Abstract
The Yatapoxvirus genus of poxviruses is comprised of Yaba monkey tumor virus (YMTV), Tanapox virus, and Yaba-like disease virus (YLDV), which all have the ability to infect primates, including humans. Unlike other poxviruses, YMTV induces formation of focalized histiocytomas upon infection. To gain a greater understanding of the Yatapoxvirus genus and the unique tumor formation properties of YMTV, we sequenced the 134,721-bp genome of YMTV. The genome of YMTV encodes at least 140 open reading frames, all of which are also found as orthologs in the closely related YLDV. However, 13 open reading frames found in YLDV are completely absent from YMTV. Common to both YLDV and YMTV are the unusually large noncoding regions between many open reading frames. To determine whether any of these noncoding regions might be functionally significant, we carried out a comparative analysis between the putative noncoding regions of YMTV and similar noncoding regions from other poxviruses. This approach identified three new gene poxvirus families, defined as orthologs of YMTV23.5L, YMTV28.5L, and YMTV120.5L, which are highly conserved in virtually all poxvirus species. Furthermore, the comparative analysis also revealed a 40-bp nucleotide sequence at approximately 14,700 bases from the left terminus that was 100% identical in the comparable intergene site within members of the Yatapoxvirus, Suipoxvirus, and Capripoxvirus genera and 95% conserved in the Leporipoxvirus genus. This conserved sequence was shown to function as a poxvirus late promoter element in transfected and infected cells, but other functions, such as an involvement in viral replication or packaging, cannot be excluded. Finally, we summarize the predicted immunomodulatory protein repertoire in the Yatapoxvirus genus as a whole.
Poxviruses are divided into two major groups, the chordopoxviruses that infect vertebrates and entomopoxviruses of insects. Chordopoxviruses contain a linear double-stranded DNA genome with covalently closed hairpin loops at either end (19). The extreme left and right termini of the poxvirus genome consist of identical, but oppositely oriented, terminal inverted repeats (TIR). Chordopoxvirus genomes can be divided into two broad domains based on the functions of the encoded gene products. The central region of the genome, which ranges in length from 80,000 to 100,000 bases, is enriched for genes that encode essential conserved functions, such as transcription, replication, and virion assembly. The regions flanking this conserved central region express an array of proteins that function to increase survival of the virus in the infected host, including proteins that determine host range, inhibit apoptosis, or mediate responses to modulate the host immune system (22).
The genome sizes of published chordopoxviruses vary from 145,000 bp for Yaba-like disease virus (YLDV) (15) up to 288,000 bp for fowlpox virus (2) and possess between 151 and 260 assigned open reading frames (ORFs). Complete genomic sequences of representative members from seven of the eight Chordopoxvirus genera have now been published, including orthopoxviruses (vaccinia virus strain Copenhagen [11], modified vaccinia virus strain Ankara [6], variola virus strain Bangladesh [16], variola virus strain India [24], variola virus strain Garcia [25], camelpox virus [1], and monkeypox virus [26]), capripoxviruses (lumpy skin disease virus [LSDV] [29],goatpox virus, and sheeppox virus [30]), leporipoxviruses (myxoma virus [8] and Shope fibroma virus [31]), suipoxviruses (swinepox virus [SPV] [3]), molluscipoxvirus (molluscum contagiosum virus [23]), avipoxviruses (fowlpox [2]), and yatapoxviruses (Yaba-like disease virus [YLDV] [15]).
The Yatapoxvirus genus of poxviruses is comprised of three virus isolates: YLDV, Tanapox virus (TPV), and Yaba monkey tumor virus (YMTV) (14). The yatapoxviruses have a narrow host range, infecting only primates, including humans. Several pieces of data suggest that TPV and YLDV may be different strains of the same virus. For example, TPV and YLDV produce a clinically indistinguishable disease, which includes a mild fever and epidermal lesions (10, 17), and the published genomic sequence of YLDV is more than 98.6% identical with the 8,300 bases of TPV sequence entered into the public database (GenBank accession no. AY253325, AF245394, and AF153912) (15). This level of sequence identity is comparable to different strains of vaccinia virus and suggests that YLDV and TPV should be considered the monkey and human versions, respectively, of the same virus.
YMTV was originally characterized to be the agent responsible for subcutaneous tumors in a rhesus monkey colony occurring in 1956 in Yaba, Nigeria (7). YMTV is one of the few poxviruses that induce substantial tumor formation upon infection (5, 12, 20, 27). In rhesus monkeys infected with YMTV, the tumors are thought to be derived from histiocytes that migrate to the site of infection. The histiocytes become infected and begin to rapidly proliferate, become multinucleated, and eventually form a polyclonal tumor (27). However, the tumors generally do not become invasive and spontaneously regress, presumably when either viral cytopathic effect kills the infected cells or cell-mediated antiviral immunity becomes sufficiently effective to clear the infection (12, 27).
The complete genomic sequence of YLDV was recently published, and a number of novel ORFs not found in other chordopoxviruses were identified (15). As well, despite the fact that the noncoding regions between ORFs in most poxviruses are typically only a few nucleotides, there were multiple identified inter-ORF regions of 200 or more nucleotides in YLDV. Typically, the minimum size for a poxvirus ORF is arbitrarily set (e.g., 30 amino acids for SPV, LSDV, molluscum contagiosum virus, and fowlpox virus [2, 3, 23, 29]; 50 amino acids for myxoma virus [8]; and 60 amino acids for YLDV [15]). If bona fide ORFs were indeed located within these assigned YLDV noncoding regions, then one would predict that these ORFs might be highly conserved between YLDV and YMTV. Therefore, in an effort to understand the clinical differences between YLDV and YMTV and to provide a closely related sequence to YLDV for a comparative genomic approach, we sequenced the genome of the YMTV and provide a comparative genomic analysis of the Yatapoxvirus genus.
MATERIALS AND METHODS
Viruses.
YMTV (VR587) was obtained from the American Type Culture Collection (Manassas, Va.) and was propagated on CV1 cells at 35°C in minimum essential medium containing 5% fetal bovine serum. Myxoma virus strain Lausanne was obtained from the American Type Culture Collection and propagated in BGMK cells at 37°C.
Isolation and sequencing of YMTV genomic fragments.
YMTV genomic DNA was isolated from infected CV1 cells and was subjected to restriction enzyme digestion with PstI, BamHI, SalI, XbaI, or EcoRI. The digested DNA was cloned into pUC19 or pBR322 vectors and sequenced by the dideoxy sequencing method (21). The remainder of the YMTV genomic sequence was cloned using overlapping PCR. Briefly, PCR was carried out using Taq polymerase, YMTV genomic DNA, and PCR primers based on the corresponding sequence of YLDV (15). The resulting PCR products were cloned into pGEMT-easy (Promega, Madison, Wis.) and were sequenced by the London Regional Genomics Centre DNA Sequencing Facility using an Applied Biosystems (Foster City, Calif.) ABI Prism 377 DNA sequencer and Big Dye terminators (Applied Biosystems). Some of the YMTV sequence was previously submitted to GenBank (accession no. AY253324, AB025319, AB018404, and AB015885).
Sequence analysis.
The sequence data were assembled using Sequencher 3.0, and ORFs were identified using MacVector 6.5.3 (Oxford Molecular Ltd.).
Cloning a conserved sequence from myxoma virus upstream of an enhanced GFP cassette.
PCR was carried out using Taq polymerase; plasmid DNA pEGFP-N1 (Clontech, Palo Alto, Calif.); the reverse PCR primer 5′ TTACGCCTTAAGATACATTG 3′, which corresponds to the 3′ end of the green fluorescent protein (GFP), and the forward PCR primers (with the start codon of GFP in boldface type) 5′ TCGCCACCATGGTGAGCAAG 3′ (PCR-GFP), 5′ TTTATTTATGTTATTAGCTAGGATTTATGTTTCATTTTTTACTCGCCACCATGGTGAGCAAG 3′ (PCR-R-GFP), and 5′ GTAAAAAATGAAACATAAATCCTAGCTAATAACATAAATAAATCGCCACCATGGTGAGCAAG 3′ (PCR-L-GFP). The resulting PCR products were cloned into pGEMT-easy (Promega) and designated GFP, R-GFP, and L-GFP.
Expression of GFP cassette in BGMK cells.
