Abstract
Restriction of murine leukemia virus (MLV) was examined in cells from a range of mammals. All nonmurine restrictions were saturable blocks to N-tropic MLV infection, and several were prior to reverse transcription. We demonstrate restriction in cells from bat and show that if we express the murine restriction factor Fv1 in human cells, then Fv1, not the human host, defines the stage at which infection is blocked.
Saturable blocks to retrovirus infection in cells from a wide range of mammals have recently been described to be active against both gamma retroviruses and lentiviruses (2, 7, 10, 14, 19, 21; reviewed in reference 4). These data indicate the existence of dominant factors, termed Ref1 and Lv1, with characteristics similar to those of the murine restriction factor Fv1 (11). The Fv1 gene is derived from endogenous retroviral sequence with no significant homology to murine leukemia virus (MLV) and is a dominant inhibitor of retroviral infection (3, 8, 17). Two Fv1 alleles allow the division of MLVs into N-tropic and B-tropic viruses, which are infectious for NIH 3T3 or BALB/3T3 cells, respectively. The viral determinant for N and B tropism is in the capsid (CA) protein, with an arginine at amino acid position CA110 specifying N tropism and a glutamate specifying B tropism (13). A number of nonmurine species are able to block N-tropic MLV infection with the same specificity for CA110 (19). Restriction of lentivirus is also targeted against the CA-p2 domain of gag (7, 21).
Restriction to both gamma retroviruses and lentiviruses can be overcome when restricting cells are challenged with high virus dose (2, 7, 9, 20). This can be explained by the fact that the CA protein in restricted virions binds and titrates out a limited pool of restriction factor. This leads to saturation of restriction at high multiplicities of infection. Here, we investigate the kinetics of restricted infection and the stage in the viral life cycle at which restriction occurs in cells from hamster, pig, cow, bat, and African green monkey (AGM). N- and B-tropic vesicular stomatitis virus G-protein-pseudotyped MLV vectors encoding green fluorescent protein (GFP) (N-GFP and B-GFP, respectively) were produced as previously described (2). N-GFP and B-GFP were used to infect cells from restricting species (Fig. 1), and the infected cells were enumerated by fluorescence-activated cell sorter (FACS) analysis at 48 h postinfection.
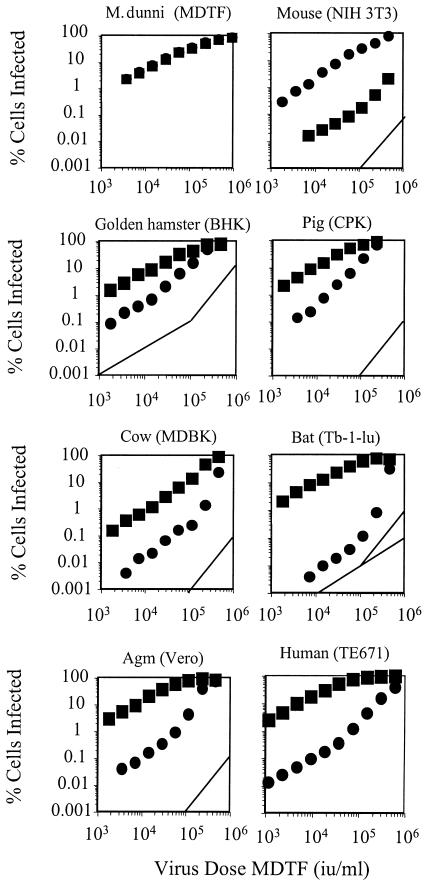
FIG. 1.
Titration of N- and B-GFP vectors. A total of 105 cells in 6-well plates were infected with twofold serial dilutions of either N-GFP (•) or B-GFP (▪) vector prepared as previously described. Virus dose was measured in terms of iu on unrestricting MDTF cells. GFP-positive cells were enumerated by FACS 48 h postinfection as previously described (22). Guides with a slope of 2 indicating two-or-greater-hit kinetics are shown, and guides with a slope of 1 indicating single-hit kinetics at low dose are shown in panels for hamster and bat cells. Data are representative of results from three independent experiments.
