Abstract
The title compound, C16H14O7·H2O, possesses a planar three-ring skeleton; its carbonyl, one of the two hydroxy and two of the three methoxy O atoms and the water molecule form hydrogen bonds, giving rise to a layer structure.
Related literature
For the antidepressant, antitumor, antimicrobial, antifungal, anti-inflammatory, antiviral, cardiotonic, hypoglycemic, antihepatotoxic and immunomodulatory activities of simple xanthones, see: Basnet et al. (1994 ▶); Fernandes et al. (1995 ▶); Karan et al. (1999 ▶); Liou et al. (1993 ▶); Miura et al. (2001 ▶); Parmar et al. (1996 ▶); Pedro et al. (2002 ▶); Sousa et al. (2002 ▶). For the crystal structures of oxygenated xanthones, see: Evans et al. (2004 ▶); Gales et al. (2001 ▶); Jiang et al. (2004 ▶); Kabaleeswaran et al. (2003 ▶); Kato et al. (2005 ▶); Kijjoa et al. (1998 ▶); Shi et al. (2004 ▶, 2005 ▶); Stout et al. (1969 ▶); Vijayalakshmi et al. (1987 ▶).
Experimental
Crystal data
C16H14O7·H2O
M r = 336.29
Monoclinic,
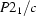
a = 10.9272 (9) Å
b = 10.4511 (8) Å
c = 14.0201 (11) Å
β = 111.6830 (10)°
V = 1487.8 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.12 mm−1
T = 298 K
0.2 × 0.1 × 0.05 mm
Data collection
Bruker SMART 1K CCD diffractometer
Absorption correction: none
8808 measured reflections
3563 independent reflections
2848 reflections with I > 2σ(I)
R int = 0.052
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.141
S = 1.06
3563 reflections
233 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.29 e Å−3
Δρmin = −0.24 e Å−3
Data collection: SMART (Bruker, 1999 ▶); cell refinement: SAINT (Bruker, 1999 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808004832/ng2424sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808004832/ng2424Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O8i—H16i⋯O2 | 0.73 (3) | 2.58 (3) | 3.091 (2) | 129.4 |
| O8i—H16i⋯O3 | 0.73 (3) | 2.46 (3) | 3.177 (2) | 167.3 |
| O8ii—H15ii⋯O6 | 0.84 (5) | 2.08 (5) | 2.923 (2) | 172.3 |
| O7—H7⋯O8iii | 0.83 | 1.88 | 2.706 (2) | 169 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported in part by the NSF-EPSCoR, USA.
supplementary crystallographic information
Comment
Xanthone compounds commonly occur in several higher plant families, such as Gentianaceae,Guttiferae, Moraceae and Polygalaceae. Simple oxygenated xanthones possess different biological activities such as antidepressant, antitumor, antimicrobial, antifungal, anti-inflammatory, antiviral, cardiotonic, hypoglycemic, antihepatotoxic and immunomodulatory (Liou et al., 1993; Basnet et al., 1994; Fernandes et al., 1995; Parmar et al., 1996; Karan et al., 1999; Miura et al., 2001; Pedro et al., 2002; Sousa et al., 2002). The majority of the xanthones isolated so far are hydroxyl or methoxy substituted in the xanthone skeleton. Up to present, only ten simple oxygenated xanthones were characterized by X-ray diffraction (Stout et al., 1969; Vijayalakshmi et al., 1987; Kijjoa et al., 1998; Gales et al., 2001; Kabaleeswaran et al., 2003; Jiang et al., 2004; Shi et al., 2004; Evans et al., 2004; Kato et al., 2005; Shi et al., 2005). 1,7-dihydroxy-2,3,4-trimethoxyxanthone (I) was first isolated from Halenia elliptica D. Don (Gentianaceae) and has antioxidant activity. Its crystal structure is reported for the first time in this paper. The structure of I (Figure 1) is similar to other xanthones reported with a planar three-ring skeleton. The asymmetric unit of crystal I contains one molecule I plus one water molecule. I forms hydrogen bonds with the water molecule through its carbonyl, one of the two hydroxyl and two of the three methoxyl O atoms (Table 1, Figure 2). The crystal structure is stabilized by the extensive hydrogen bond network.
