Abstract
Hepatitis C virus (HCV) is a positive-strand RNA virus that replicates exclusively in the cytoplasm of infected cells. The viral envelope glycoproteins, E1 and E2, appear to be retained in the endoplasmic reticulum, where viral budding is thought to occur. Surprisingly, we found that the expression system used to generate HCV envelope glycoproteins influences their subcellular localization and processing. These findings have important implications for optimizing novel HCV fusion and entry assays as well as for budding and virus particle formation.
It is estimated that 170 million people worldwide are infected with the hepatitis C virus (HCV) and are at risk of developing chronic hepatitis or cirrhosis, the latter often leading to hepatocellular carcinoma (1, 11, 29, 30). Basic understanding of HCV replication and pathogenesis remains poor, primarily due to a lack of experimental models and key reagents. The HCV genome is a 9.4-kb single-stranded positive RNA that replicates exclusively in the cytoplasm of infected cells (47). The genomic RNA encodes a ∼3,000-amino-acid polyprotein that is processed to generate at least 10 proteins, termed C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (47). The C protein constitutes the nucleocapsid; E1 and E2 are transmembrane envelope glycoproteins; p7 is a membrane-spanning protein of unknown function; and the various nonstructural (NS) proteins have replication functions (2, 37).
Translocation of the HCV structural proteins, C-E1-E2-p7, into the endoplasmic reticulum (ER) is accompanied by cleavage of internal signal sequences by ER-resident signal peptidases. Proper folding of the envelope glycoproteins, E1 and E2, is dependent on their cotranslation and processing within the context of the HCV polyprotein (7, 13, 14, 16, 34, 36, 40). The transmembrane (TM) domains of E1 and E2 are also the signal sequences (SS) of E2 and p7, respectively, and topological reorientation of cleaved E1 and E2 C termini toward the cytosol converts these regions into single membrane-spanning domains (5, 9). The TM domains of E1 and E2 mediate their noncovalent heterodimerization (13, 15, 38, 39, 45, 51). The apparent absence of E1/E2 heterodimers on the cell surface as well as the lack of N-glycan modifications by Golgi enzymes suggest that the HCV envelope glycoproteins are retained in the ER (16, 32, 36, 40, 51). The TM domains of E1 and E2 mediate ER retention as well as E1/E2 heterodimerization (6, 8, 10, 19, 20), which has made it difficult to generate cell surface-expressed E1/E2 heterodimers for use in cell fusion and viral pseudotype assays.
The initial goal of our studies was to create recombinant cell surface-expressed HCV envelope glycoproteins that would be incorporated onto pseudovirions and mediate entry into HCV target cells. We chose a strategy wherein the ectodomains of HCV E1 and E2 were fused to the TM domains of E1 and E2 from a related alphavirus, the Semliki Forest virus (SFV). The SFV envelope glycoproteins form cell surface-associated heterodimers that efficiently pseudotype heterologous viral nucleocapsids in order to mediate their entry into host cells. Chimeric HCV-SFV envelope glycoproteins were similar to unmodified HCV envelope glycoproteins in size and posttranslational processing and were expressed on the cell surface. During these experiments, however, we detected unmodified, wild-type HCV E1 and E2 on the cell surface, and this unexpected observation was investigated further.
Constructs were generated for expression of full-length unmodified HCV E1 (E1), E2 (E2), and E1/E2 (E1-E2). DNA encoding HCV envelope glycoproteins was derived by PCR amplification of p90/HCV FL-long pU carrying the full-length genome of an infectious HCV isolate H77 (26). The first nucleotide of the C start codon is defined as position +1 in the HCV coding sequence. E1 therefore comprises nucleotides 511 to 1149, E2 comprises nucleotides 1111 to 2238, and E1-E2 includes nucleotides 511 to 2238 of HCV. Start and stop translation codons were introduced at the beginning and end of every construct, which were cloned into the pcDNA3.1+ expression vector (Invitrogen) and verified by DNA sequencing. Transient expression of HCV envelope glycoproteins was achieved by lipofection of the different expression plasmids into HeLa cells. Alternatively, HeLa cells were infected with a vaccinia virus vector expressing the T7 polymerase (vTF7.3 [17]) followed by lipofection with HCV E1/E2-expression plasmids (resulting in cytoplasmic transcription from the T7 promoter in pcDNA3.1+).