There is no restriction in Mus dunni tail fibroblast (MDTF)cells, and consequently the titers and kinetics of N-GFP and B-GFP are equal and single hit and the curves for N- and B-GFP infection are superimposed. While infection kinetics of unrestricted virus are linear and single hit in all species tested, in restricted infection the slope of the infection curve is 1 at low viral dose and steepens as the dose increases. The reason for this characteristic bend is not clear but is presumably related to restriction factor saturation. The restriction is overcome at high virus dose, and slopes representing restricted infection are equal to or greater than 2 at high dose, indicating multiple-hit infection kinetics. This suggests that exposure to an initial, restricted virion facilitates infection by a second, restricted virion.
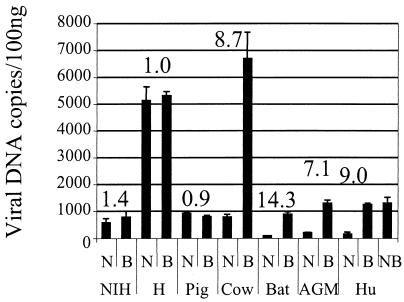
We next investigated the stage at which restriction occurs. The Fv1-mediated block to infection in murine cells occurs after completion of reverse transcription (12, 23), whereas in human cells, Ref1 inhibits N-tropic reverse transcription (19). Restriction to lentivirus also occurs before reverse transcription in all cases examined (2, 7, 14). We examined the appearance of newly synthesized viral DNA, consisting of newly synthesized linear molecules as well as integrated provirus, by quantitative TaqMan PCR as previously described (2, 22) (Fig. 2). Triplicate samples of cells were infected with equivalent doses (MDTF infectious doses [iu]) of N- and B-GFP chosen such that the multiplicity of infection for each target cell line would be between 0.15 and 0.25 (Fig. 2). Total DNA was prepared 6 h postinfection for two samples, and the third sample was subjected to FACS analysis to determine the percentage of cells infected. The percentage of infection and the viral dose expressed in MDTF iu per milliliter (19) for each cell line are shown in Table 1. Quantitative PCR was performed on 100 ng of the extracted DNA as previously described (2, 22) (Fig. 2). These data provide information on the relative ability of each cell line to support MLV reverse transcription as well as indicate the difference in reverse transcription efficiencies between restricted and unrestricted infection. NIH 3T3 cells restrict infection by B-GFP, but not by N-GFP (Table 1), but reverse transcription is not blocked, as previously described (12, 23). BHK cells support particularly high levels of reverse transcription after infection by both N- and B-GFP but are able to block N-GFP infection by four- to fivefold. Pig CPK cells support less reverse transcription than do BHK cells and also appear to block N-GFP infection by fivefold despite similar levels of N- and B-GFP reverse transcription. This suggests that the block in hamster and pig cells occurs after reverse transcription, as is the case in mouse cells. In cells from human, cow, AGM, and bat, there is a significant reduction, around 10-fold, in the level of DNA synthesis after restricted N-GFP infection compared to what is seen with unrestricted B-GFP infection. Cow MDBK cells are particularly permissive to B-GFP reverse transcription, and here restriction is able to reduce N-GFP DNA synthesis significantly. We included unrestricted NB-GFP infection of TE671 cells as a control in the experiment whose results are shown in Fig. 3. In summary, the block to infection occurs before reverse transcription in cells from cows, bats, and primates but after reverse transcription in cells from mice, pigs, and hamsters. While this might suggest that the interaction between the restriction factor and the incoming virus occurs later (after reverse transcription) in these latter species, this does not appear to be the case in mice, as an MLV lacking the pol gene product, and unable to make DNA, has been shown to be able to saturate Fv1 restriction (1). We assume that the difference in the abilities to block reverse transcription is a consequence of the specific factor-virus interaction or a difference in the efficiencies of this interaction. Furthermore, we note that the magnitude of the block to infection as measured by GFP expression (Table 1) tends to be greater than the block to DNA synthesis measured 6 h postinfection (Fig. 2). It is therefore possible that the weaker blocks in cells from pig and hamster are not strong enough to reveal an inhibition of DNA synthesis.
FIG. 2.