Twelve-well dishes of BGMK cells approximately 90% confluent growing in minimum essential medium-5% fetal bovine serum were either infected with myxoma virus at a multiplicity of infection of 10 or mock infected. The cells were incubated at 37°C for 2 h, and this was followed by transfection with GFP, R-GFP, or L-GFP plasmid DNA using Lipofectamine Plus (Invitrogen, Burlington, Ontario, Canada) per the manufacturer's protocol. The cells were subsequently incubated at 37°C for 48 h. Cells expressing the GFP construct were detected using a fluorescence microscope.
Nucleotide sequence accession number.
Sequence data from this article have been deposited in GenBank under accession number AY386371.
RESULTS
Genome structure of YMTV.
The genome of YMTV was sequenced through the subcloning of genomic fragments into plasmid vectors, and clones were individually sequenced. In addition, regions of the genome not represented in the cloned fragments were isolated using PCR, and a minimum of three independent PCR products for each primer set were sequenced. After assembling the sequence files, a single continuous sequence of 134,721 bases was generated, making YMTV the smallest poxvirus genome yet sequenced. This deduced sequence lacks the terminal hairpin region, but evidence suggests that all the coding ORFs have been fully sequenced and only the very extreme hairpin termini of the genome were not included. In particular, the putative YMTV concatemer resolution sequence was obtained, which is typically found very close to the molecular hairpin loop at the termini (18). Published reports also confirm that the YMTV genome size is indeed approximately 135,000 bases (4).
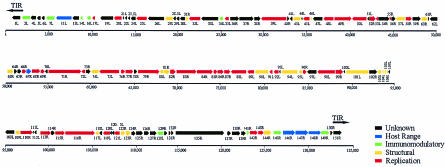
The YMTV genome has an A+T content of 70.2% and encodes at least 140 ORFs (Table 1; Fig. 1), of which 139 are single copies and 1 is repeated in each copy of the TIR. In comparison, YLDV has been assigned 151 ORFs (15). YMTV and YLDV is closely related viruses with approximately 75% identity between the viruses at the nucleotide level overall, which is typical for chordopoxvirus members from a single genus. Furthermore, all the ORFs identified in YMTV have a corresponding ortholog in YLDV, but YMTV has lost 13 ORFs that are present in YLDV (Table 2), which accounts for the 10 kb of sequence loss in YMTV. Since YMTV and YLDV are so similar, and to avoid unnecessary confusion, we have adopted the proposed YLDV nomenclature (15) for naming orthologous YMTV ORFs.
TABLE 1.
YMTV ORFs
| ORF | Codon
|
No. of aaf | TOEg | Predicted structure or functionh | YLDVa
|
SPVb
|
Myxc
|
LSDVd
|
VVe
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | ORF | BLASTP2 score | % Iden- tity | ORF | % Iden- tity | ORF | % Iden- tity | ORF | % Iden- tity | ORF | % Iden- tity | ||||
| 1L | 1808 | 804 | 334 | E | A52R family | 1L | 489 | 72 | LSDV007 | 35 | C10L | 26 | ||||
| 2L | 2963 | 1938 | 341 | ? | vTNF-α bp, SP | 2L | 497 | 71 | SPV003 | 34 | ||||||
| 4L | 3722 | 3003 | 239 | ? | α-Amanitin sensitivity | 4L | 340 | 71 | SPV007 | 28 | LSDV009 | 35 | N2L | 31 | ||
| 5L | 4232 | 3762 | 156 | L | LAP/PHD finger, TM | 5L | 200 | 58 | SPV009 | 36 | M153R | 32 | LSDV010 | 48 | ||
| 6L | 4732 | 4274 | 152 | E | Unknown | 6L | 265 | 81 | SPV001/150 | 37 | M003.1 | 28 | LSDV001/156 | 34 | B15R | 41 |
| 7L | 5840 | 4800 | 346 | E? | vCCR8 | 7L | 265 | 70 | SPV005 | LSDV011 | 39 | |||||
| 11L | 8306 | 6393 | 637 | ? | 14 ankyrin domains | 11L | 1061 | 79 | ||||||||
| 12L | 8574 | 8308 | 88 | E | eIF2α mimic | 12L | 111 | 64 | SPV010 | 36 | M156R | 32 | LSDV014 | 35 | K3L | 31 |
| 13L | 9474 | 8614 | 286 | L | Monoglyceride lipase | 13L | 406 | 68 | K6L | 48 | ||||||
| 14L | 9917 | 9504 | 137 | I? | IL-18 bp, SP | 14L | 156 | 55 | SPV011 | 30 | LSDV015 | 37 | ||||
| 16L | 10584 | 10066 | 172 | L? | Inhibition of apoptosis | 16L | 207 | 64 | SPV012 | 29 | LSDV017 | 30 | I1L | 24 | ||
| 17L | 11066 | 10635 | 143 | L? | dUTPase | 17L | 215 | 75 | SPV013 | 52 | M012L | 48 | LSDV018 | 52 | F2L | 46 |
| 19L | 12774 | 11200 | 524 | L? | Kelch-like protein | 19L | 820 | 74 | SPV015 | 35 | M014L | 32 | LSDV019 | 34 | F3L | 25 |
| 20L | 13779 | 12802 | 325 | L | Ribonucleotide reduc- tase (small subunit) | 20L | 611 | 91 | SPV016 | 76 | M015L | 75 | LSDV020 | 79 | F4L | 76 |
| 21L | 14054 | 13806 | 82 | ? | SP, TM | 21L | 114 | 64 | SPV017 | 33 | M016L | 42 | LSDV021 | 31 | ||
| 22L | 14325 | 14098 | 75 | E | Unknown | 22L | 45 | 37 | ||||||||
| 23.5L | 14742 | 14530 | 70 | E | Unknown | 23.5L | 88 | 86 | M018L | 45 | LSDV023 | 57 | F8L | 43 | ||
| 24L | 15443 | 14799 | 214 | L | TM | 24L | 312 | 74 | SPV021 | 45 | M019L | 43 | LSDV024 | 43 | F9L | 48 |
| 25L | 16758 | 15421 | 445 | L | Ser/Thr protein kinase | 25L | 845 | 90 | SPV022 | 77 | M020L | 74 | LSDV025 | 77 | F10L | 72 |
| 26L | 18711 | 16786 | 642 | L | TM | 26L | 861 | 65 | SPV024 | 42 | M021L | 37 | LSDV027 | 40 | F12L | 31 |
| 27L | 19845 | 18736 | 369 | L | EEV envelope protein | 27L | 657 | 87 | SPV025 | 70 | M022L | 69 | LSDV028 | 72 | F13L | 57 |
| 28.5L | 20085 | 19909 | 58 | L | Unknown | 28.5L | ||||||||||
| 29L | 20563 | 20117 | 148 | ? | Unknown | 29L | 263 | 82 | SPV027 | 60 | M024L | 49 | LSDV029 | 62 | F15L | 56 |
| 30L | 21275 | 20628 | 215 | ? | Unknown | 30L | 320 | 73 | SPV028 | 38 | M025L | 30 | LSDV030 | 36 | F16L | 37 |
| 31R | 21335 | 21649 | 104 | L | DNA binding phosphoprotein | 31R | 169 | 79 | SPV029 | 62 | M026L | 68 | LSDV031 | 63 | F17R | 59 |
| 32L | 23058 | 21646 | 470 | ? | Poly(A) polymerase | 32L | 832 | 88 | SPV030 | 67 | M027L | 68 | LSDV032 | 68 | E1L | 64 |
| 33L | 25120 | 23072 | 683 | ? | Unknown | 33L | 1006 | 71 | SPV031 | 44 | M028L | 40 | LSDV033 | 40 | E2L | 37 |