Experimental
Halenia elliptica D. Don was collected in DALI, Yunnan Province, in April 2005 and was identified by Professor Xiaokuang Ma, Department of Pharmacognosy, School of Pharmacy, DALI University, People's Republic of China.
Extraction and isolation: 1,7-dihydroxy-2,3,4-trimethoxyxanthone (I) was isolated from ethyl acetate fraction of the ethanol extract of the aerial parts of Halenia elliptica with other four 1,7-dihydroxy substituted xanthones by silica gel column chromatography with gradient mixtures of petroleum ether and ethyl acetate. Yellow crystals of I were obtained by slow evaporation of a solution in EtOH. M.p.164–165°C. ESI-MS m/z (rel. %): 319[M+H]+.1H-NMR (400 MHz, CDCl3), d12.60 (1H, s, OH-1), 7.60 (1H, d, J=3.0 Hz, H-8), 7.49 (1H, d, J=9.3 Hz, H-5), 7.34 (1H, dd, J=3.0, 9.0 Hz, H-6), 5.32 (1H, br. s, OH-7), 4.15 (3H, s, OCH3), 3.96 (3H, s, OCH3), 3.95 (3H, s, OCH3).
Refinement
H atoms attached to O atoms were located in a difference map and refined with bond restraints O—H = 0.82 (2) Å. C-bound H atoms were positioned geometrically (C—H 0.93 - 0.96 Å). All H atoms were refined as riding, with Uiso(H) = 1.2 - 1.5 Ueq of the parent atom.
Figures
Fig. 1.
The molecular structure of (I). Displacement ellipsoids are drawn at the 50% probability level. Arbitrary atom numbering.
Fig. 2.
The packing of (I), viewed down the b axis.
Crystal data
| C16H14O7·H2O | F000 = 704 |
| Mr = 336.29 | Dx = 1.501 Mg m−3 |
| Monoclinic, P21/c | Melting point: 437 K |
| Hall symbol: -P 2ybc | Mo Kα radiation λ = 0.71073 Å |
| a = 10.9272 (9) Å | Cell parameters from 3563 reflections |
| b = 10.4511 (8) Å | θ = 2.5–28.3º |
| c = 14.0201 (11) Å | µ = 0.12 mm−1 |
| β = 111.6830 (10)º | T = 298 K |
| V = 1487.8 (2) Å3 | Block, yellow |
| Z = 4 | 0.2 × 0.1 × 0.05 mm |
Data collection
| Bruker SMART 1K CCD diffractometer | 2848 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.053 |
| Monochromator: graphite | θmax = 28.3º |
| T = 298 K | θmin = 2.5º |
| Thin–slice ω scans | h = −14→14 |
| Absorption correction: none | k = −8→13 |
| 8808 measured reflections | l = −18→18 |
| 3563 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.046 | w = 1/[σ2(Fo2) + (0.0743P)2 + 0.2855P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.142 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.29 e Å−3 |
