Abstract
In the title compound, [Cu(C10H8N2)(C18H15P)2]NO2·CHCl3·0.5H2O, the Cu atom is tetrahedrally coordinated by a bidentate 2,2′-bipyridine ligand and two PPh3 ligands. The Cu—N and Cu—P distances are similar to those observed in similar compounds. The range of coordination angles shows a moderate distortion from ideal tetrahedral geometry. The bipyridine ligand is twisted [14.2 (4)°] about the ring–ring C—C bond. The nitrate anion and the water and chloroform molecules of solvation are disordered. In the crystal structure, there are O(water)—H⋯O(nitrate), C—H⋯O(water) and C—H⋯O(nitrate) hydrogen bonds.
Related literature
For related literature, see: Allen et al. (1987 ▶); Engelhardt et al. (1985 ▶); Hirshfeld (1976 ▶); Navarro et al. (2003 ▶). 
Experimental
Crystal data
[Cu(C10H8N2)(C18H15P)2]NO3·CHCl3·0.5H2O
M r = 934.65
Triclinic,

a = 10.754 (2) Å
b = 12.672 (3) Å
c = 17.464 (4) Å
α = 99.100 (5)°
β = 99.279 (4)°
γ = 101.229 (5)°
V = 2259.4 (9) Å3
Z = 2
Mo Kα radiation
μ = 0.78 mm−1
T = 296 (2) K
0.56 × 0.51 × 0.40 mm
Data collection
Rigaku AFC7S Mercury diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005 ▶) T min = 0.584, T max = 0.733
25842 measured reflections
8576 independent reflections
6719 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.136
S = 1.06
8576 reflections
549 parameters
20 restraints
H-atom parameters constrained
Δρmax = 0.49 e Å−3
Δρmin = −0.53 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: PLATON (Spek, 2003 ▶) and WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808006260/bg2166sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808006260/bg2166Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Cu1—N1 | 2.070 (2) |
| Cu1—N12 | 2.103 (3) |
| Cu1—P2 | 2.2600 (9) |
| Cu1—P1 | 2.2659 (9) |
| N1—Cu1—N12 | 79.71 (10) |
| N1—Cu1—P2 | 111.18 (8) |
| N12—Cu1—P2 | 111.86 (7) |
| N1—Cu1—P1 | 111.46 (8) |
| N12—Cu1—P1 | 108.68 (7) |
| P2—Cu1—P1 | 124.89 (3) |
| N1—C6—C7—N12 | 14.2 (4) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O3Si | 0.93 | 2.56 | 3.317 (9) | 139 |
| C5—H5⋯O1Wi | 0.93 | 2.34 | 3.182 (15) | 151 |
| C8—H8⋯O2Sii | 0.93 | 2.38 | 3.242 (11) | 155 |
| C45—H45⋯O3S | 0.93 | 2.59 | 3.493 (10) | 163 |
| C1S—H1S⋯O1Siii | 0.98 | 2.25 | 3.204 (7) | 165 |
| O1W⋯O2Siv | 2.663 (19) | |||
| O1W⋯O2Sv | 2.667 (19) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv) 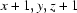 ; (v)
; (v)  .
.
Acknowledgments
Financial support from the Fondo Nacional de Ciencia, Tecnología e Investigación (FONACIT) of Venezuela, project Lab-199700821, is gratefully acknowledged.
supplementary crystallographic information
Comment
The title compound (I) was prepared within our program of studies of copper complexes containing N-bidentated aromatic ligands, focused on the search for drugs with biological activity, especially against parasitic diseases (Navarro et al., 2003).
The structure analysis showed that, in addition to the complex cation and the (partially disordered) nitrate anion the crystals contain disordered molecules of water and chloroform of solvation.
In the cation (Fig. 1), the metal atom is tetrahedrally coordinated to a bidentated 2,2'-bipyridine (bipy) ligand and to two PPh3 moieties. The Cu—N and Cu—P (Table 1) distances are comparable to Cu—N, 2.056 (8), 2.113 (9) Å and Cu—P, 2.246 (3), 2.256 (3) Å observed in the same cation in [Cu(PPh3)2(bipy)]ClO4 (Engelhardt et al., 1985). All bond lengths and angles in the organic ligands are within normal values (Allen et al., 1987).
The range of coordination angles (Table 1) shows a moderate distortion from the ideal tetrahedral geometry. As expected, the bite angle is the smaller one, while the P—Cu—P angle is the largest, due to the bulkiness of the PPh3 moieties. The five-member metallacycle, Cu1—N1—C6—C7—N2, can be described as an envelope on C6, albeit a flat one [C6 is 0.148 (4) Å out of the plane]. The bipy ligand is twisted about C6—C7 (Table 1); the dihedral angle between both heterocycles is 17.0 (2)°.
Although the water's H atoms could not be located (see Refinement Section), short contacts with the nitrate anion [O1w···O2s(1 + x, y, z), 2.663 (19) Å; O1w···O2s(1 - x, 2 - y, -z), 2.667 (19) Å] indicate hydrogen bonds between these two groups. Further evidence of the feasibility of these links is given by the corresponding O2s···O1w···O2s angle [146.7 (5)°]. Several C—H···O bonds may also contribute to the crystal packing (Fig. 2 and Supplementary material).
Experimental
The title compound (I) was synthesized by the reaction of copper nitrate with bipyridine. To a solution of Cu(PPh3)2NO3 (100 mg, 0.15 mmol) in dichloromethane (10 ml) was added 2,2'-bipyridine (24 mg, 0.15 mmol). The solution, initially transparent, became yellow. It was stirred for 1 h at room temperature and then was added to hexane (50 ml). The light yellow solid formed was filtered and dried (122 mg, 98%). All operations were carried out under inert atmosphere. Crystals suitable for X-ray analysis were obtained by slow evaporation of a chloroform solution.
Refinement
The hydrogen atoms were placed in calculated positions using a riding atom model with fixed C—H distances [0.93 Å for C(sp2), 0.98 Å for C(sp3) in CHCl3] and Uiso = 1.2 Ueq(parent atom).