| 34L | 25700 | 25146 | 185 | L | dsRNA bp | 34L | 217 | 57 | SPV032 | 44 | M029L | 57 | LSDV034 | 42 | E3L | 38 |
| 35L | 26311 | 25745 | 188 | L | RNA polymerase sub- unit RPO30 | 35L | 326 | 82 | SPV033 | 66 | M030L | 64 | LSDV036 | 63 | E4L | 67 |
| 36R | 26444 | 27472 | 342 | L? | Unknown | 36R | 512 | 71 | M031R | 31 | LSDV035 | 33 | E5R | 25 | ||
| 37R | 27498 | 29201 | 567 | L? | Unknown | 37R | 1006 | 85 | SPV034 | 65 | M032R | 61 | LSDV037 | 67 | E6R | 60 |
| 38R | 29219 | 30025 | 268 | L | ER-localized protein, TM | 38R | 521 | 93 | SPV035 | 76 | M033R | 75 | LSDV038 | 78 | E8R | 70 |
| 39L | 33042 | 30022 | 1006 | ? | DNA polymerase | 39L | 1689 | 81 | SPV036 | 64 | M034L | 66 | LSDV039 | 66 | E9L | 63 |
| 40R | 33075 | 33359 | 94 | L | Redox protein | 40R | 181 | 88 | SPV037 | 67 | M035R | 69 | LSDV040 | 71 | E10R | 67 |
| 41L | 33770 | 33393 | 125 | L | TM | 41L | 200 | 74 | LSDV041 | 53 | E11L | 48 | ||||
| 43L | 34881 | 33955 | 308 | L | DNA binding protein | 43L | 472 | 77 | SPV039 | 60 | M038L | 64 | LSDV043 | 66 | I1L | 60 |
| 44L | 35103 | 34882 | 73 | L | TM | 44L | 116 | 77 | SPV040 | 48 | M039L | 52 | LSDV044 | 51 | I2L | 45 |
| 45L | 35898 | 35104 | 264 | E? | DNA binding phosphoprotein | 45L | 437 | 84 | SPV041 | 60 | M040L | 61 | LSDV045 | 58 | I3L | 56 |
| 46L | 36227 | 35988 | 79 | L | IMV protein, SP, TM | 46L | 138 | 86 | SPV043 | 55 | M041L | 48 | LSDV046 | 69 | 15L | 45 |
| 47L | 37402 | 36245 | 385 | L | TM | 47L | 625 | 79 | SPV044 | 51 | M042L | 52 | LSDV047 | 53 | 16L | 54 |
| 48L | 38688 | 37399 | 429 | L | Virion core protein | 48L | 773 | 87 | SPV045 | 69 | M043L | 69 | LSDV048 | 71 | 17L | 64 |
| 49R | 38694 | 40730 | 678 | ? | NPH-II, RNA helicase | 49R | 1133 | 80 | SPV046 | 58 | M044R | 54 | LSDV049 | 59 | 18R | 54 |
| 50L | 42496 | 40727 | 590 | L | Metalloproteinase | 50L | 963 | 78 | SPV047 | 59 | M045L | 55 | LSDV050 | 57 | G1L | 51 |
| 51L | 42828 | 42493 | 111 | L | TM | 51L | 167 | 71 | SPV049 | 54 | M046L | 48 | LSDV052 | 47 | G3L | 41 |
| 52R | 42822 | 43490 | 222 | ? | Transcriptional elongation factor | 52R | 349 | 78 | SPV048 | 45 | M047R | 44 | LSDV051 | 46 | G2R | 47 |
| 53L | 43834 | 43457 | 125 | L | Glutaredoxin 2 | 53L | 255 | 99 | SPV050 | 64 | M048L | 69 | LSDV053 | 75 | G4L | 45 |
| 54R | 43837 | 45156 | 439 | ? | Unknown | 54R | 672 | 76 | SPV051 | 49 | M049R | 44 | LSDV054 | 49 | G5R | 43 |
| 55R | 45159 | 45350 | 63 | ? | RNA polymerase subunit, RPO7 | 55R | 125 | 96 | SPV052 | 84 | M050R | 85 | LSDV055 | 85 | G5.5R | 79 |
| 56R | 45350 | 45895 | 181 | L | TM | 56R | 291 | 79 | SPV053 | 53 | M051R | 57 | LSDV056 | 54 | G6R | 47 |
| 57L | 46964 | 45864 | 366 | L | Virion core protein, TM | 57L | 580 | 78 | SPV054 | 53 | M052L | 52 | LSDV057 | 55 | G7L | 48 |
| 58R | 46994 | 47776 | 260 | L | Late transcription factor, VLTF-1, TM | 58R | 511 | 97 | SPV055 | 88 | M053R | 83 | LSDV058 | 86 | G8R | 83 |
| 59R | 47808 | 48806 | 332 | L | Myristylated protein | 59R | 528 | 78 | SPV056 | 52 | M054R | 53 | LSDV059 | 57 | G9R | 45 |
| 60R | 48807 | 49550 | 247 | L | Myristylated IMV envelope protein | 60R | 452 | 91 | SPV057 | 82 | M055R | 75 | LSDV060 | 80 | L1R | 69 |
| 61R | 49565 | 49840 | 91 | ? | TM | 61R | 99 | 57 | SPV058 | 32 | LSDV061 | 34 | ||||
| 62L | 50763 | 49816 | 315 | L | Unknown | 62L | 518 | 80 | SPV059 | 59 | M057L | 54 | LSDV062 | 60 | L3L | 51 |
| 63R | 50788 | 51546 | 252 | L | DNA binding protein | 63R | 455 | 92 | SPV060 | 76 | M058R | 77 | LSDV063 | 79 | L4R | 60 |
| 64R | 51566 | 51964 | 132 | L | TM | 64R | 181 | 68 | SPV061 | 46 | M059R | 44 | LSDV064 | 50 | L5R | 44 |
| 65R | 51906 | 52385 | 159 | L | Unknown | 65R | 275 | 83 | SPV062 | 58 | M060R | 58 | LSDV065 | 65 | J1R | 47 |
| 66R | 52382 | 52927 | 181 | E? | Thymidine kinase | 66R | 287 | 78 | SPV063 | 62 | M061R | 61 | LSDV066 | 58 | J2R | 61 |
| 67R | 52968 | 53471 | 167 | L | Host range protein | 67R | 283 | 81 | SPV064 | 45 | M062R | 38 | LSDV067 | 44 | C7L | 37 |
| 68R | 53549 | 54550 | 333 | ? | Poly(A) polymerase | 68R | 583 | 85 | SPV065 | 69 | M065R | 70 | LSDV068 | 72 | J3R | 67 |
| 69R | 54465 | 55022 | 185 | ? | RNA polymerase subunit, RPO22 | 69R | 317 | 91 | SPV066 | 75 | M066R | 72 | LSDV069 | 78 | J4R | 72 |
| 70L | 55412 | 54999 | 137 | L | Unknown | 70L | 251 | 82 | SPV067 | 62 | M067L | 62 | LSDV070 | 64 | J5L | 60 |
| 71R | 55509 | 59366 | 1285 | L | RNA polymerase subunit, RPO147 | 71R | 2382 | 91 | SPV068 | 81 | M068R | 82 | LSDV071 | 82 | J6R | 78 |
| 72L | 59872 | 59363 | 169 | L | Protein tyrosine phosphatase | 72L | 317 | 88 | SPV069 | 71 | M069L | 74 | LSDV072 | 77 | H1L | 63 |
| 73R | 59887 | 60456 | 189 | L? | TM | 73R | 340 | 84 | SPV070 | 67 | M070R | 65 | LSDV073 | 67 | H2R | 61 |
| 74L | 61420 | 60458 | 320 | L | IMV envelope protein, TM | 74L | 498 | 79 | SPV071 | 55 | M071L | 50 | LSDV074 | 53 | H3L | 36 |
| 75L | 63814 | 61421 | 797 | L | RNA polymerase-associated protein, RAP94 | 75L | 1388 | 86 | SPV072 | 71 | M072L | 70 | LSDV075 | 72 | H4L | 64 |
| 76R | 64007 | 64549 | 180 | L? | Late transcription factor VLTF-4 | 76R | 205 | 61 | SPV073 | 41 | M073R | 40 | LSDV076 | 34 | H5R | 34 |
| 77R | 64560 | 65507 | 315 | ? | DNA topoisomerase | 77R | 536 | 83 | SPV074 | 62 | M074R | 64 | LSDV077 | 68 | H6R | 63 |
| 78R | 65515 | 65967 | 150 | L | Unknown | 78R | 246 | 80 | SPV075 | 54 | M075R | 53 | LSDV078 | 50 | H7R | 36 |
| 79R | 65982 | 68504 | 840 | L | mRNA capping enzyme (large subunit) | 79R | 1462 | 85 | SPV076 | 65 | M076R | 65 | LSDV079 | 68 | D1R | 63 |
| 80L | 68927 | 68466 | 153 | L | Virion protein | 80L | 224 | 69 | SPV077 | 39 | M077L | 38 | LSDV080 | 33 | D2L | 42 |
| 81R | 68926 | 69663 | 245 | ? | Virion protein | 81R | 332 | 64 | SPV078 | 31 | M078R | 28 | LSDV081 | 38 | D3R | 32 |