| 3563 reflections | Δρmin = −0.24 e Å−3 |
| 233 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0085 (18) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O5 | 0.62447 (9) | 0.47253 (9) | 0.17678 (8) | 0.0374 (2) | |
| O1 | 0.44791 (10) | 0.90069 (10) | 0.12081 (9) | 0.0441 (3) | |
| H1 | 0.3758 | 0.8590 | 0.0943 | 0.066* | |
| C11 | 0.52749 (12) | 0.68367 (13) | 0.14635 (9) | 0.0315 (3) | |
| O6 | 0.29738 (10) | 0.70239 (10) | 0.06007 (9) | 0.0492 (3) | |
| O2 | 0.69716 (11) | 0.99477 (10) | 0.21570 (8) | 0.0437 (3) | |
| C2 | 0.67515 (14) | 0.86433 (13) | 0.20640 (10) | 0.0361 (3) | |
| C12 | 0.63679 (13) | 0.60211 (13) | 0.18592 (10) | 0.0325 (3) | |
| C10 | 0.39784 (13) | 0.63185 (13) | 0.09340 (10) | 0.0343 (3) | |
| C13 | 0.50227 (13) | 0.41989 (13) | 0.12579 (10) | 0.0336 (3) | |
| C4 | 0.76454 (13) | 0.64776 (14) | 0.23766 (11) | 0.0370 (3) | |
| C8 | 0.26918 (14) | 0.43164 (14) | 0.02687 (11) | 0.0371 (3) | |
| H8 | 0.1934 | 0.4798 | −0.0049 | 0.045* | |
| O3 | 0.89800 (11) | 0.84290 (12) | 0.29620 (11) | 0.0599 (4) | |
| C3 | 0.78279 (13) | 0.78037 (15) | 0.24786 (11) | 0.0385 (3) | |
| C9 | 0.38925 (13) | 0.49338 (13) | 0.08075 (10) | 0.0331 (3) | |
| C1 | 0.54949 (13) | 0.81769 (13) | 0.15801 (10) | 0.0331 (3) | |
| O7 | 0.14973 (11) | 0.23466 (11) | −0.03034 (10) | 0.0517 (3) | |
| H7 | 0.0882 | 0.2864 | −0.0560 | 0.078* | |
| C7 | 0.26315 (14) | 0.30003 (14) | 0.02086 (11) | 0.0386 (3) | |
| O4 | 0.86845 (10) | 0.56302 (11) | 0.26990 (9) | 0.0500 (3) | |
| C5 | 0.49582 (15) | 0.28688 (14) | 0.12202 (11) | 0.0398 (3) | |
| H5 | 0.5711 | 0.2383 | 0.1543 | 0.048* | |
| C6 | 0.37723 (16) | 0.22793 (14) | 0.07010 (11) | 0.0421 (3) | |
| H6 | 0.3727 | 0.1391 | 0.0677 | 0.051* | |
| C15 | 1.02209 (16) | 0.7842 (2) | 0.32996 (18) | 0.0673 (5) | |
| H15A | 1.0321 | 0.7309 | 0.3881 | 0.101* | |
| H15B | 1.0895 | 0.8486 | 0.3492 | 0.101* | |
| H15C | 1.0297 | 0.7329 | 0.2756 | 0.101* | |
| C14 | 0.7299 (2) | 1.04398 (18) | 0.13328 (16) | 0.0595 (5) | |
| H14A | 0.8034 | 0.9974 | 0.1290 | 0.089* | |
| H14B | 0.7526 | 1.1328 | 0.1453 | 0.089* | |
| H14C | 0.6557 | 1.0349 | 0.0700 | 0.089* | |
| C16 | 0.8790 (2) | 0.4892 (2) | 0.35909 (16) | 0.0701 (6) | |
| H16A | 0.8795 | 0.5457 | 0.4132 | 0.105* | |
| H16B | 0.9592 | 0.4406 | 0.3811 | 0.105* | |