The nitrate anion and the water and chloroform molecules of solvation were found to be disordered. The NO3- showed severe disorder, difficult to model satisfactorily; in the final refinement the four largest residual electron density peaks were close (0.64–1.07 e/Å3) to NO3 atoms. O2s and O3s were split in two positions, with complementary occupancies, and refined isotropically to final occupancies of 0.505 (14) and 0.719 (12) respectively. To obtain better geometries, restraints were applied: SADI to all N—O bonds and to all O···O distances, and FLAT to both NO3 groups. An attempt at splitting N1s, as suggested by its elongated ADP, gave meaningless results. The O1w atom of the water molecule was given an occupancy of 1/2, since it is disordered between two centrosymmetrically related positions, which are mutually exclusive [O1w···O1w(2 - x, 2 - y, -x), 1.53 (3) Å]. The corresponding H atoms could not be found. Each of the Cl atoms of chloroform was split in two alternative positions, with complementary occupancies. The main positions [final occupancy 0.911 (6) for all three] were refined anisotropically, while the alternative ones were given isotropic displacement parameters.
Both Cu—P bonds, with ΔU/σ = 7.21 for Cu1—P1 and 7.13 for Cu1—P2, failed to pass the standard rigid-bond test (e.g.: ΔU/σ≤ 5, Spek, 1998; Hirshfeld, 1976) even after applying a DELU restraint (Sheldrick, 2008). This was probably due to an unfavorable specimen morphology which caused a poor absorption correction.
Figures
Fig. 1.
Structure of the complex cation [Cu(PPh3)2(bipy)]+ showing the atomic numbering. Displacement parameters are drawn at 50% probability level.
Fig. 2.
Crystal structure of (I). For clarity, alternative positions for disordered atoms and H atoms were omitted. Possible hydrogen bonds are shown as dashed lines.
Crystal data
| [Cu(C10H8N2)(C18H15P)2]NO3·CHCl3·0.5H2O | Z = 2 |
| Mr = 934.65 | F000 = 962 |
| Triclinic, P1 | Dx = 1.374 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71070 Å |
| a = 10.754 (2) Å | Cell parameters from 15269 reflections |
| b = 12.672 (3) Å | θ = 1.7–28.0º |
| c = 17.464 (4) Å | µ = 0.78 mm−1 |
| α = 99.100 (5)º | T = 296 (2) K |
| β = 99.279 (4)º | Irregular, green |
| γ = 101.229 (5)º | 0.56 × 0.51 × 0.40 mm |
| V = 2259.4 (9) Å3 |
Data collection
| Rigaku AFC7S Mercury diffractometer | 8576 independent reflections |
| Radiation source: fine-focus sealed tube | 6719 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.031 |
| Detector resolution: 14.63 pixels mm-1 | θmax = 28.0º |
| T = 296(2) K | θmin = 1.2º |
| ω scans | h = −13→13 |
| Absorption correction: multi-scan(CrystalClear; Rigaku/MSC, 2005) | k = −16→15 |
| Tmin = 0.584, Tmax = 0.733 | l = −21→21 |
| 25842 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters constrained |
| wR(F2) = 0.136 | w = 1/[σ2(Fo2) + (0.0572P)2 + 1.3813P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 8576 reflections | Δρmax = 0.49 e Å−3 |
| 549 parameters | Δρmin = −0.52 e Å−3 |
| 20 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. Refinement details for disordered atoms are given in the Refimnement section. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cu1 | 0.56159 (3) | 0.26669 (3) | 0.22887 (2) | 0.04336 (12) | |
| P1 | 0.40007 (8) | 0.34553 (6) | 0.18315 (5) | 0.04467 (19) | |
| P2 | 0.61841 (7) | 0.24899 (6) | 0.35575 (4) | 0.04036 (18) | |
| N1 | 0.5630 (3) | 0.1260 (2) | 0.15089 (15) | 0.0517 (6) | |
| C2 | 0.4854 (4) | 0.0263 (3) | 0.1418 (2) | 0.0661 (10) | |
| H2 | 0.4128 | 0.0195 | 0.1648 | 0.079* | |
| C3 | 0.5102 (5) | −0.0673 (3) | 0.0989 (3) | 0.0839 (13) | |
| H3 | 0.4550 | −0.1356 | 0.0934 | 0.101* | |
| C4 | 0.6161 (6) | −0.0569 (4) | 0.0654 (3) | 0.0951 (15) | |