| 82R | 69660 | 70319 | 219 | ? | Uracil DNA glycosylase | 82R | 394 | 82 | SPV079 | 69 | M079R | 71 | LSDV082 | 70 | D4R | 67 |
| 83R | 70393 | 72753 | 786 | L | NTPase, TM | 83R | 1465 | 91 | SPV080 | 74 | M080R | 75 | LSDV083 | 74 | D5R | 66 |
| 84R | 72750 | 74657 | 635 | L | Early transcription factor VETFs, TM | 84R | 1227 | 95 | SPV081 | 87 | M081R | 87 | LSDV084 | 88 | D6R | 80 |
| 85R | 74690 | 75172 | 160 | L | RNA polymerase subunit RPO18 | 85R | 309 | 94 | SPV082 | 71 | M082R | 77 | LSDV085 | 78 | D7R | 73 |
| 86R | 75194 | 75859 | 221 | ? | mutT motif | 86R | 367 | 87 | SPV083 | 62 | M084R | 56 | LSDV086 | 64 | D9R | 55 |
| 87R | 75856 | 76572 | 239 | L | mutT motif | 87R | 431 | 89 | SPV084 | 60 | M085R | 61 | LSDV087 | 62 | D10 | 50 |
| 88L | 78481 | 76586 | 631 | L | NPH-1, transcription termination factor | 88L | 1159 | 90 | SPV085 | 69 | M086L | 67 | LSDV088 | 71 | D11L | 69 |
| 89L | 79373 | 78510 | 287 | L | mRNA capping enzyme, VITF | 89L | 528 | 91 | SPV086 | 78 | M087L | 73 | LSDV089 | 74 | D12L | 70 |
| 90L | 81059 | 79398 | 553 | L | Rifampin resistance protein | 90L | 1055 | 93 | SPV087 | 79 | M088L | 77 | LSDV090 | 80 | D13L | 73 |
| 91L | 81531 | 81076 | 151 | L | Late transcription factor, VLTF-2 | 91L | 266 | 86 | SPV088 | 62 | M089L | 68 | LSDV091 | 64 | A1L | 62 |
| 92L | 82229 | 81555 | 224 | ? | Late transcription factor, VLTF-3 | 92L | 442 | 95 | SPV089 | 83 | M090L | 86 | LSDV092 | 85 | A2L | 84 |
| 93L | 82453 | 82226 | 75 | L | Unknown | 93L | 139 | 84 | SPV090 | 55 | M091L | 69 | LSDV093 | 63 | A2.5L | 53 |
| 94L | 84440 | 82467 | 657 | L | Virion core protein | 94L | 1195 | 90 | SPV091 | 73 | M092L | 71 | LSDV094 | 70 | A3L | 63 |
| 95L | 84946 | 84500 | 148 | L | Virion core protein | 95L | 201 | 70 | SPV092 | 37 | M093L | 33 | LSDV095 | 37 | ||
| 96R | 84986 | 85483 | 165 | L | RNA polymerase subunit RPO19 | 96R | 260 | 80 | SPV093 | 54 | M094R | 52 | LSDV096 | 56 | A5R | 58 |
| 97L | 86595 | 85480 | 371 | L | Unknown | 97L | 673 | 91 | SPV094 | 70 | M095L | 70 | LSDV097 | 75 | A6L | 56 |
| 98L | 88760 | 86619 | 713 | L? | Early transcription factor, VETF1 | 98L | 1273 | 89 | SPV095 | 74 | M096L | 73 | LSDV098 | 74 | A7L | 68 |
| 99R | 88817 | 89692 | 291 | E? | Intermediate transcription factor VITF-3 | 99R | 528 | 90 | SPV096 | 64 | M097R | 68 | LSDV099 | 64 | A8R | 61 |
| 100L | 89932 | 89693 | 79 | L | IMV membrane protein, SP, TM | 100L | 146 | 91 | SPV097 | 82 | M098L | 72 | LSDV100 | 74 | A9L | 71 |
| 101L | 92641 | 89933 | 902 | L | Virion core protein P4a | 101L | 1575 | 87 | SPV098 | 64 | M099L | 57 | LSDV101 | 62 | A10L | 50 |
| 102R | 92656 | 93600 | 314 | L | Unknown | 102R | 521 | 85 | SPV099 | 72 | M100R | 69 | LSDV102 | 71 | A11R | 52 |
| 103L | 94104 | 93601 | 167 | L | Virion core protein | 103L | 249 | 77 | SPV100 | 55 | M101L | 61 | LSDV103 | 55 | A12L | 46 |
| 104L | 94357 | 94151 | 68 | L | IMV membrane protein, TM | 104L | 122 | 83 | SPV101 | 51 | M102L | 47 | LSDV104 | 58 | A13L | 35 |
| 105L | 94685 | 94404 | 93 | L | IMV membrane protein SP, TM | 105L | 160 | 82 | SPV102 | 72 | M103L | 64 | LSDV105 | 65 | A14L | 45 |
| 106L | 94864 | 94676 | 62 | L | Virulence factor, SP | 106L | 46 | 86 | SPV103 | 76 | M104L | 81 | LSDV106 | 67 | ||
| 107L | 95138 | 94854 | 94 | L | Unknown | 107L | 167 | 78 | SPV104 | 51 | M105L | 52 | LSDV107 | 55 | A15L | 49 |
| 108L | 96267 | 95122 | 381 | L | Myristylated membrane protein, TM | 108L | 629 | 78 | SPV105 | 58 | M106L | 54 | LSDV108 | 60 | A16L | 50 |
| 109L | 96847 | 96278 | 189 | L | Phosphorylated IMV membrane protein, TM | 109L | 299 | 80 | SPV106 | 59 | M107L | 51 | LSDV109 | 50 | A17L | 37 |
| 110R | 96862 | 98298 | 478 | ? | DNA helicase, TM | 110R | 839 | 85 | SPV107 | 61 | M108R | 62 | LSDV110 | 57 | A18R | 55 |
| 111L | 98503 | 98279 | 74 | L | Unknown | 111L | 102 | 68 | SPV108 | 69 | M109L | 81 | LSDV111 | 75 | A19L | 58 |
| 112L | 98840 | 98508 | 110 | L | TM | 112L | 167 | 71 | SPV110 | 49 | M110L | 44 | LSDV113 | 47 | A21L | 44 |
| 113R | 98839 | 100119 | 426 | ? | DNA polymerase processivity factor | 113R | 668 | 75 | SPV109 | 48 | M111R | 46 | LSDV112 | 51 | A20R | 46 |
| 114R | 100126 | 100602 | 158 | L | DNA processing | 114R | 259 | 77 | SPV111 | 66 | M112R | 60 | LSDV114 | 65 | A22R | 63 |
| 115R | 100625 | 101773 | 382 | L | Intermediate transcription factor VITF-3 | 115R | 614 | 80 | SPV112 | 59 | M113R | 58 | LSDV115 | 61 | A23R | 59 |
| 116R | 101775 | 105269 | 1164 | L | RNA polymerase subunit RPO132 | 116R | 2179 | 92 | SPV113 | 84 | M114R | 83 | LSDV116 | 85 | A24R | 79 |
| 117L | 105724 | 105272 | 150 | L | Fusion protein SP, TM | 117L | 125 | 48 | SPV114 | 39 | M115L | 25 | LSDV117 | 31 | A27L | 56 |
| 118L | 106150 | 105725 | 141 | L | 118L | 215 | 70 | SPV115 | 57 | M116L | 52 | LSDV118 | 57 | A28L | 48 | |
| 119L | 107065 | 106163 | 300 | ? | RNA polymerase subunit RPO35 | 119L | 528 | 85 | SPV116 | 64 | M117L | 62 | LSDV119 | 64 | A29L | 56 |
| 120L | 107261 | 107034 | 75 | L | Virion protein | 120L | 99 | 72 | SPV117 | 45 | M118L | 46 | LSDV120 | 45 | A30L | 51 |
| 120.5L | 107421 | 107287 | 44 | ? | Unknown | 120.5L | SPV117.5 | M119L | LSDV118.5 | A30.5L | ||||||
| 121L | 108224 | 107460 | 254 | L | DNA packaging | 121L | 466 | 90 | SPV118 | 79 | M120L | 81 | LSDV121 | 84 | A32L | 60 |
| 122R | 108278 | 108826 | 182 | L? | EEV glycoprotein, TM | 122R | 208 | 58 | SPV119 | 30 | M121R | 36 | LSDV122 | 32 | A33R | 27 |
| 123R | 108849 | 109361 | 170 | L | EEV protein | 123R | 271 | 75 | SPV120 | 57 | M122R | 51 | LSDV123 | 48 | A34R | 45 |
| 124R | 109364 | 109942 | 192 | ? | Unknown | 124R | 266 | 70 | SPV121 | 40 | M123R | 43 | LSDV124 | 36 | A35R | 38 |