| H16C | 0.8052 | 0.4321 | 0.3425 | 0.105* | |
| O8 | 0.92795 (15) | 0.37653 (18) | 0.88918 (14) | 0.0695 (4) | |
| H15 | 0.862 (4) | 0.361 (4) | 0.905 (3) | 0.123 (12)* | |
| H16 | 0.912 (3) | 0.436 (3) | 0.860 (2) | 0.104 (12)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O5 | 0.0319 (5) | 0.0313 (5) | 0.0448 (5) | 0.0041 (4) | 0.0093 (4) | 0.0004 (4) |
| O1 | 0.0363 (5) | 0.0311 (5) | 0.0567 (6) | 0.0041 (4) | 0.0076 (5) | 0.0031 (4) |
| C11 | 0.0310 (6) | 0.0309 (6) | 0.0311 (6) | 0.0016 (5) | 0.0098 (5) | 0.0008 (5) |
| O6 | 0.0318 (5) | 0.0347 (5) | 0.0694 (7) | 0.0041 (4) | 0.0048 (5) | 0.0046 (5) |
| O2 | 0.0460 (6) | 0.0323 (5) | 0.0512 (6) | −0.0056 (4) | 0.0160 (5) | −0.0051 (4) |
| C2 | 0.0382 (7) | 0.0318 (6) | 0.0374 (7) | −0.0016 (5) | 0.0128 (6) | −0.0024 (5) |
| C12 | 0.0329 (6) | 0.0314 (6) | 0.0328 (6) | 0.0024 (5) | 0.0116 (5) | −0.0001 (5) |
| C10 | 0.0322 (6) | 0.0318 (6) | 0.0359 (6) | 0.0017 (5) | 0.0089 (5) | 0.0031 (5) |
| C13 | 0.0348 (7) | 0.0328 (7) | 0.0336 (6) | 0.0019 (5) | 0.0132 (5) | −0.0006 (5) |
| C4 | 0.0297 (6) | 0.0379 (7) | 0.0396 (7) | 0.0051 (5) | 0.0086 (5) | 0.0001 (5) |
| C8 | 0.0352 (7) | 0.0341 (7) | 0.0393 (7) | −0.0005 (5) | 0.0107 (6) | 0.0001 (5) |
| O3 | 0.0317 (5) | 0.0457 (7) | 0.0854 (9) | −0.0031 (5) | 0.0018 (5) | −0.0102 (6) |
| C3 | 0.0318 (6) | 0.0401 (7) | 0.0401 (7) | −0.0028 (5) | 0.0091 (5) | −0.0042 (6) |
| C9 | 0.0345 (7) | 0.0314 (6) | 0.0328 (6) | 0.0006 (5) | 0.0119 (5) | 0.0011 (5) |
| C1 | 0.0342 (6) | 0.0310 (6) | 0.0333 (6) | 0.0024 (5) | 0.0113 (5) | 0.0014 (5) |
| O7 | 0.0443 (6) | 0.0396 (6) | 0.0659 (7) | −0.0102 (5) | 0.0139 (5) | −0.0092 (5) |
| C7 | 0.0422 (7) | 0.0356 (7) | 0.0398 (7) | −0.0061 (6) | 0.0175 (6) | −0.0047 (6) |
| O4 | 0.0329 (5) | 0.0435 (6) | 0.0661 (7) | 0.0095 (4) | 0.0095 (5) | 0.0028 (5) |
| C5 | 0.0433 (7) | 0.0315 (7) | 0.0449 (7) | 0.0061 (6) | 0.0166 (6) | 0.0003 (6) |
| C6 | 0.0519 (8) | 0.0294 (6) | 0.0483 (8) | −0.0010 (6) | 0.0222 (7) | −0.0033 (6) |
| C15 | 0.0329 (8) | 0.0638 (12) | 0.0911 (14) | −0.0019 (8) | 0.0063 (8) | −0.0113 (10) |
| C14 | 0.0677 (11) | 0.0416 (9) | 0.0790 (12) | −0.0009 (8) | 0.0387 (10) | 0.0066 (8) |
| C16 | 0.0575 (11) | 0.0644 (12) | 0.0714 (12) | 0.0185 (9) | 0.0038 (9) | 0.0198 (10) |