| H4 | 0.6346 | −0.1187 | 0.0372 | 0.114* | |
| C5 | 0.6960 (5) | 0.0441 (4) | 0.0730 (2) | 0.0810 (12) | |
| H5 | 0.7687 | 0.0517 | 0.0500 | 0.097* | |
| C6 | 0.6661 (3) | 0.1351 (3) | 0.11567 (19) | 0.0555 (8) | |
| C7 | 0.7456 (3) | 0.2477 (3) | 0.12513 (19) | 0.0549 (8) | |
| C8 | 0.8316 (4) | 0.2757 (4) | 0.0758 (2) | 0.0813 (12) | |
| H8 | 0.8441 | 0.2222 | 0.0367 | 0.098* | |
| C9 | 0.8968 (4) | 0.3815 (5) | 0.0854 (3) | 0.0913 (14) | |
| H9 | 0.9547 | 0.4007 | 0.0531 | 0.110* | |
| C10 | 0.8769 (4) | 0.4593 (4) | 0.1426 (3) | 0.0830 (13) | |
| H10 | 0.9203 | 0.5322 | 0.1495 | 0.100* | |
| C11 | 0.7911 (3) | 0.4281 (3) | 0.1902 (2) | 0.0625 (9) | |
| H11 | 0.7791 | 0.4811 | 0.2299 | 0.075* | |
| N12 | 0.7239 (2) | 0.3238 (2) | 0.18134 (15) | 0.0496 (6) | |
| C21 | 0.3703 (3) | 0.3373 (3) | 0.07633 (18) | 0.0506 (7) | |
| C22 | 0.2493 (4) | 0.3103 (3) | 0.0274 (2) | 0.0727 (10) | |
| H22 | 0.1753 | 0.2938 | 0.0484 | 0.087* | |
| C23 | 0.2388 (5) | 0.3081 (4) | −0.0532 (2) | 0.0900 (14) | |
| H23 | 0.1573 | 0.2884 | −0.0859 | 0.108* | |
| C24 | 0.3442 (5) | 0.3339 (3) | −0.0849 (2) | 0.0841 (13) | |
| H24 | 0.3351 | 0.3340 | −0.1387 | 0.101* | |
| C25 | 0.4636 (5) | 0.3597 (3) | −0.0381 (2) | 0.0769 (11) | |
| H25 | 0.5364 | 0.3766 | −0.0601 | 0.092* | |
| C26 | 0.4774 (4) | 0.3610 (3) | 0.0419 (2) | 0.0605 (9) | |
| H26 | 0.5599 | 0.3780 | 0.0733 | 0.073* | |
| C31 | 0.2417 (3) | 0.2894 (3) | 0.20327 (19) | 0.0506 (7) | |
| C32 | 0.1871 (3) | 0.1791 (3) | 0.1751 (2) | 0.0676 (10) | |
| H32 | 0.2279 | 0.1370 | 0.1431 | 0.081* | |
| C33 | 0.0709 (4) | 0.1305 (4) | 0.1943 (3) | 0.0825 (12) | |
| H33 | 0.0345 | 0.0563 | 0.1753 | 0.099* | |
| C34 | 0.0109 (4) | 0.1929 (5) | 0.2412 (3) | 0.0854 (14) | |
| H34 | −0.0662 | 0.1608 | 0.2544 | 0.103* | |
| C35 | 0.0637 (4) | 0.3019 (4) | 0.2687 (3) | 0.0831 (13) | |
| H35 | 0.0221 | 0.3438 | 0.3001 | 0.100* | |
| C36 | 0.1787 (3) | 0.3507 (3) | 0.2502 (2) | 0.0633 (9) | |
| H36 | 0.2140 | 0.4251 | 0.2694 | 0.076* | |
| C41 | 0.4297 (3) | 0.4929 (2) | 0.22167 (19) | 0.0509 (7) | |
| C42 | 0.5256 (4) | 0.5401 (3) | 0.2877 (2) | 0.0698 (10) | |
| H42 | 0.5735 | 0.4964 | 0.3119 | 0.084* | |
| C43 | 0.5515 (5) | 0.6512 (4) | 0.3184 (3) | 0.0949 (15) | |
| H43 | 0.6155 | 0.6819 | 0.3633 | 0.114* | |
| C44 | 0.4819 (5) | 0.7157 (4) | 0.2822 (3) | 0.0934 (14) | |
| H44 | 0.5004 | 0.7907 | 0.3022 | 0.112* | |
| C45 | 0.3853 (5) | 0.6713 (3) | 0.2168 (3) | 0.0797 (12) | |
| H45 | 0.3376 | 0.7158 | 0.1934 | 0.096* | |
| C46 | 0.3594 (4) | 0.5600 (3) | 0.1862 (2) | 0.0618 (9) | |
| H46 | 0.2947 | 0.5298 | 0.1417 | 0.074* | |
| C51 | 0.4918 (3) | 0.1884 (2) | 0.40385 (17) | 0.0422 (6) | |
| C52 | 0.3641 (3) | 0.1788 (3) | 0.3692 (2) | 0.0585 (9) | |
| H52 | 0.3439 | 0.2004 | 0.3211 | 0.070* | |
| C53 | 0.2666 (4) | 0.1371 (4) | 0.4059 (3) | 0.0776 (12) | |
| H53 | 0.1810 | 0.1313 | 0.3824 | 0.093* | |
| C54 | 0.2936 (4) | 0.1043 (3) | 0.4756 (2) | 0.0666 (10) | |
| H54 | 0.2268 | 0.0769 | 0.4999 | 0.080* | |
| C55 | 0.4182 (4) | 0.1115 (3) | 0.5100 (2) | 0.0630 (9) | |
| H55 | 0.4366 | 0.0882 | 0.5576 | 0.076* | |
| C56 | 0.5182 (3) | 0.1533 (3) | 0.4745 (2) | 0.0583 (8) | |
| H56 | 0.6034 | 0.1578 | 0.4983 | 0.070* | |
| C61 | 0.6926 (3) | 0.3802 (2) | 0.42251 (17) | 0.0446 (7) | |
| C62 | 0.8032 (3) | 0.4444 (3) | 0.4071 (2) | 0.0597 (9) | |
| H62 | 0.8394 | 0.4181 | 0.3652 | 0.072* | |
| C63 | 0.8595 (4) | 0.5465 (3) | 0.4535 (3) | 0.0743 (11) | |
| H63 | 0.9344 | 0.5877 | 0.4434 | 0.089* | |
| C64 | 0.8059 (4) | 0.5877 (3) | 0.5145 (3) | 0.0793 (12) | |
| H64 | 0.8435 | 0.6570 | 0.5453 | 0.095* | |
| C65 | 0.6963 (4) | 0.5260 (3) | 0.5297 (2) | 0.0742 (11) | |
| H65 | 0.6595 | 0.5539 | 0.5708 | 0.089* | |
| C66 | 0.6399 (3) | 0.4228 (3) | 0.4846 (2) | 0.0575 (8) | |