| 125R | 109969 | 110826 | 285 | I? | TM | 125R | 436 | 75 | SPV122 | 36 | M124R | 39 | LSDV125 | 36 | ||
| 126R | 110872 | 111366 | 164 | ? | EEV glycoprotein, TM | 126R | 69 | 29 | ||||||||
| 127R | 111457 | 112257 | 266 | E/L? | TM | 127R | 379 | 70 | SPV124 | 37 | M126R | 33 | LSDV127 | 37 | A37R | 26 |
| 128L | 113060 | 112260 | 266 | L? | CD47 | 128L | 263 | 52 | SPV125 | 28 | M128L | 26 | LSDV128 | 26 | ||
| 129R | 113065 | 113481 | 138 | L? | 129R | 202 | 75 | M129R | 40 | E7R | 26 | |||||
| 131R | 113605 | 113856 | 83 | L | 131R | 49 | 38 | |||||||||
| 132R | 113894 | 114151 | 85 | E | Unknown | 132R | 92 | 59 | SPV127 | 35 | LSDV130 | 40 | ||||
| 135R | 114315 | 120002 | 1895 | L? | 8 TM, SP | 135R | 2805 | 72 | SPV131 | 56 | M134R | 52 | LSDV134 | 43 | ||
| 137R | 120468 | 120932 | 154 | E | A52R family | 137R | 195 | 62 | SPV133 | 32 | M136R | 31 | LSDV136 | 34 | C6L | 28 |
| 138R | 120962 | 121981 | 339 | L | Unknown | 138R | 453 | 64 | SPV134 | 36 | M137R | 31 | LSDV137 | 39 | A51R | 34 |
| 139R | 122047 | 122631 | 194 | ? | A52R family | 139R | 253 | 68 | SPV135 | 43 | M139R | 43 | (LSDV136) | 28 | A52R | 34 |
| 141R | 122895 | 123254 | 119 | E? | Ox-2 mimic | 141R | 172 | 72 | M141R | 38 | LSDV138 | 51 | ||||
| 142R | 123296 | 124225 | 309 | ? | Ser/Thr protein kinase | 142R | 560 | 84 | SPV137 | 57 | M142R | 57 | LSDV139 | 59 | B1R | 47 |
| 143R | 124262 | 124972 | 236 | L | Host range RING finger protein | 143R | 403 | 80 | SPV138 | 43 | M143R | 47 | LSDV140 | 40 | ||
| 144R | 125037 | 125843 | 268 | L | CD46 mimic | 144R | 283 | 65 | SPV139 | 48 | M144R | 37 | LSDV141 | 43 | C3L | 37 |
| 145R | 126011 | 127000 | 329 | ? | vCCR8 | 145R | 375 | 60 | SPV146 | 30 | LSDV011 | 31 | ||||
| 146R | 127424 | 128494 | 356 | ? | Ankyrin repeat | 146R | 534 | 73 | SPV142 | 35 | M149R | 33 | LSDV147 | 37 | B4R | 24 |
| 147R | 128524 | 130017 | 497 | ? | Ankyrin repeat | 147R | 727 | 72 | SPV143 | 28 | M148R | 26 | LSDV148 | 30 | B4R | 22 |
| 148R | 130014 | 131465 | 483 | ? | Ankyrin repeat | 148R | 588 | 62 | SPV144 | 25 | M149R | 24 | LSDV152 | 24 | B4R | 16 |
| 149R | 131527 | 132456 | 310 | E | Serpin/SPI-2 ortholog | 149R | 490 | 75 | SPV145 | 37 | M151R | 40 | LSDV149 | 40 | C12L | 29 |
| 150R | 132492 | 132812 | 106 | L | Unknown | 150R | 152 | 74 | SPV147 | 33 | M004.1 | 29 | LSDV153 | 26 | ||
| 151R | 132914 | 133918 | 334 | E | A52R family | 151R | 491 | 72 | LSDV007 | 34 | C10L | 26 | ||||
Ortholog from YLDV (accession no. AJ293568).
Ortholog from SPV (accession no. AF410153).
Ortholog from myxoma virus (accession no. AF170726).
Ortholog from LSDV (accession no. AF325528).
Ortholog from vaccinia virus strain Copenhagen (accession no. M35027).
aa, amino acids.
Predicted promoters (early [E], intermediate [I], and late [L]) were determined (15). ?, uncertain or unknown; TOE, time of expression.
Predicted functions were determined by identifying YMTV orthologs from YLDV and SPV. Abbreviations: vTNF-α, viral tumor necrosis factor alpha; SP, signal peptide; TM, transmembrane domain; eIF2α, eukaryotic initiation factor 2α; IL-18, interleukin-18; EEV, extracellular enveloped virions; bp, binding protein. BLASTP2 scores were determined by performing BLAST searches at http://www.ncbi.nlm.nih.gov/BLAST/.
FIG. 1.
YMTV genomic map. The assigned ORFs of YMTV are displayed, with an arrow indicating the direction of transcription. Each ORF is colored to indicate a general functional category. In addition, the black arrows above the ORFs at either end of the genome indicate the TIR.
TABLE 2.
Immune evasion ORFs and ORFs absent from YMTV but present in YLDV
| YMTV ORF | YLDV ORF | Putative function(s)a |
|---|---|---|
| 2L | 2L | vTNF bp |
| 3L | A52R ortholog, TLR signaling inhibitor | |
| 7L | 7L | vCCR8 |
| 8L | 4 ankyrin domains | |
| 9L | Ortholog of vv M2L | |
| 10L | Secreted serpin, myxoma virus SERP-1 ortholog | |
| 12L | 12L | eIF2α mimic |
| 14L | 14L | vIL-18 bp |
| 15L | EGF domain | |
| 16L | 16L | Inhibition of apoptosis, ortholog of myxoma virus M11L |
| 18L | Ortholog of myxoma virus M013L | |
| 23L | Unknown | |
| 28R | Unknown | |
| 34L | 34L | dsRNA bp |
| 42L | Ortholog of vv O1L | |
| 128L | 128L | CD47 mimic |
| 130L | Unknown | |
| 133L | 133L | 3β-HSD |
| 134R | vIL-10 | |
| 136R | IFN-α/β binding protein | |
| 140R | Ortholog of vv A54R, kelch-like protein | |
| 141R | 141R | Ox-2 mimic |
| 144R | 144R | CD46 mimic |
| 145R | 145R | CCR8 mimic |
| 149R | 149R | Intracellular serpin, SPI-2 ortholog |
Abbreviations: vTNF, viral tumor necrosis factor; vCCR8, viral CCR8 ortholog; vv, vaccinia virus; eIF2α, eukaryotic initiation factor 2α; vIL-18, viral interleukin-18; EGF, epidermal growth factor; dsRNA, double-stranded RNA; vIL-10, viral interleukin-10; IFN-α/β, alpha/beta interferon; bp, binding protein.
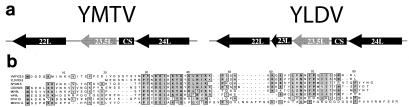
The TIR of YMTV are 1,962 bases long and contain a single ORF designated 1L/151R. The noncoding region in the TIR of YMTV and YLDV is relatively large, with 804 and 755 bases (15), respectively, between the terminal ORF and the concatemer resolution sequence. In comparison, closely related genera, such as members of the Capripoxvirus, Leporipoxvirus, and Suipoxvirus genera, have noncoding regions in their termini ranging from 159 to 366 bases (3, 8, 29). Analysis of the noncoding region from YMTV and YLDV revealed a nucleotide sequence in each that exhibited striking similarity to that of the SPV002 gene (Fig. 2). However, both the YMTV and YLDV sequences lack an initiating methionine (ATG) codon, suggesting that either the large noncoding sequence in the TIR of yatapox viruses has evolved into a pseudogene of SPV002, or else the yatapox virus orthologs utilize a nonstandard initiator codon.