| O8 | 0.0527 (8) | 0.0662 (10) | 0.0874 (11) | 0.0122 (7) | 0.0232 (7) | 0.0162 (8) |
Geometric parameters (Å, °)
| O5—C12 | 1.3623 (16) | O3—C3 | 1.3567 (17) |
| O5—C13 | 1.3753 (17) | O3—C15 | 1.402 (2) |
| O1—C1 | 1.3523 (16) | O7—C7 | 1.3640 (17) |
| O1—H1 | 0.8561 | O7—H7 | 0.8336 |
| C11—C12 | 1.4040 (18) | C7—C6 | 1.401 (2) |
| C11—C1 | 1.4204 (19) | O4—C16 | 1.437 (2) |
| C11—C10 | 1.4402 (18) | C5—C6 | 1.375 (2) |
| O6—C10 | 1.2601 (16) | C5—H5 | 0.9300 |
| O2—C2 | 1.3820 (17) | C6—H6 | 0.9300 |
| O2—C14 | 1.426 (2) | C15—H15A | 0.9600 |
| C2—C1 | 1.3765 (19) | C15—H15B | 0.9600 |
| C2—C3 | 1.409 (2) | C15—H15C | 0.9600 |
| C12—C4 | 1.3979 (19) | C14—H14A | 0.9600 |
| C10—C9 | 1.4568 (19) | C14—H14B | 0.9600 |
| C13—C5 | 1.3919 (19) | C14—H14C | 0.9600 |
| C13—C9 | 1.3920 (19) | C16—H16A | 0.9600 |
| C4—O4 | 1.3779 (16) | C16—H16B | 0.9600 |
| C4—C3 | 1.400 (2) | C16—H16C | 0.9600 |
| C8—C7 | 1.3781 (19) | O8—H15 | 0.85 (4) |
| C8—C9 | 1.4057 (19) | O8—H16 | 0.73 (4) |
| C8—H8 | 0.9300 | ||
| C12—O5—C13 | 119.33 (10) | O1—C1—C11 | 120.54 (12) |
| C1—O1—H1 | 109.5 | C2—C1—C11 | 120.10 (12) |
| C12—C11—C1 | 118.04 (12) | C7—O7—H7 | 109.5 |
| C12—C11—C10 | 120.44 (12) | O7—C7—C8 | 123.07 (14) |
| C1—C11—C10 | 121.51 (12) | O7—C7—C6 | 117.39 (13) |
| C2—O2—C14 | 111.49 (12) | C8—C7—C6 | 119.54 (13) |
| C1—C2—O2 | 120.20 (12) | C4—O4—C16 | 114.94 (13) |
| C1—C2—C3 | 120.75 (13) | C6—C5—C13 | 119.48 (14) |
| O2—C2—C3 | 119.05 (12) | C6—C5—H5 | 120.3 |
| O5—C12—C4 | 115.67 (12) | C13—C5—H5 | 120.3 |
| O5—C12—C11 | 121.75 (12) | C5—C6—C7 | 120.84 (13) |
| C4—C12—C11 | 122.58 (13) | C5—C6—H6 | 119.6 |
| O6—C10—C11 | 121.83 (12) | C7—C6—H6 | 119.6 |
| O6—C10—C9 | 121.87 (12) | O3—C15—H15A | 109.5 |
| C11—C10—C9 | 116.31 (12) | O3—C15—H15B | 109.5 |
| O5—C13—C5 | 116.43 (12) | H15A—C15—H15B | 109.5 |
| O5—C13—C9 | 122.92 (12) | O3—C15—H15C | 109.5 |
| C5—C13—C9 | 120.65 (13) | H15A—C15—H15C | 109.5 |
| O4—C4—C12 | 119.65 (13) | H15B—C15—H15C | 109.5 |
| O4—C4—C3 | 122.26 (12) | O2—C14—H14A | 109.5 |
| C12—C4—C3 | 117.91 (12) | O2—C14—H14B | 109.5 |
| C7—C8—C9 | 120.33 (13) | H14A—C14—H14B | 109.5 |
| C7—C8—H8 | 119.8 | O2—C14—H14C | 109.5 |
| C9—C8—H8 | 119.8 | H14A—C14—H14C | 109.5 |
| C3—O3—C15 | 124.13 (14) | H14B—C14—H14C | 109.5 |
| O3—C3—C4 | 126.78 (13) | O4—C16—H16A | 109.5 |