| H66 | 0.5660 | 0.3817 | 0.4960 | 0.069* | |
| C71 | 0.7379 (3) | 0.1653 (2) | 0.37355 (18) | 0.0444 (7) | |
| C72 | 0.7141 (3) | 0.0629 (3) | 0.3242 (2) | 0.0583 (8) | |
| H72 | 0.6405 | 0.0402 | 0.2843 | 0.070* | |
| C73 | 0.7991 (4) | −0.0051 (3) | 0.3339 (2) | 0.0678 (10) | |
| H73 | 0.7824 | −0.0733 | 0.3005 | 0.081* | |
| C74 | 0.9088 (3) | 0.0279 (3) | 0.3931 (2) | 0.0657 (10) | |
| H74 | 0.9666 | −0.0173 | 0.3993 | 0.079* | |
| C75 | 0.9312 (3) | 0.1275 (3) | 0.4421 (2) | 0.0645 (9) | |
| H75 | 1.0039 | 0.1494 | 0.4827 | 0.077* | |
| C76 | 0.8469 (3) | 0.1965 (3) | 0.4323 (2) | 0.0558 (8) | |
| H76 | 0.8643 | 0.2646 | 0.4659 | 0.067* | |
| C1S | −0.0946 (4) | 0.2653 (3) | 0.6980 (3) | 0.0822 (12) | |
| H1S | −0.1093 | 0.2748 | 0.7524 | 0.099* | |
| Cl1S | 0.07297 (19) | 0.27094 (17) | 0.69940 (16) | 0.1137 (7) | 0.911 (6) |
| Cl2S | −0.18878 (19) | 0.13990 (18) | 0.64402 (13) | 0.1167 (7) | 0.911 (6) |
| Cl3S | −0.1396 (2) | 0.37078 (17) | 0.65417 (17) | 0.1141 (9) | 0.911 (6) |
| Cl1T | 0.0416 (19) | 0.2325 (18) | 0.7246 (11) | 0.094 (6)* | 0.089 (6) |
| Cl2T | −0.162 (2) | 0.191 (3) | 0.6233 (15) | 0.130 (8)* | 0.089 (6) |
| Cl3T | −0.0968 (14) | 0.3998 (12) | 0.7051 (13) | 0.083 (5)* | 0.089 (6) |
| N1S | 0.1383 (10) | 0.8269 (6) | 0.1199 (3) | 0.157 (3) | |
| O1S | 0.1002 (5) | 0.7275 (5) | 0.1184 (3) | 0.1440 (16) | |
| O2S | 0.0559 (12) | 0.8453 (8) | 0.0668 (6) | 0.146 (5)* | 0.505 (14) |
| O2S' | 0.1575 (9) | 0.9082 (8) | 0.0846 (5) | 0.123 (4)* | 0.495 (14) |
| O3S | 0.2184 (9) | 0.8712 (7) | 0.1718 (6) | 0.182 (4)* | 0.719 (12) |
| O3S' | 0.0415 (18) | 0.8635 (15) | 0.1616 (12) | 0.162 (9)* | 0.281 (12) |
| O1W | 0.9283 (12) | 0.9778 (13) | −0.0008 (10) | 0.240 (6)* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0457 (2) | 0.0458 (2) | 0.0419 (2) | 0.01544 (16) | 0.01346 (16) | 0.00713 (15) |
| P1 | 0.0477 (4) | 0.0513 (4) | 0.0405 (4) | 0.0202 (3) | 0.0129 (3) | 0.0096 (3) |
| P2 | 0.0363 (4) | 0.0465 (4) | 0.0409 (4) | 0.0123 (3) | 0.0109 (3) | 0.0093 (3) |
| N1 | 0.0652 (17) | 0.0463 (14) | 0.0434 (15) | 0.0186 (13) | 0.0071 (13) | 0.0042 (11) |
| C2 | 0.080 (2) | 0.052 (2) | 0.059 (2) | 0.0102 (18) | 0.0028 (19) | 0.0029 (16) |
| C3 | 0.116 (4) | 0.049 (2) | 0.074 (3) | 0.016 (2) | −0.004 (3) | 0.0004 (18) |
| C4 | 0.136 (4) | 0.068 (3) | 0.084 (3) | 0.048 (3) | 0.018 (3) | −0.006 (2) |
| C5 | 0.109 (3) | 0.081 (3) | 0.065 (3) | 0.052 (3) | 0.027 (2) | 0.003 (2) |
| C6 | 0.070 (2) | 0.062 (2) | 0.0399 (18) | 0.0328 (17) | 0.0117 (16) | 0.0060 (14) |
| C7 | 0.0579 (19) | 0.070 (2) | 0.0459 (19) | 0.0287 (17) | 0.0176 (15) | 0.0137 (15) |
| C8 | 0.089 (3) | 0.107 (3) | 0.067 (3) | 0.040 (3) | 0.042 (2) | 0.022 (2) |
| C9 | 0.082 (3) | 0.117 (4) | 0.090 (3) | 0.019 (3) | 0.045 (3) | 0.041 (3) |
| C10 | 0.073 (3) | 0.085 (3) | 0.097 (3) | 0.005 (2) | 0.026 (2) | 0.044 (3) |
| C11 | 0.059 (2) | 0.059 (2) | 0.069 (2) | 0.0074 (17) | 0.0163 (18) | 0.0153 (17) |
| N12 | 0.0491 (14) | 0.0550 (15) | 0.0500 (16) | 0.0185 (12) | 0.0147 (12) | 0.0125 (12) |
| C21 | 0.062 (2) | 0.0529 (17) | 0.0414 (18) | 0.0228 (15) | 0.0111 (15) | 0.0106 (13) |
| C22 | 0.068 (2) | 0.101 (3) | 0.057 (2) | 0.035 (2) | 0.0095 (19) | 0.023 (2) |
| C23 | 0.095 (3) | 0.121 (4) | 0.052 (3) | 0.034 (3) | −0.010 (2) | 0.021 (2) |
| C24 | 0.128 (4) | 0.083 (3) | 0.049 (2) | 0.035 (3) | 0.015 (3) | 0.022 (2) |
| C25 | 0.105 (3) | 0.077 (3) | 0.059 (2) | 0.023 (2) | 0.031 (2) | 0.027 (2) |
| C26 | 0.071 (2) | 0.067 (2) | 0.049 (2) | 0.0191 (18) | 0.0166 (17) | 0.0155 (16) |
| C31 | 0.0467 (17) | 0.063 (2) | 0.0479 (19) | 0.0194 (15) | 0.0093 (14) | 0.0190 (15) |
| C32 | 0.058 (2) | 0.078 (3) | 0.066 (2) | 0.0139 (19) | 0.0105 (18) | 0.0127 (19) |
| C33 | 0.063 (2) | 0.086 (3) | 0.091 (3) | −0.003 (2) | 0.005 (2) | 0.031 (2) |