FIG. 2.
YMTV and YLDV each contain an apparent pseudogene within the noncoding region of the termini. An alignment of the assigned SPV002 ORF (3) with a portion of the noncoding region of the YMTV and YLDV termini is shown.
Identification of putative orthologs of YMTV23.5L in multiple poxviruses.
An unusual number of large gaps occur between ORFs in YMTV (Table 1) and YLDV (15). Our assumption is that if these presumptive noncoding regions between yatapox virus ORFs have important functions, then they would likely be conserved between YLDV and YMTV. The largest inter-ORF gap in YLDV is 376 bases and maps between 23L and 24L (15). The corresponding region in YMTV is a 474-bp gap between 22L and 24L, with YMTV lacking any obvious ortholog of 23L. As Table 1 illustrates, a small ORF between 22L and 24L of YMTV was identified and designated 23.5L (Table 1; Fig. 3a). Orthologs of 23.5L were previously reported in myxoma virus, LSDV, and vaccinia virus (8, 13, 29).
FIG. 3.
Comparative analysis of the noncoding region between YMTV22L and YMTV24L. (a) Schematic arrangement of ORFs in the region of the 23.5L orthologs in YMTV and YLDV. A 42-base conserved sequence (CS) is shown as a black box. (b) Alignment of orthologs of 23.5L in YMTV, YLDV, LSDV (LSDV023), myxoma virus (M018L), vaccinia virus (vv F8L), fowlpox (FPV113), and molluscum contagiosum virus (MC014.1L). The SPV (SPV20.5) sequence is also shown, although it lacks a start codon in the published sequence (3).
Since YMTV23.5L was present in a number of divergent poxvirus species, we wanted to examine whether YLDV and SPV might carry a 23.5L version in their genomes. We examined the large noncoding region between YLDV23L and YLDV24L and identified a 153-bp orthologous ORF, which we have designated 23.5L in YLDV (Fig. 3a). This YLDV ORF was classified as a predicted ORF in the annotated sequence of YLDV (accession no. AJ293568) but was not classified as an authentic ORF in the published sequence (15). Interestingly, an ortholog of YMTV23.5L was also found in SPV between positions 13229 and 13445 on the genomic sequence map (3) that had significant similarity to other versions of 23.5L (Fig. 3b). However, this potential ORF in SPV lacks a canonical start codon (ATG) (Fig. 3b), suggesting that the SPV version is either a pseudogene or a sequencing error that resulted in the insertion of an extra nucleotide between a potential upstream start ATG codon in an alternative reading frame six codons upstream of the assigned codon for the first lysine residue.
Unusual conserved promoter-like sequence found in yatapox, suipox, capripox, and leporipox viruses.
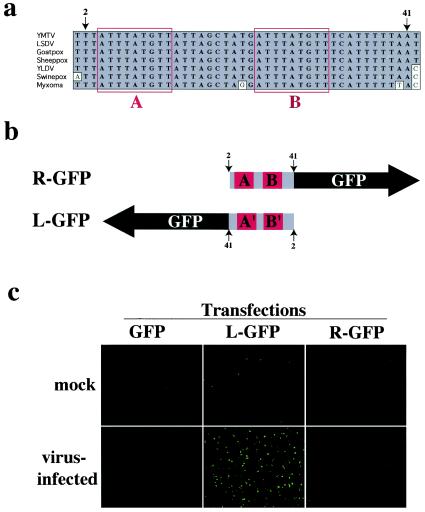
The identification of the 212-bp gene YMTV23.5L greatly reduced the amount of assigned noncoding sequence in the region between 22L and 24L. Nevertheless, when we continued our analysis of the noncoding sequence in this region between the ORFs 23.5L and 24L in YMTV and YLDV, we noticed a striking 42-bp sequence that was 100% identical between YMTV and YLDV (Fig. 4a).
FIG. 4.
Analysis of the conserved promoter-like sequence between YMTV23.5L and YMTV24L. (a) Alignment of a conserved sequence found between orthologs of YMTV23.5L and YMTV24L. The sequence within the red boxes labeled “A” and “B” represents the 9-bp repeat. The numbers above the grey box indicate the nucleotide positions. (b) Schematic of the orientation of the promoterless GFP with respect to the orientation of the cloned myxoma virus conserved sequence. R-GFP contains the conserved sequence from bases 2 to 41. L-GFP contains the reverse complement of the conserved sequence. The red boxes show the location of the two repeats in panel a. (c) Cells were either mock infected or infected with myxoma virus and subsequently transfected with either a promoterless GFP, R-GFP, or L-GFP construct. Forty-eight hours postinfection, the cells were visualized using a fluorescence microscope.
To determine whether this sequence was conserved in other poxviruses, we examined the region between the orthologs of 23.5L and 24L in SPV (SPV020.5 and SPV021), LSDV (LSDV023 and LSDV024), myxoma virus (M018L and M019L), and vaccinia virus (F8L and F9L). Figure 4a demonstrates that this identical nucleotide sequence is found in SPV, LSDV, goatpox virus, and sheeppox virus and was 95% conserved in myxoma virus but is not present in vaccinia virus or other orthopoxviruses. The unusually high degree of sequence conservation (i.e., 100% identity between positions 2 through 41 [Fig. 4a] for YMTV, YLDV, SPV, and LSDV) suggests that the sequence may have an important and conserved function.
Analysis of the sequence identified two 9-bp repeats separated by 10 bases (Fig. 4a). Since one turn of the DNA double helix is 10.4 bp, this suggests that the two repeats are registered on the same face of the DNA molecule. One possible function for this type of sequence arrangement is the binding of transcription factors to the DNA sequence, and indeed the sequence does resemble a tandem repeat of a canonical poxvirus late promoter (9). To test whether the conserved sequence might function as a viral promoter element, we inserted the conserved 42-bp sequence (derived from myxoma virus) in either the forward (R-GFP) or the reverse complement (L-GFP) orientations in front of a promoterless GFP construct. Cells were either mock infected or infected with myxoma virus and then transfected with promoterless GFP, R-GFP, or L-GFP constructs. The L-GFP but not the R-GFP sequence was able to drive some detectable GFP expression in the absence of virus infection, but a myxoma virus coinfection greatly increased the level of expression of the L-GFP construct (Fig. 4c). From these data we conclude that the conserved sequence could act as a late promoter element for the gene 23.5L; however, other potential functions such as an involvement in viral replication or packaging cannot be excluded. The reason for the unusual conservation of this promoter sequence across four genera of poxviruses remains to be determined.
Identification of two new conserved poxvirus gene families.
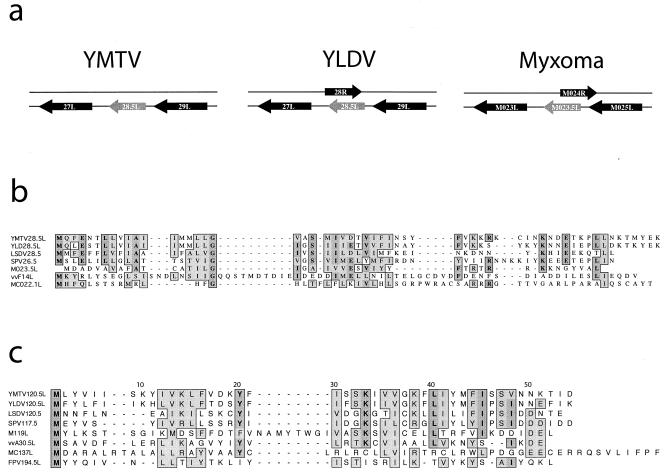
The central region of the poxvirus genome is inevitably enriched for genes that are highly conserved among all poxviruses. In YMTV, this conserved region maps between YMTV24L and YMTV124R. However, inspection of the genomic sequences from a number of poxviruses revealed that the region between YMTV ORFs 27L and 29L and between ORFs 120L and 121L were unexpectedly divergent (Fig. 5a). Analysis of the region between YMTV27L and YMTV29L identified an ORF, designated 28.5L, which encodes a 58-amino-acid protein (Table 1). Initially we examined the region between 27L and 29L in YLDV, where the previously assigned 28R gene is present. Analysis of the YLDV sequence revealed a clear ortholog of 28.5L (Fig. 6a) which overlaps extensively with 28R. Based on the fact that there are no other reported poxvirus versions of YLDV 28R in the database and there is typically only minor overlap of poxvirus ORFs with each other, we postulate that 28.5L represents the true yatapox virus ORF that maps between 27L and 29L for both YMTV and YLDV and that the slightly longer 28R encoded in the opposite polarity originally annotated for YLDV might not be expressed.