Measurement of DNA synthesis after restricted and unrestricted infection by TaqMan quantitative PCR. Abbreviations: NIH, murine NIH 3T3; H, hamster BHK; Pig, porcine CPK; Cow, bovine MDBK; Bat, Bat Tb-1-lu; AGM, AGM CV1; Hu, human TE671. A total of 105 cells were infected with equal doses of DNase-treated N- or B-GFP vector at a multiplicity of unrestricted infection of 0.15 to 0.25. Viral doses required to achieve these multiplicities were determined independently for each line and are recorded in Table 1. Cells were infected for 6 h, and then total DNA was extracted by DNAeasy (Qiagen, Chatsworth, Calif.). Template copy number values were assigned with reference to a standard curve generated by PCR of plasmid dilutions as previously described (22). One hundred nanograms of DNA was subjected to PCR using primers and probe specific for GFP as previously described (2). Total DNA includes newly synthesized linear and circular molecules as well as integrated provirus. Values of B/N copy numbers are recorded above each pair of columns. Data are representative of results from two independent experiments, and error bars indicate standard errors of the mean.
TABLE 1.
Input doses of N-, B-, and NB-tropic viruses and percentages of infection
| Cell line (species) | Input dose (MDTF iu/ml)a | % Infectionb
|
||
|---|---|---|---|---|
| N | B | NB | ||
| NIH 3T3 (mouse) | 3.9 × 104 | 16 | <1 | NTc |
| BHK (hamster) | 2.8 × 104 | 6 | 27 | NT |
| CPK (pig) | 2.3 × 104 | 4 | 20 | NT |
| MDBK (cow) | 7.9 × 104 | <1 | 14 | NT |
| Tb-1-lu (bat) | 3.5 × 104 | <1 | 20 | NT |
| Vero (AGM) | 4 × 104 | <1 | 17 | NT |
| TE671 (human) | 5.6 × 103 | <1 | 24 | 29 |
Values are in MDTF iu determined by infection of Fv1-null MDTF cells
Shown are percentages of infection for experiments performed in parallel. Equal MDTF iu of N- and B-GFP were used to infect 105 target cells. Infected cells were enumerated by FACS 48 h postinfection.
NT, not tested.
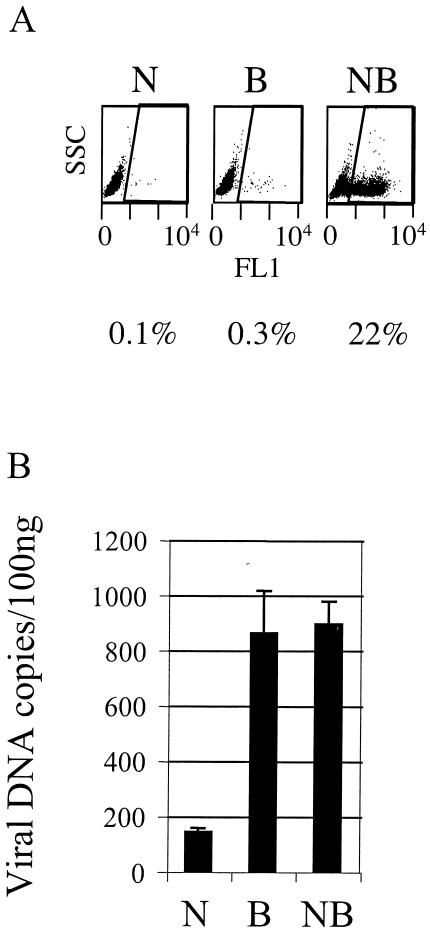
FIG. 3.
Restriction in human TE671 cells overexpressing Fv1 N (TEN cells). Duplicate samples of 105 cells were infected with 6 × 103 MDTF iu of DNase-treated N-, B-, and NB-GFP vectors. (A) GFP-positive cells were enumerated by FACS 48 h after infection. Plots are side scatter versus green fluorescence in FL1. The region denoting positive cells and percentage of infection are shown. (B) Quantitative TaqMan PCR was performed, using DNAeasy (Qiagen), on 100 ng of total DNA purified from TEN cells (TE671 overexpressing Fv1 N) 6 h after infection. PCR was performed using primers and probe to GFP as previously described (2). Data are representative of results for two independent TEN clones and two independent experiments. Error bars indicate standard errors of the mean.