| C34 | 0.053 (2) | 0.132 (4) | 0.084 (3) | 0.020 (3) | 0.021 (2) | 0.051 (3) |
| C35 | 0.071 (3) | 0.123 (4) | 0.080 (3) | 0.044 (3) | 0.037 (2) | 0.042 (3) |
| C36 | 0.061 (2) | 0.080 (2) | 0.063 (2) | 0.0306 (19) | 0.0269 (18) | 0.0231 (18) |
| C41 | 0.0577 (19) | 0.0502 (17) | 0.0527 (19) | 0.0214 (15) | 0.0208 (15) | 0.0117 (14) |
| C42 | 0.079 (3) | 0.060 (2) | 0.066 (2) | 0.0253 (19) | 0.006 (2) | −0.0021 (18) |
| C43 | 0.106 (4) | 0.073 (3) | 0.090 (3) | 0.025 (3) | 0.001 (3) | −0.015 (2) |
| C44 | 0.124 (4) | 0.058 (2) | 0.098 (4) | 0.025 (3) | 0.036 (3) | −0.002 (2) |
| C45 | 0.110 (3) | 0.066 (2) | 0.091 (3) | 0.045 (2) | 0.053 (3) | 0.031 (2) |
| C46 | 0.072 (2) | 0.063 (2) | 0.063 (2) | 0.0302 (18) | 0.0256 (18) | 0.0188 (17) |
| C51 | 0.0431 (15) | 0.0408 (15) | 0.0455 (17) | 0.0129 (12) | 0.0112 (13) | 0.0100 (12) |
| C52 | 0.0426 (17) | 0.075 (2) | 0.063 (2) | 0.0106 (16) | 0.0104 (15) | 0.0297 (18) |
| C53 | 0.0441 (19) | 0.102 (3) | 0.095 (3) | 0.010 (2) | 0.022 (2) | 0.043 (3) |
| C54 | 0.061 (2) | 0.067 (2) | 0.081 (3) | 0.0106 (18) | 0.035 (2) | 0.0257 (19) |
| C55 | 0.077 (3) | 0.066 (2) | 0.054 (2) | 0.0147 (19) | 0.0265 (19) | 0.0225 (17) |
| C56 | 0.0505 (18) | 0.075 (2) | 0.053 (2) | 0.0146 (17) | 0.0109 (15) | 0.0230 (17) |
| C61 | 0.0445 (16) | 0.0490 (16) | 0.0411 (16) | 0.0137 (13) | 0.0068 (13) | 0.0091 (13) |
| C62 | 0.0518 (19) | 0.064 (2) | 0.059 (2) | 0.0031 (16) | 0.0134 (16) | 0.0092 (16) |
| C63 | 0.065 (2) | 0.064 (2) | 0.081 (3) | −0.0072 (19) | 0.009 (2) | 0.009 (2) |
| C64 | 0.083 (3) | 0.056 (2) | 0.082 (3) | 0.003 (2) | 0.001 (2) | −0.003 (2) |
| C65 | 0.081 (3) | 0.065 (2) | 0.071 (3) | 0.022 (2) | 0.016 (2) | −0.0115 (19) |
| C66 | 0.0544 (19) | 0.0541 (18) | 0.063 (2) | 0.0131 (15) | 0.0170 (16) | 0.0014 (15) |
| C71 | 0.0399 (15) | 0.0529 (17) | 0.0478 (18) | 0.0174 (13) | 0.0156 (13) | 0.0164 (13) |
| C72 | 0.0558 (19) | 0.0559 (19) | 0.065 (2) | 0.0204 (16) | 0.0077 (17) | 0.0125 (16) |
| C73 | 0.079 (3) | 0.056 (2) | 0.078 (3) | 0.0294 (19) | 0.022 (2) | 0.0162 (18) |
| C74 | 0.055 (2) | 0.071 (2) | 0.093 (3) | 0.0326 (18) | 0.030 (2) | 0.041 (2) |
| C75 | 0.0427 (18) | 0.080 (3) | 0.079 (3) | 0.0223 (17) | 0.0105 (17) | 0.030 (2) |
| C76 | 0.0440 (17) | 0.064 (2) | 0.061 (2) | 0.0180 (15) | 0.0082 (15) | 0.0113 (16) |
| C1S | 0.106 (3) | 0.072 (3) | 0.070 (3) | 0.032 (2) | 0.015 (2) | 0.008 (2) |
| Cl1S | 0.1039 (12) | 0.0881 (11) | 0.1527 (17) | 0.0357 (9) | 0.0224 (11) | 0.0201 (11) |
| Cl2S | 0.1263 (13) | 0.0855 (12) | 0.1194 (14) | 0.0151 (10) | 0.0038 (10) | −0.0035 (10) |
| Cl3S | 0.1557 (17) | 0.1091 (12) | 0.123 (2) | 0.0812 (12) | 0.0695 (16) | 0.0501 (12) |
| N1S | 0.281 (9) | 0.163 (6) | 0.063 (3) | 0.120 (6) | 0.025 (5) | 0.049 (4) |
| O1S | 0.138 (4) | 0.184 (5) | 0.110 (3) | 0.059 (4) | 0.013 (3) | 0.012 (3) |
Geometric parameters (Å, °)
| Cu1—N1 | 2.070 (2) | C42—C43 | 1.381 (5) |
| Cu1—N12 | 2.103 (3) | C42—H42 | 0.9300 |
| Cu1—P2 | 2.2600 (9) | C43—C44 | 1.370 (7) |
| Cu1—P1 | 2.2659 (9) | C43—H43 | 0.9300 |
| P1—C21 | 1.824 (3) | C44—C45 | 1.373 (6) |
| P1—C31 | 1.825 (3) | C44—H44 | 0.9300 |
| P1—C41 | 1.828 (3) | C45—C46 | 1.383 (5) |
| P2—C51 | 1.821 (3) | C45—H45 | 0.9300 |
| P2—C61 | 1.825 (3) | C46—H46 | 0.9300 |
| P2—C71 | 1.839 (3) | C51—C52 | 1.382 (4) |
| N1—C2 | 1.341 (4) | C51—C56 | 1.382 (4) |
| N1—C6 | 1.347 (4) | C52—C53 | 1.377 (5) |
| C2—C3 | 1.395 (5) | C52—H52 | 0.9300 |
| C2—H2 | 0.9300 | C53—C54 | 1.353 (5) |
| C3—C4 | 1.356 (7) | C53—H53 | 0.9300 |
| C3—H3 | 0.9300 | C54—C55 | 1.357 (5) |
| C4—C5 | 1.370 (6) | C54—H54 | 0.9300 |
| C4—H4 | 0.9300 | C55—C56 | 1.385 (5) |
| C5—C6 | 1.393 (5) | C55—H55 | 0.9300 |
| C5—H5 | 0.9300 | C56—H56 | 0.9300 |
| C6—C7 | 1.483 (5) | C61—C66 | 1.386 (4) |