| O3—C3—C2 | 112.65 (13) | O4—C16—H16B | 109.5 |
| C4—C3—C2 | 120.57 (12) | H16A—C16—H16B | 109.5 |
| C13—C9—C8 | 119.08 (12) | O4—C16—H16C | 109.5 |
| C13—C9—C10 | 119.11 (12) | H16A—C16—H16C | 109.5 |
| C8—C9—C10 | 121.79 (12) | H16B—C16—H16C | 109.5 |
| O1—C1—C2 | 119.35 (12) | H15—O8—H16 | 106 (3) |
| C14—O2—C2—C1 | −93.37 (16) | O5—C13—C9—C8 | 177.67 (12) |
| C14—O2—C2—C3 | 87.18 (17) | C5—C13—C9—C8 | −3.1 (2) |
| C13—O5—C12—C4 | −179.21 (11) | O5—C13—C9—C10 | −4.2 (2) |
| C13—O5—C12—C11 | 1.25 (19) | C5—C13—C9—C10 | 175.03 (12) |
| C1—C11—C12—O5 | −179.12 (11) | C7—C8—C9—C13 | 1.5 (2) |
| C10—C11—C12—O5 | −0.6 (2) | C7—C8—C9—C10 | −176.60 (13) |
| C1—C11—C12—C4 | 1.4 (2) | O6—C10—C9—C13 | −175.33 (13) |
| C10—C11—C12—C4 | 179.86 (12) | C11—C10—C9—C13 | 4.52 (19) |
| C12—C11—C10—O6 | 177.61 (13) | O6—C10—C9—C8 | 2.7 (2) |
| C1—C11—C10—O6 | −4.0 (2) | C11—C10—C9—C8 | −177.40 (12) |
| C12—C11—C10—C9 | −2.24 (18) | O2—C2—C1—O1 | −0.8 (2) |
| C1—C11—C10—C9 | 176.19 (12) | C3—C2—C1—O1 | 178.70 (12) |
| C12—O5—C13—C5 | −178.02 (12) | O2—C2—C1—C11 | 178.48 (12) |
| C12—O5—C13—C9 | 1.24 (19) | C3—C2—C1—C11 | −2.1 (2) |
| O5—C12—C4—O4 | 3.85 (19) | C12—C11—C1—O1 | 179.59 (11) |
| C11—C12—C4—O4 | −176.61 (12) | C10—C11—C1—O1 | 1.1 (2) |
| O5—C12—C4—C3 | 179.11 (12) | C12—C11—C1—C2 | 0.37 (19) |
| C11—C12—C4—C3 | −1.4 (2) | C10—C11—C1—C2 | −178.10 (12) |
| C15—O3—C3—C4 | 10.8 (3) | C9—C8—C7—O7 | −179.92 (13) |
| C15—O3—C3—C2 | −169.68 (17) | C9—C8—C7—C6 | 0.9 (2) |
| O4—C4—C3—O3 | −5.7 (2) | C12—C4—O4—C16 | −75.54 (18) |
| C12—C4—C3—O3 | 179.12 (14) | C3—C4—O4—C16 | 109.41 (18) |
| O4—C4—C3—C2 | 174.75 (13) | O5—C13—C5—C6 | −178.43 (12) |
| C12—C4—C3—C2 | −0.4 (2) | C9—C13—C5—C6 | 2.3 (2) |
| C1—C2—C3—O3 | −177.47 (13) | C13—C5—C6—C7 | 0.2 (2) |
| O2—C2—C3—O3 | 1.98 (19) | O7—C7—C6—C5 | 179.03 (13) |
| C1—C2—C3—C4 | 2.1 (2) | C8—C7—C6—C5 | −1.8 (2) |
| O2—C2—C3—C4 | −178.45 (12) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O8i—H16i···O2 | 0.73 (3) | 2.58 (3) | 3.091 (2) | 129.4 |
| O8i—H16i···O3 | 0.73 (3) | 2.46 (3) | 3.177 (2) | 167.3 |
| O8ii—H15ii···O6 | 0.84 (5) | 2.08 (5) | 2.923 (2) | 172.3 |
| O7—H7···O8iii | 0.83 | 1.88 | 2.706 (2) | 169 |
Symmetry codes: (i) x, −y+3/2, z−1/2; (ii) −x+1, −y+1, −z+1; (iii) x−1, y, z−1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG2424).