To investigate this further and to test whether the restriction factor or the intracellular environment determines the stage of block (before or after reverse transcription), we expressed the murine restriction factor Fv1 N in human cells and measured GFP expression and DNA synthesis after infection with N-, B-, or NB-GFP. NB-GFP vector was made using Moloney MLV-derived gag sequence as previously described (19), and this vector is insensitive to either Fv1 or Ref1. Human TE671 cells restrict N-tropic MLV through endogenously expressed Ref1, and Fv1 N restricts B-tropic MLV. We transduced cells with a retroviral vector expressing Fv1 N as previously described (5) and derived single-cell clones positive for Fv1 N expression by limiting dilution. We called these cells TEN cells. Figure 3A shows that after infection with equal doses of N-, B-, and NB-GFP, these cells restrict N- and B-GFP but not NB-GFP. We then measured viral DNA synthesis in DNA purified 6 h after infection of TEN cells (Fig. 3B). N-GFP DNA synthesis was blocked by Ref1 before reverse transcription as previously described (19), and although B-GFP is now also restricted by the exogenously expressed Fv1 N, B-GFP DNA synthesis remains at the level of unrestricted NB-GFP (compare with the TE671 data shown in Fig. 2). These data indicate that although Fv1 N can restrict B-GFP in human cells, it cannot block reverse transcription. The block to infection by Fv1 N occurs after reverse transcription, as is the case in murine cells (12, 23). This demonstrates that the ability of Fv1 to block infection, without blocking reverse transcription, is a property of the restriction factor rather than of the cellular background. It appears that one cell may simultaneously mediate blocks at either of two steps in the viral life cycle.
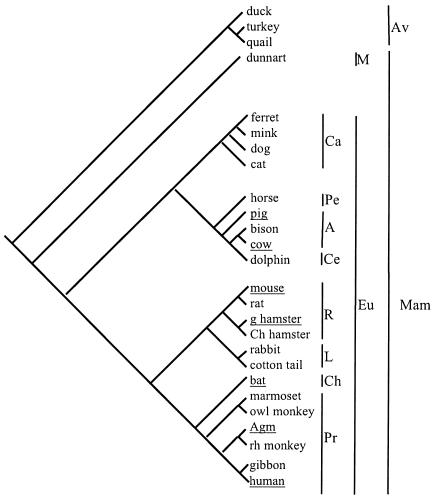
Southern blotting of DNA from cell lines (Fig. 1) by using labeled Fv1 cDNA as a probe indicated that there were no sequences in any of the restricting cells closely related to Fv1 (data not shown). In Fig. 4, all previously reported restricting species (19), nine new species that do not restrict (data not shown), and a new restrictor (bat) are placed in a phylogenetic tree. All underlined species are able to restrict N-tropic MLV, including mouse, which is also able to restrict B-tropic MLV (11). This tree shows that restriction is spread among many mammalian orders but is not uniformly present in any. Even relatively closely related species pairs, such as mouse and rat or the two species of hamster, differentially restrict. Some cell lines may have lost the restrictive abilities of the species, but it is unlikely that all negative data are cell line artifacts as the vast majority of cell lines isolated from Fv1-positive mice have the appropriate restriction phenotype. Furthermore, all human lines tested restricted N-GFP to some degree (data not shown). The data in Fig. 4 suggest that the ability to restrict has been convergently acquired many times during mammalian evolution. In mouse, in which the only animal studies on restriction have been performed, a saturable block is enough to protect the animal from disease (16). Accordingly, we would expect that the restrictions seen in primates, cows, pigs, hamsters, and bats would also confer protection.
FIG. 4.