| C7—N12 | 1.345 (4) | C61—C62 | 1.394 (4) |
| C7—C8 | 1.396 (5) | C62—C63 | 1.379 (5) |
| C8—C9 | 1.357 (6) | C62—H62 | 0.9300 |
| C8—H8 | 0.9300 | C63—C64 | 1.371 (6) |
| C9—C10 | 1.362 (7) | C63—H63 | 0.9300 |
| C9—H9 | 0.9300 | C64—C65 | 1.370 (6) |
| C10—C11 | 1.382 (5) | C64—H64 | 0.9300 |
| C10—H10 | 0.9300 | C65—C66 | 1.382 (5) |
| C11—N12 | 1.349 (4) | C65—H65 | 0.9300 |
| C11—H11 | 0.9300 | C66—H66 | 0.9300 |
| C21—C22 | 1.384 (5) | C71—C76 | 1.373 (4) |
| C21—C26 | 1.388 (5) | C71—C72 | 1.392 (4) |
| C22—C23 | 1.389 (5) | C72—C73 | 1.382 (5) |
| C22—H22 | 0.9300 | C72—H72 | 0.9300 |
| C23—C24 | 1.347 (6) | C73—C74 | 1.383 (5) |
| C23—H23 | 0.9300 | C73—H73 | 0.9300 |
| C24—C25 | 1.355 (6) | C74—C75 | 1.363 (5) |
| C24—H24 | 0.9300 | C74—H74 | 0.9300 |
| C25—C26 | 1.378 (5) | C75—C76 | 1.386 (5) |
| C25—H25 | 0.9300 | C75—H75 | 0.9300 |
| C26—H26 | 0.9300 | C76—H76 | 0.9300 |
| C31—C32 | 1.382 (5) | C1S—H1S | 0.9800 |
| C31—C36 | 1.384 (5) | C1S—Cl1S | 1.786 (5) |
| C32—C33 | 1.398 (5) | C1S—Cl2S | 1.741 (4) |
| C32—H32 | 0.9300 | C1S—Cl3S | 1.750 (4) |
| C33—C34 | 1.371 (6) | C1S—Cl1t | 1.617 (19) |
| C33—H33 | 0.9300 | C1S—Cl2t | 1.47 (3) |
| C34—C35 | 1.363 (6) | C1S—Cl3t | 1.694 (15) |
| C34—H34 | 0.9300 | N1S—O3S | 1.131 (10) |
| C35—C36 | 1.382 (5) | N1S—O1S | 1.241 (8) |
| C35—H35 | 0.9300 | N1S—O2S | 1.259 (12) |
| C36—H36 | 0.9300 | N1S—O2S' | 1.282 (10) |
| C41—C42 | 1.382 (5) | N1S—O3S' | 1.47 (2) |
| C41—C46 | 1.394 (5) | ||
| N1—Cu1—N12 | 79.71 (10) | C42—C41—C46 | 118.5 (3) |
| N1—Cu1—P2 | 111.18 (8) | C42—C41—P1 | 119.2 (3) |
| N12—Cu1—P2 | 111.86 (7) | C46—C41—P1 | 122.2 (3) |
| N1—Cu1—P1 | 111.46 (8) | C43—C42—C41 | 121.0 (4) |
| N12—Cu1—P1 | 108.68 (7) | C43—C42—H42 | 119.5 |
| P2—Cu1—P1 | 124.89 (3) | C41—C42—H42 | 119.5 |
| C21—P1—C31 | 104.14 (15) | C44—C43—C42 | 119.4 (4) |
| C21—P1—C41 | 102.77 (14) | C44—C43—H43 | 120.3 |
| C31—P1—C41 | 104.43 (15) | C42—C43—H43 | 120.3 |
| C21—P1—Cu1 | 114.07 (11) | C43—C44—C45 | 121.1 (4) |
| C31—P1—Cu1 | 115.68 (10) | C43—C44—H44 | 119.5 |
| C41—P1—Cu1 | 114.29 (11) | C45—C44—H44 | 119.5 |
| C51—P2—C61 | 103.12 (13) | C44—C45—C46 | 119.4 (4) |
| C51—P2—C71 | 101.93 (13) | C44—C45—H45 | 120.3 |
| C61—P2—C71 | 103.69 (14) | C46—C45—H45 | 120.3 |
| C51—P2—Cu1 | 118.06 (10) | C45—C46—C41 | 120.5 (4) |
| C61—P2—Cu1 | 112.74 (10) | C45—C46—H46 | 119.7 |
| C71—P2—Cu1 | 115.48 (10) | C41—C46—H46 | 119.7 |
| C2—N1—C6 | 118.3 (3) | C52—C51—C56 | 118.5 (3) |
| C2—N1—Cu1 | 127.0 (2) | C52—C51—P2 | 118.9 (2) |
| C6—N1—Cu1 | 113.9 (2) | C56—C51—P2 | 122.6 (2) |
| N1—C2—C3 | 122.1 (4) | C53—C52—C51 | 120.0 (3) |
| N1—C2—H2 | 118.9 | C53—C52—H52 | 120.0 |
| C3—C2—H2 | 118.9 | C51—C52—H52 | 120.0 |
| C4—C3—C2 | 118.7 (4) | C54—C53—C52 | 121.0 (4) |
| C4—C3—H3 | 120.6 | C54—C53—H53 | 119.5 |
| C2—C3—H3 | 120.6 | C52—C53—H53 | 119.5 |
| C3—C4—C5 | 120.3 (4) | C53—C54—C55 | 119.9 (3) |
| C3—C4—H4 | 119.8 | C53—C54—H54 | 120.0 |
| C5—C4—H4 | 119.8 | C55—C54—H54 | 120.0 |
| C4—C5—C6 | 118.7 (4) | C54—C55—C56 | 120.3 (3) |
| C4—C5—H5 | 120.7 | C54—C55—H55 | 119.8 |
| C6—C5—H5 | 120.7 | C56—C55—H55 | 119.8 |
| N1—C6—C5 | 121.8 (3) | C51—C56—C55 | 120.2 (3) |
| N1—C6—C7 | 116.0 (3) | C51—C56—H56 | 119.9 |
| C5—C6—C7 | 122.2 (3) | C55—C56—H56 | 119.9 |
| N12—C7—C8 | 121.4 (3) | C66—C61—C62 | 118.1 (3) |
| N12—C7—C6 | 115.8 (3) | C66—C61—P2 | 123.4 (2) |
| C8—C7—C6 | 122.7 (3) | C62—C61—P2 | 118.4 (2) |
| C9—C8—C7 | 119.6 (4) | C63—C62—C61 | 120.7 (3) |
| C9—C8—H8 | 120.2 | C63—C62—H62 | 119.7 |
| C7—C8—H8 | 120.2 | C61—C62—H62 | 119.7 |
| C8—C9—C10 | 119.7 (4) | C64—C63—C62 | 120.5 (4) |
| C8—C9—H9 | 120.1 | C64—C63—H63 | 119.7 |
| C10—C9—H9 | 120.1 | C62—C63—H63 | 119.7 |
| C9—C10—C11 | 118.8 (4) | C65—C64—C63 | 119.4 (3) |