FIG. 5.
Alignment of two conserved ORF clusters in a variety of poxvirus genera. (a) Alignment of predicted ORFs from representative members from seven of the eight poxvirus genera. Two regions of the genome are shown, the orthologous region between YMTV027L and YMTV029L and the region between YMTV120L and YMTV121L. Orthologous ORFs share the same color. (b) Analysis of the region between YMTV027L/029L and YMTV120L/121L revealed two new conserved gene families. The proposed arrangements of these ORFs are shown, highlighting the arrangement of the two new gene families YMTV028.5L and YMTV120.5L.
FIG. 6.
Alignments of the predicted YMTV28.5L and YMTV120.5L protein families. (a) Arrangement of YMTV 28.5L orthologs in YMTV, YLDV, and myxoma virus. (b) Alignment of orthologs of YMTV28.5L, including YLDV28.5L, LSDV (LSDV28.5), myxoma virus (M023.5L), vaccinia virus (vvF14L), SPV (SPV26.5), and molluscum contagiosum virus (MC022.1L). (c) Alignment of orthologs of YMTV120.5L, including YLDV120.5L, LSDV (LSDV120.5), myxoma virus (M119L), vaccinia virus (vvA30.5L), swinepox virus (SPV117.5), fowlpox virus (FPV194.5L), and molluscum contagiosum virus (MC137L).
Since 28.5L appeared to be present in both sequenced members of the Yatapoxvirus genus, we examined members of other poxvirus genera to determine if orthologs of this gene could be identified. We examined the noncoding sequence between the orthologs of 27L and 29L in myxoma virus, SPV, LSDV, vaccinia virus, molluscum contagiosum virus, and fowlpox virus to determine if a previously unreported version of 28.5L existed in these genomes. Surprisingly, we found closely related orthologs of 28.5L in all poxvirus species examined, with the exception of fowlpox virus (Fig. 5b and 6b). Interestingly, the deduced ortholog of 23.5L in myxoma virus overlaps extensively with M024R (which bears no similarity with YLDV 28R [Fig. 6a]). This conservation of 23.5L in so many poxvirus genera and the lack of any other orthologs for M024R has led us to conclude that M23.5L, rather than the annotated M024R (8), may be the correct ORF that maps between M023L and M025L.
We next examined the 199-bp noncoding region between YMTV ORFs 120L and 121L. A single 44-amino-acid ORF designated YMTV120.5L was identified which lacked sequence similarity to any gene in the published database. Therefore, as in the case of the gap between 27L and 29L of YMTV, we examined the sequence gap between YMTV120L and YMTV121L (Fig. 5a) and looked for other poxvirus ORFs in this conserved region. This approach yielded clear orthologs of YMTV120.5L in all poxvirus species examined (Fig. 5b and 6c). Interestingly, versions of YMTV120.5L were previously identified in myxoma virus and molluscum contagiosum virus, although originally no relationship was reported between them, presumably because the small gene size made determination of significant identity difficult. However, the position of the conserved ORF in the genomes, the sequence similarities, and the similar gene sizes all indicate that these ORFs are part of an ancestrally evolved gene cluster that is conserved across multiple poxvirus genera.
DISCUSSION
In this work we report the complete sequence of the YMTV genome and have identified three ORFs previously unidentified in most poxviruses. The YMTV genome size of 134,721 bases represents the smallest poxvirus genome yet sequenced. In contrast, the closely related YLDV genome is approximately 144,575 bases long (15). The difference in genome sizes between YMTV and YLDV is due to the complete deletion of 13 ORFs found in YLDV but absent from YMTV. The bulk of the YMTV deleted ORFs are found at the left end of the genome and represent determinants of immune evasion, host range, or genes of unknown function (Table 2). Clinically, YMTV and YLDV produce distinct diseases, with YMTV producing histiocyte-filled tumors upon infection, whereas YLDV infection resembles a mild form of smallpox (5, 20, 27). It is possible that the absence of various YLDV gene products might, in some way, contribute to the tumorigenic phenotype produced upon YMTV infection, but the contribution of these 13 deleted genes to disease phenotype awaits further study.
The data presented here highlight the utility of using a comparative genomic approach when analyzing viral genomes for predicted genes. One of the difficulties in whether to assign a nucleotide sequence as an annotated ORF, particularly for small ORFs of less than 150 nucleotides, is that there is no way to confirm that a predicted ORF is actually expressed until the translated protein or mRNA is detected experimentally. However, we reasoned that if a putative ORF actually encodes a protein, it would be conserved in at least some other poxvirus genus members. Therefore, we examined the tentatively assigned noncoding regions between ORFs in poxvirus genomic sequences to identify yatapoxvirus ORFs with demonstrable similarity in terms of size, sequence, and presence of contiguous orthologs. This approach identified three new yatapoxvirus gene families (23.5L, 28.5L, and 120.5L) that are clearly conserved throughout many genera of poxviruses (Table 3). These three gene families all appear to encode unique proteins with no significant similarity with any other viral or cellular proteins in the sequence database, but which are clearly conserved in most of the known poxvirus genera. With the renewed interest in variola virus, the causative agent of smallpox, it is particularly relevant to identify new families of conserved viral genes that may have important conserved roles in poxvirus replication or pathogenesis.
TABLE 3.
Members of three new poxvirus gene families
| Virus | YMTV23.5 family
|
YMTV28.5 family
|
YMTV120.5 family
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Start | Stop | Gene | Start | Stop | Gene | Start | Stop | |
| YMTV | 23.5L | 14742 | 14530 | 28.5L | 20085 | 19909 | 120.5L | 107287 | 107421 |
| YLDV | 23.5L | 17960 | 17808 | 28.5L | 23539 | 23366 | 120.5L | 113015 | 112881 |
| SPV | 20.5 | 13430 | 13229 | 26.5 | 20113 | 19949 | 117.5 | 110742 | 110617 |
| LSDV | 023 | 15949 | 15734 | 28.5 | 22161 | 22012 | 120.5 | 112519 | 112394 |
| Goatpox virus strain G20-LKV | 023 | 15430 | 15211 | 28.5 | 21620 | 21470 | 120.5 | 111927 | 111807 |
| Sheeppox virus | 023 | 15557 | 15342 | 28.5 | 21685 | 21536 | 120.5 | 112134 | 112009 |
| Myxoma virus | 018L | 18513 | 18316 | 23.5L | 23834 | 23703 | 119L | 114993 | 114844 |
| Shope fibroma virus | 018L | 17726 | 17526 | 23.5L | 23037 | 22906 | 119L | 114122 | 114003 |
| Vaccinia virus | |||||||||
| Ankara | 037L | 30731 | 30534 | 044L | 37105 | 36884 | 141.5L | 133014 | 132889 |
| Tian Tan | TF8L | 35166 | 35318 | TF14L | 41537 | 41758 | TA30.5L | 141666 | 141540 |
| Copenhagen | F8L | 38878 | 38684 | F14L | 45318 | 45100 | A30.5L | 141046 | 140918 |
| WR | VACWR047 | 35577 | 35774 | VACWR053 | 41967 | 42188 | VACWR153.5 | 142061 | 141933 |
| Variola virus | |||||||||
| Garcia | E8L | 27400 | 27579 | E14L | 33818 | 34039 | A34.5L | 133824 | 133696 |
| Bangladesh 1975 | C12L | 27031 | 27228 | C18L | 33457 | 33678 | A33.5L | 133432 | 133313 |
| India 1967 | E8L | 27597 | 27400 | E14L | 34039 | 33818 | A33.5L | 132818 | 132690 |
| Ectromelia virus | EVM031 | 44331 | 44528 | EVM037 | 50749 | 50964 | 132.5 | 150491 | 150363 |
| Camelpox virus | CMLV043 | 38437 | 38634 | CMLV049 | 44853 | 45074 | CMLV170.5 | 144313 | 144185 |
| Monkeypox virus | C14L | 36022 | 35828 | C20L | 42461 | 42240 | A31.5L | 141603 | 141484 |
| Cowpox virus | CPXV055 | 52234 | 52431 | CPXV062 | 58648 | 58869 | CPVX165.5 | 159066 | 158938 |
| Fowlpox virus | 113 | 134861 | 135058 | 194.5L | 227787 | 227667 | |||
| Molluscum contagiosum virus | 014.1L | 18646 | 18897 | MC22.1L | 28628 | 28807 | 137L | 158648 | 158812 |
An unexpected finding from these observations was that several small ORFs that turned out to be members of conserved poxvirus gene families were originally characterized as unique. For example, fowlpox virus gene FPV113 (2) and molluscum contagiosum virus gene MC014.1L (23) were identified as unique genes, but our comparative analysis demonstrated that they are instead part of a larger poxvirus gene family that includes the vaccinia virus F8L gene. The reason that FPV113 and MC014.1L were not identified as being related to vaccinia virus F8L was likely that it is difficult to reach a level of statistical significance with computer database searches when the raw similarity score is reduced because of their small sizes.