Phylogenetic distribution of restriction among vertebrates. Underlined species restrict, and species not underlined do not. Cell lines were tested by infecting 105 cells with equal doses of N- and B-GFP vectors as previously described (19). The cell lines were from duck (duck embryo), turkey (Tur8), quail (QT6 and QT35), dunnart (SC300), ferret (MPF), mink (Mv-1-lu), dog, (CF2S+L−, D17, A72, DK), cat (FEA, AH927, CRFK), horse (NBL6, EDERM), pig (PAE, ST IOWA, SKL, CPK, MPK, PK15), bison (bu IMR-31), cow (MDBK), dolphin (Db-1-tes), mouse (NIH 3T3, BALB/3T12, MDTF), rat (HSN, NRK) golden hamster (BHK), Chinese hamster (CHO, A23), rabbit (SIRC), cotton tail (EREp), bat (Tb-1-lu), owl monkey (OMK), AGM (CV1, Vero), rhesus monkey (LLC-MK2, FRhK4), gibbon (MLA144), squirrel monkey (Pindak), and human (TE671, HeLa, MCF, MRC5, HT1080, NP2, Hos) and were obtained from the Centro Substrati Cellulari, Brescia, Italy, and the American Type Culture Collection. The tree was drawn according to published literature (6, 15, 18). Abbreviations: Mam, mammals; Av, birds; M, marsupials; Eu, Eutheria. Eutherian orders: Ca, Carnivora; Pe, Perissodactyla; A, Artiodactyla; Ce, Cetacea; R, Rodentia; L, Lagomorpha; Ch, Chiroptera; Pr, Primates.
These data extend our observations of widespread restrictions to MLV in mammals to bats and characterize the blocks as having multiple-hit infection kinetics at high virus dose in all cases. We also show that at least in the case of Fv1 the ability to block infection after reverse transcription is a property of the restriction factor rather than of the target cell. This observation suggests that there might be mechanistic differences between restriction factors like Fv1 and Ref1 or Lv1 which block infection after reverse transcription (2, 19). Sensitivity to restriction in all species able to restrict MLV-N is dependent on CA amino acid 110 (19) (data not shown for bat cells). The significance of this exquisite specificity is still unclear but may represent a structural homology between the factors responsible or similarities in the mechanism of restriction itself despite slightly different timings. The selection force for restriction could be pathogenic retroviruses that are transmitted between species and cause disease. Certainly, further characterization and identification of restriction factors will help the understanding of the interplay between retroviruses and hosts and provide further information on a step in the viral life cycle susceptible to effective inhibition.
Acknowledgments
We thank Paul Clapham, Akio Fukusho, Jonathan Scammel, and Jo Martin for cell lines.
This work was funded by career development fellowship no. 064257 to G.J.T. from the Wellcome Trust and the Medical Research Council UK. S.P.G. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bassin, R. H., B. I. Gerwin, J. G. Levin, G. Duran-Troise, B. M. Benjers, and A. Rein. 1980. Macromolecular requirements for abrogation of Fv-1 restriction by murine leukemia viruses. J. Virol. 35:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 5.Bock, M., K. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, Y., A. Janke, P. J. Waddell, M. Westerman, O. Takenaka, S. Murata, N. Okada, S. Paabo, and M. Hasegawa. 1998. Conflict among individual mitochondrial proteins in resolving the phylogeny of Eutherian orders. J. Mol. Evol. 47:307-322. [DOI] [PubMed] [Google Scholar]
- 7.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goff, S., P. 1996. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell 86:691-693. [DOI] [PubMed] [Google Scholar]
- 9.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67-122. [DOI] [PubMed] [Google Scholar]
- 12.Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33:183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 14.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy, W. J., E. Eizirik, W. E. Johnson, Y. P. Zhang, O. A. Ryder, and S. J. O'Brien. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409:614-618. [DOI] [PubMed] [Google Scholar]
- 16.Odaka, T. 1969. Inheritance of susceptibility to Friend mouse leukemia virus. V. Introduction of a gene responsible for susceptibility in the genetic complement of resistant mice. J. Virol. 3:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pincus, T., W. P. Rowe, and F. Lilly. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to Friend murine leukemia virus. J. Exp. Med. 133:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimamura, M., H. Yasue, K. Ohshima, H. Abe, H. Kato, T. Kishiro, M. Goto, I. Munechika, and N. Okada. 1997. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature 388:666-670. [DOI] [PubMed] [Google Scholar]
- 19.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med., in press. [DOI] [PubMed]
- 22.Towers, G. J., D. Stockholm, V. Labrousse-Najburg, F. Carlier, O. Danos, and J. C. Pages. 1999. One step screening of retroviral producer clones by real time quantitative PCR. J. Gene Med. 1:352-359. [DOI] [PubMed] [Google Scholar]
- 23.Yang, W. K., J. O. Kiggans, D. M. Yang, C. Y. Ou, R. W. Tennant, A. Brown, and R. H. Bassin. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA 77:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]