| C9—C10—H10 | 120.6 | C65—C64—H64 | 120.3 |
| C11—C10—H10 | 120.6 | C63—C64—H64 | 120.3 |
| N12—C11—C10 | 122.7 (4) | C64—C65—C66 | 120.8 (4) |
| N12—C11—H11 | 118.6 | C64—C65—H65 | 119.6 |
| C10—C11—H11 | 118.6 | C66—C65—H65 | 119.6 |
| C7—N12—C11 | 117.7 (3) | C65—C66—C61 | 120.5 (3) |
| C7—N12—Cu1 | 113.2 (2) | C65—C66—H66 | 119.8 |
| C11—N12—Cu1 | 128.0 (2) | C61—C66—H66 | 119.8 |
| C22—C21—C26 | 117.8 (3) | C76—C71—C72 | 118.5 (3) |
| C22—C21—P1 | 125.0 (3) | C76—C71—P2 | 124.5 (2) |
| C26—C21—P1 | 117.2 (3) | C72—C71—P2 | 117.0 (2) |
| C21—C22—C23 | 119.7 (4) | C73—C72—C71 | 120.5 (3) |
| C21—C22—H22 | 120.1 | C73—C72—H72 | 119.7 |
| C23—C22—H22 | 120.1 | C71—C72—H72 | 119.7 |
| C24—C23—C22 | 121.3 (4) | C72—C73—C74 | 120.2 (4) |
| C24—C23—H23 | 119.3 | C72—C73—H73 | 119.9 |
| C22—C23—H23 | 119.3 | C74—C73—H73 | 119.9 |
| C23—C24—C25 | 119.8 (4) | C75—C74—C73 | 119.3 (3) |
| C23—C24—H24 | 120.1 | C75—C74—H74 | 120.3 |
| C25—C24—H24 | 120.1 | C73—C74—H74 | 120.3 |
| C24—C25—C26 | 120.3 (4) | C74—C75—C76 | 120.8 (3) |
| C24—C25—H25 | 119.9 | C74—C75—H75 | 119.6 |
| C26—C25—H25 | 119.9 | C76—C75—H75 | 119.6 |
| C25—C26—C21 | 121.0 (4) | C71—C76—C75 | 120.7 (3) |
| C25—C26—H26 | 119.5 | C71—C76—H76 | 119.7 |
| C21—C26—H26 | 119.5 | C75—C76—H76 | 119.7 |
| C32—C31—C36 | 118.8 (3) | Cl2S—C1S—Cl1S | 110.6 (2) |
| C32—C31—P1 | 118.3 (3) | Cl2S—C1S—Cl3S | 109.0 (3) |
| C36—C31—P1 | 122.7 (3) | Cl3S—C1S—Cl1S | 109.3 (3) |
| C31—C32—C33 | 120.4 (4) | Cl1S—C1S—H1S | 109.3 |
| C31—C32—H32 | 119.8 | Cl2S—C1S—H1S | 109.3 |
| C33—C32—H32 | 119.8 | Cl3S—C1S—H1S | 109.3 |
| C34—C33—C32 | 119.6 (4) | Cl2t—C1S—Cl1t | 107.6 (12) |
| C34—C33—H33 | 120.2 | Cl2t—C1S—Cl3t | 117.1 (11) |
| C32—C33—H33 | 120.2 | Cl3t—C1S—Cl1t | 118.8 (9) |
| C35—C34—C33 | 120.2 (4) | Cl1t—C1S—H1S | 91.7 |
| C35—C34—H34 | 119.9 | Cl2t—C1S—H1S | 134.5 |
| C33—C34—H34 | 119.9 | Cl3t—C1S—H1S | 85.6 |
| C34—C35—C36 | 120.6 (4) | O3S—N1S—O1S | 114.4 (7) |
| C34—C35—H35 | 119.7 | O3S—N1S—O2S | 141.2 (10) |
| C36—C35—H35 | 119.7 | O1S—N1S—O2S | 103.1 (9) |
| C35—C36—C31 | 120.4 (4) | O1S—N1S—O2S' | 151.1 (7) |
| C35—C36—H36 | 119.8 | O1S—N1S—O3S' | 96.4 (10) |
| C31—C36—H36 | 119.8 | O2S'—N1S—O3S' | 96.0 (9) |
| N1—Cu1—P1—C21 | −37.02 (14) | C21—P1—C31—C32 | 67.5 (3) |
| N12—Cu1—P1—C21 | 48.98 (14) | C41—P1—C31—C32 | 175.0 (3) |
| P2—Cu1—P1—C21 | −175.39 (11) | Cu1—P1—C31—C32 | −58.5 (3) |
| N1—Cu1—P1—C31 | 83.75 (15) | C21—P1—C31—C36 | −117.3 (3) |
| N12—Cu1—P1—C31 | 169.75 (14) | C41—P1—C31—C36 | −9.8 (3) |
| P2—Cu1—P1—C31 | −54.62 (13) | Cu1—P1—C31—C36 | 116.7 (3) |
| N1—Cu1—P1—C41 | −154.89 (14) | C36—C31—C32—C33 | −0.5 (5) |
| N12—Cu1—P1—C41 | −68.88 (14) | P1—C31—C32—C33 | 174.9 (3) |
| P2—Cu1—P1—C41 | 66.75 (12) | C31—C32—C33—C34 | 0.1 (6) |
| N1—Cu1—P2—C51 | −87.33 (13) | C32—C33—C34—C35 | 0.4 (6) |
| N12—Cu1—P2—C51 | −174.41 (13) | C33—C34—C35—C36 | −0.5 (7) |
| P1—Cu1—P2—C51 | 51.13 (11) | C34—C35—C36—C31 | 0.2 (6) |
| N1—Cu1—P2—C61 | 152.51 (14) | C32—C31—C36—C35 | 0.3 (5) |
| N12—Cu1—P2—C61 | 65.43 (13) | P1—C31—C36—C35 | −174.8 (3) |
| P1—Cu1—P2—C61 | −69.03 (11) | C21—P1—C41—C42 | −140.2 (3) |
| N1—Cu1—P2—C71 | 33.56 (14) | C31—P1—C41—C42 | 111.3 (3) |
| N12—Cu1—P2—C71 | −53.51 (13) | Cu1—P1—C41—C42 | −16.1 (3) |
| P1—Cu1—P2—C71 | 172.03 (11) | C21—P1—C41—C46 | 39.2 (3) |
| N12—Cu1—N1—C2 | 174.9 (3) | C31—P1—C41—C46 | −69.3 (3) |
| P2—Cu1—N1—C2 | 65.3 (3) | Cu1—P1—C41—C46 | 163.3 (2) |
| P1—Cu1—N1—C2 | −79.0 (3) | C46—C41—C42—C43 | 0.2 (6) |
| N12—Cu1—N1—C6 | 5.6 (2) | P1—C41—C42—C43 | 179.6 (3) |
| P2—Cu1—N1—C6 | −104.0 (2) | C41—C42—C43—C44 | −0.8 (7) |
| P1—Cu1—N1—C6 | 111.7 (2) | C42—C43—C44—C45 | 1.3 (8) |
| C6—N1—C2—C3 | 1.6 (5) | C43—C44—C45—C46 | −1.2 (7) |
| Cu1—N1—C2—C3 | −167.3 (3) | C44—C45—C46—C41 | 0.6 (6) |
