroscopy. J. Mol. Biol. 333:721-745. [DOI] [PubMed]
- 17.Tischer, I., H. Gelderblom, W. Vetterman, and M. A. Koch. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64-66. [DOI] [PubMed] [Google Scholar]
- 18.Tischer, I., D. Peters, R. Rasch, and S. Pociuli. 1987. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch. Virol. 96:39-57. [DOI] [PubMed] [Google Scholar]
- 19.Todd, D., J. L. Creelan, D. P. Mackie, F. Rixon, and M. S. McNulty. 1990. Purification and biochemical characterization of chicken anaemia agent. J. Gen. Virol. 71:819-823. [DOI] [PubMed] [Google Scholar]
- 20.Todd, D., F. D. Niagro, B. W. Ritchie, W. L. Curran, G. M. Allan, P. D. Lukert, K. S. Latimer, W. L. Steffens III, and M. S. McNulty. 1991. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch. Virol. 117:129-135. [DOI] [PubMed] [Google Scholar]
- 21.Walker, I. W., C. A. Konoby, V. A. Jewhurst, I. McNair, F. McNeilly, B. M. Meehan, T. S. Cottrell, J. A. Ellis, and G. M. Allan. 2000. Development and application of a competitive enzyme-linked immunosorbent assay for the detection of serum antibodies to porcine circovirus type 2. J. Vet. Diagn. Investig. 12:400-405. [DOI] [PubMed] [Google Scholar]