References
- Basnet, P., Kadota, S., Shimizu, M. & Namba, T. (1994). Planta Med.60, 507–511. [DOI] [PubMed]
- Bruker (1999). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Evans, I. R., Howard, J. A. K., Šavikin-Fodulović, K. & Menković, N. (2004). Acta Cryst. E60, o1557–o1559.
- Fernandes, E. R., Carvalho, F. D., Remiao, F. G., Bastos, M. L., Pinto, M. M. & Gottlieb, O. R. (1995). Pharm. Res.12, 1756–1760. [DOI] [PubMed]
- Gales, L., de Sousa, M. E., Pinto, M. M. M., Kijjoa, A. & Damas, A. M. (2001). Acta Cryst. C57, 1319–1323. [DOI] [PubMed]
- Jiang, K. J., Zhang, Y. H., Shao, Z. Y., Chen, J. J. & Kuai, B. K. (2004). Chin. J. Struct. Chem.23, 262–266.
- Kabaleeswaran, V., Malathi, R. & Rajan, S. S. (2003). J. Chem. Crystallogr.33, 233–237.
- Karan, M., Vasisht, K. & Handa, S. S. (1999). Phytother. Res.13, 95–101. [DOI] [PubMed]
- Kato, L., De Oliveira, C. M. A., Vencato, I. & Lauriucci, C. (2005). J. Chem. Crystallogr.35, 23–26.
- Kijjoa, A., Jose, M., Gonzalez, T. G., Pinto, M. M. M., Damas, A. M., Mondranondra, I. O., Silva, A. M. S. & Herz, W. (1998). Phytochemistry, 49, 2159–2162.
- Liou, S. S., Shieh, W. L., Cheng, T. H., Won, S. J. & Lin, C. N. (1993). J. Pharm. Pharmacol.45, 791–794. [DOI] [PubMed]
- Miura, T., Ichiki, H., Hashimoto, I., Iwamoto, N., Kato, M., Kubo, M., Ishihara, E., Komatsu, Y., Okada, M., Ishida, T. & Tanigawa, K. (2001). Phytomedicine, 8, 85–87. [DOI] [PubMed]
- Parmar, V. S., Bisht, K. S., Jain, R., Singh, S., Sharma, S. K., Gupta, S., Malhotra, S., Tyagi, O. D. & Vardhan, A. (1996). Indian J. Chem.35B, 220–232.
- Pedro, M., Cerqueira, F., Sousa, M. E., Nascimento, M. S. & Pinto, M. (2002). Bioorg. Med. Chem.10, 3725–3730. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, G.-F., Lu, R.-H., Yang, Y.-S., Li, C.-L., Yang, A.-M. & Cai, L.-X. (2004). Acta Cryst. E60, o878–o880.
- Shi, G. F., Lu, R. H., Yang, Y. S., Li, C. L., Yang, A. M. & Cai, L. X. (2005). J. Chem. Crystallogr.35, 135–139.
- Sousa, E. P., Silva, A. M. S., Pinto, M. M. M., Pedro, M. M., Cerqueira, F. A. M. & Nascimento, M. S. J. (2002). Helv. Chim. Acta, 85, 2862–2876.
- Stout, G. H., Lin, T. S. & Singh, I. (1969). Tetrahedron, 25, 1975–1983.
- Vijayalakshmi, J., Rajan, S. S. & Srinivasan, R. (1987). Acta Cryst. C43, 2108–2110.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808004832/ng2424sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808004832/ng2424Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