| N1—C2—C3—C4 | 0.0 (6) | C42—C41—C46—C45 | −0.2 (5) |
| C2—C3—C4—C5 | −0.8 (7) | P1—C41—C46—C45 | −179.6 (3) |
| C3—C4—C5—C6 | 0.2 (7) | C61—P2—C51—C52 | 110.3 (3) |
| C2—N1—C6—C5 | −2.3 (5) | C71—P2—C51—C52 | −142.3 (3) |
| Cu1—N1—C6—C5 | 168.0 (3) | Cu1—P2—C51—C52 | −14.7 (3) |
| C2—N1—C6—C7 | 177.7 (3) | C61—P2—C51—C56 | −68.6 (3) |
| Cu1—N1—C6—C7 | −12.0 (3) | C71—P2—C51—C56 | 38.8 (3) |
| C4—C5—C6—N1 | 1.4 (6) | Cu1—P2—C51—C56 | 166.4 (2) |
| C4—C5—C6—C7 | −178.5 (4) | C56—C51—C52—C53 | 1.4 (5) |
| N1—C6—C7—N12 | 14.2 (4) | P2—C51—C52—C53 | −177.6 (3) |
| C5—C6—C7—N12 | −165.8 (3) | C51—C52—C53—C54 | −0.5 (6) |
| N1—C6—C7—C8 | −161.7 (3) | C52—C53—C54—C55 | −0.6 (6) |
| C5—C6—C7—C8 | 18.3 (5) | C53—C54—C55—C56 | 0.7 (6) |
| N12—C7—C8—C9 | 1.2 (6) | C52—C51—C56—C55 | −1.2 (5) |
| C6—C7—C8—C9 | 176.8 (4) | P2—C51—C56—C55 | 177.7 (3) |
| C7—C8—C9—C10 | −0.5 (7) | C54—C55—C56—C51 | 0.2 (6) |
| C8—C9—C10—C11 | 0.6 (7) | C51—P2—C61—C66 | −10.6 (3) |
| C9—C10—C11—N12 | −1.4 (6) | C71—P2—C61—C66 | −116.6 (3) |
| C8—C7—N12—C11 | −1.9 (5) | Cu1—P2—C61—C66 | 117.8 (3) |
| C6—C7—N12—C11 | −177.9 (3) | C51—P2—C61—C62 | 173.9 (3) |
| C8—C7—N12—Cu1 | 167.0 (3) | C71—P2—C61—C62 | 67.9 (3) |
| C6—C7—N12—Cu1 | −9.0 (4) | Cu1—P2—C61—C62 | −57.7 (3) |
| C10—C11—N12—C7 | 2.0 (5) | C66—C61—C62—C63 | 1.2 (5) |
| C10—C11—N12—Cu1 | −164.9 (3) | P2—C61—C62—C63 | 176.9 (3) |
| N1—Cu1—N12—C7 | 2.2 (2) | C61—C62—C63—C64 | −1.5 (6) |
| P2—Cu1—N12—C7 | 111.0 (2) | C62—C63—C64—C65 | 0.8 (7) |
| P1—Cu1—N12—C7 | −107.2 (2) | C63—C64—C65—C66 | 0.3 (7) |
| N1—Cu1—N12—C11 | 169.6 (3) | C64—C65—C66—C61 | −0.6 (6) |
| P2—Cu1—N12—C11 | −81.5 (3) | C62—C61—C66—C65 | −0.1 (5) |
| P1—Cu1—N12—C11 | 60.3 (3) | P2—C61—C66—C65 | −175.6 (3) |
| C31—P1—C21—C22 | 9.1 (3) | C51—P2—C71—C76 | −99.3 (3) |
| C41—P1—C21—C22 | −99.6 (3) | C61—P2—C71—C76 | 7.6 (3) |
| Cu1—P1—C21—C22 | 136.1 (3) | Cu1—P2—C71—C76 | 131.4 (2) |
| C31—P1—C21—C26 | −171.6 (3) | C51—P2—C71—C72 | 79.9 (3) |
| C41—P1—C21—C26 | 79.7 (3) | C61—P2—C71—C72 | −173.2 (2) |
| Cu1—P1—C21—C26 | −44.6 (3) | Cu1—P2—C71—C72 | −49.4 (3) |
| C26—C21—C22—C23 | −0.1 (6) | C76—C71—C72—C73 | −0.5 (5) |
| P1—C21—C22—C23 | 179.1 (3) | P2—C71—C72—C73 | −179.8 (3) |
| C21—C22—C23—C24 | −1.4 (7) | C71—C72—C73—C74 | 0.1 (5) |
| C22—C23—C24—C25 | 1.9 (7) | C72—C73—C74—C75 | 0.8 (6) |
| C23—C24—C25—C26 | −0.8 (7) | C73—C74—C75—C76 | −1.3 (5) |
| C24—C25—C26—C21 | −0.7 (6) | C72—C71—C76—C75 | 0.0 (5) |
| C22—C21—C26—C25 | 1.1 (5) | P2—C71—C76—C75 | 179.2 (3) |
| P1—C21—C26—C25 | −178.2 (3) | C74—C75—C76—C71 | 0.9 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O3Si | 0.93 | 2.56 | 3.317 (9) | 139 |
| C5—H5···O1Wi | 0.93 | 2.34 | 3.182 (15) | 151 |
| C8—H8···O2Sii | 0.93 | 2.38 | 3.242 (11) | 155 |
| C45—H45···O3S | 0.93 | 2.59 | 3.493 (10) | 163 |
| C1S—H1S···O1Siii | 0.98 | 2.25 | 3.204 (7) | 165 |
| O1W—?···O2Siv | ? | ? | 2.663 (19) | ? |
| O1W—?···O2Sv | ? | ? | 2.667 (19) | ? |
Symmetry codes: (i) x, y−1, z; (ii) −x+1, −y+1, −z; (iii) −x, −y+1, −z+1; (iv) x+1, y, z+1; (v) −x+1, −y+2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BG2166).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–S19.
- Engelhardt, L. M., Pakawatchai, C., White, A. H. & Healy, P. C. (1985). J. Chem. Soc. Dalton Trans. pp. 125–133.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Hirshfeld, F. L. (1976). Acta Cryst. A32, 239–244.
- Navarro, M., Cisneros-Fajardo, E., Fernandez-Mestre, M., Arriechi, D. & Marchan, E. (2003). J. Inorg. Biochem.97, 364–369. [DOI] [PubMed]
- Rigaku/MSC (2005). CrystalClear Rigaku/MSC., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808006260/bg2166sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808006260/bg2166Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