Comparing genomic sequences from different poxviruses in this fashion can provide insight into the evolutionary history of these viruses. For example, comparing the presumptive noncoding regions of both YLDV and YMTV with the same region of SPV revealed a potential pseudogene in YLDV and YMTV that had significant sequence similarity with the SPV002 gene. The presence of the same pseudogene in both YMTV and YLDV but of a functional copy of the gene in SPV implies that the pseudogene arose after the split of the suipox viruses from the yatapox viruses. In this way, we can develop an evolutionary timeline for some of the major events that differentiated members of the diverse poxvirus genera.
In addition to the identification of potential ORFs, the comparative genomic approach resulted in the unexpected identification of a 40-nucleotide stretch of YMTV sequence that was 100% conserved across members of the Yatapoxvirus, Suipoxvirus, and Capripoxvirus genera. This domain represents the most highly conserved sequence yet described among these poxviruses. Even the highly conserved concatemer resolution sequence, which is involved in the essential elements of poxvirus replication at the termini, is only 81% conserved between these species. This conserved sequence maps in the noncoding region between YMTV ORFs 23.5L and 24L. Although we demonstrated that this sequence can function as a late promoter element (Fig. 4), it is not yet clear if that is the actual function of this sequence during a viral infection. For example, the poxvirus concatemer resolution sequence can function as a poxvirus late promoter element (TAAAT) sequence (28); however, its primary role appears to be in resolving concatemers during viral replication (19). One way to test the potential function of this conserved promoter-like sequence would be to generate virus deletion mutants in any one of the virus members that contain a copy of the sequence.
The data presented here have illustrated some of the potential applications of taking a comparative approach to analyze poxvirus genomics. Through the comparison of poxvirus genomes across genera we identified three new gene families that had previously been overlooked because of their small size. In addition, conserved sequences that do not encode an ORF but that potentially play an important role in poxvirus replication were also identified. The comparative genomic analysis that we undertook was originally made possible due to the sequencing of the YMTV genome and the ability to compare its sequence to that of another relatively close species, YLDV (15). However, in theory, the comparative approach that we took could be applied to any viral family and may be particularly valuable when trying to predict whether small potential ORFs truly encode a protein.
Acknowledgments
We thank Karim Essani and Koji Ishii for critical reading of the manuscript.
This work was supported by the National Cancer Institute of Canada and by Viron Therapeutics, Inc.
REFERENCES
- 1.Afonso, C., E. Tulman, Z. Lu, L. Zsak, N. Sandybaev, U. Kerembekova, V. Zaitsev, G. Kutish, and D. Rock. 2002. The genome of camelpox virus. Virology 295:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osario, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano, H., Y. Ueda, and T. Miyamura. 1995. Identification and characterization of the thymidine kinase gene of Yaba virus. J. Gen. Virol. 76:1109-1115. [DOI] [PubMed] [Google Scholar]
- 5.Ambrus, J. L., E. T. Feltz, J. T. Grace, Jr., and G. Owens. 1963. A virus-induced tumor in primates. Natl. Cancer Inst. Monogr. 10:447-458. [Google Scholar]
- 6.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-395. [DOI] [PubMed] [Google Scholar]
- 7.Bearcroft, W. G. C., and M. F. Jamieson. 1958. An outbreak of subcutaneous tumours in rhesus monkeys. Nature 182:195-196. [DOI] [PubMed] [Google Scholar]
- 8.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J.-X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., and B. Moss. 1989. Structure of vaccinia virus late promoters. J. Mol. Biol. 210:771-784. [DOI] [PubMed] [Google Scholar]
- 10.Downie, A. W., and C. Espana. 1972. Comparison of Tanapox virus and Yaba-like viruses causing epidemic disease in monkeys. J. Hyg. 70:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 12.Grace, T. T. J., and E. A. Mirand. 1963. Human susceptibility to a simian tumor virus. Ann. N. Y. Acad. Sci. 108:1123-1128. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology 196:381-401. [DOI] [PubMed] [Google Scholar]
- 14.Knight, J. C., F. J. Novembre, D. R. Brown, C. S. Goldsmith, and J. J. Esposito. 1989. Studies on Tanapox virus. Virology 172:116-124. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H.-J., K. Essani, and G. L. Smith. 2001. The genome sequence of Yaba-like disease virus, a Yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 16.Massung, R. F., L. I. Liu, J. Qi, J. C. Knight, T. E. Yuran, A. R. Kerlavage, J. M. Parsons, J. C. Venter, and J. J. Esposito. 1994. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 201:215-240. [DOI] [PubMed] [Google Scholar]
- 17.McNulty, W. P., W. C. Lobitz, F. Hu, C. A. Maruffo, and A. S. Hall. 1968. A pox disease in monkeys transmitted to man. Clinical and histological features. Arch. Dermatol. 97:286-293. [PubMed] [Google Scholar]
- 18.Merchlinsky, M., and B. Moss. 1989. Nucleotide sequence required for resolution of the concatemer junction of vaccinia virus DNA. J. Virol. 63:4354-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Niven, J. S. F., J. A. Armstrong, C. H. Andrews, H. G. Pereira, and R. C. Valentine. 1961. Subcutaneous ‘growths' in monkeys produced by a poxvirus. J. Pathol. Bacteriol. 81:1-14. [PubMed] [Google Scholar]
- 21.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain- terminating inhibitors. Proc. Natl. Acad. Sci. USA 93:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 23.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 24.Shchelkunov, S. N., R. F. Massung, and J. J. Esposito. 1995. Comparison of the genome DNA sequences of Bangladesh-1975 and India-1967 variola viruses. Virus Res. 36:107-118. [DOI] [PubMed] [Google Scholar]
- 25.Shchelkunov, S. N., A. V. Totmenin, V. N. Loparev, P. F. Safronov, V. V. Gutorov, V. E. Chizhikov, J. C. Knight, J. M. Parsons, R. F. Massung, and J. J. Esposito. 2000. Alastrim smallpox variola minor virus genome DNA sequences. Virology 266:361-386. [DOI] [PubMed] [Google Scholar]
- 26.Shchelkunov, S. N., A. V. Totmenin, P. F. Safronov, M. V. Mikheev, V. V. Gutorov, O. I. Ryazankina, N. A. Petrov, I. V. Babkin, E. A. Uvarova, L. S. Sandakhchiev, J. R. Sisler, J. J. Esposito, I. K. Damon, P. B. Jahrling, and B. Moss. 2002. Analysis of the monkeypox virus genome. Virology 297:172-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sproul, E. E., R. S. Metzgar, and J. T. J. Grace. 1963. The pathogenesis of Yaba virus-induced histiocytomas in primates. Cancer Res. 23:671-675. [PubMed] [Google Scholar]
- 28.Stuart, D., K. Graham, M. Schreiber, C. Macaulay, and G. McFadden. 1991. The target DNA sequence for resolution of poxvirus replicative intermediates is an active late promoter. J. Virol. 65:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2001. Genome of lumpy skin disease virus. J. Virol. 75:7122-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, J.-H. Sur, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genomes of sheeppox and goatpox viruses. J. Virol. 76:6054-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willer, D., G. McFadden, and D. H. Evans. 1999. The complete genome sequence of Shope (rabbit) fibroma virus. Virology 264:319-343. [DOI] [PubMed] [Google Scholar]