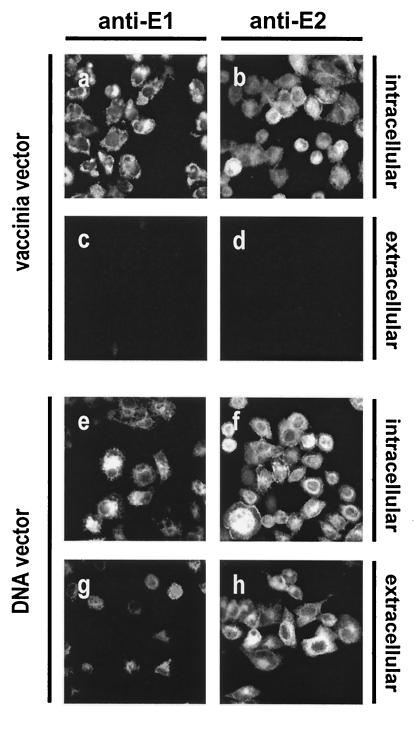
Intracellular but not cell surface-associated E1 and E2 were detected by immunofluorescence after vaccinia virus-driven expression of E1-E2 (Fig. 1a to d). In contrast, E1 and E2 were detected both intracellularly and on the cell surface following plasmid-based expression of the E1-E2 construct (Fig. 1 f to h). Intracellular staining of envelope glycoproteins was comparable in the two expression systems (Fig. 1a, b, e and f), indicating that similar levels of E1 and E2 were being generated but transport to the cell surface was not occurring in the vaccinia virus-based expression system. Differences in expression levels were not observed between envelope glycoproteins expressed as single proteins (E1 and E2) or as part of an E1-E2 polyprotein (E1-E2) (data not shown). Similar expression patterns were obtained after transient expression of E1 and E2 by vaccinia- and plasmid-based systems in a hepatoma cell line, HepG2 (data not shown).
FIG. 1.
Cell surface expression of E1 and E2. Unmodified HCV envelope glycoproteins were transiently expressed in HeLa cells. Briefly, cells were seeded overnight on glass coverslips and were infected with 5 PFU of recombinant vaccinia virus vTF7.3 per cell for 1 h at 37°C, followed by lipofection (Invitrogen) with the E1-E2 construct. Protein expression was analyzed 24 h postinfection (a to d). Alternatively, cells were only lipofected with the E1-E2 construct and protein expression was analyzed 24 h postlipofection (e to h). Cells were either fixed in 3% formaldehyde for 20 min at room temperature or fixed and permeabilized with methanol for 20 min at −20°C, followed by washing with 2% gelatin in phosphate-buffered saline. Cells were incubated with anti-E1 MAb A4 (1:100) (a, c, e, and g) or anti-E2 MAb H53 (1:100) (b, d, f, and h), followed by washing and incubation with a phycoerythrin (PE)-labeled goat anti-mouse immunoglobulin G secondary antibody (1:100) (Pierce). Coverslips were mounted on slides with Mowiol and were observed under a fluorescence microscope.
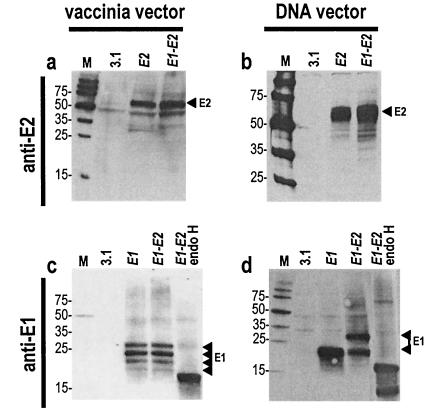
Vaccinia vector- and plasmid vector-based expression of unmodified HCV envelope glycoproteins invariably generated a major E2 protein species of 62-kDa apparent molecular size (Fig. 2a and b). Vaccinia expression of unmodified E1 and E1-E2 constructs generated four E1 protein species with apparent molecular sizes of 18, 21, 24, and 27 kDa (Fig. 2c). Treatment of cell lysates by endoglycosidase H (endo H) generated a single small-molecular-size band corresponding to the deglycosylated E1 protein core (Fig. 2c). The 27-kDa species therefore corresponds to the fully glycosylated E1 protein, whereas the lower-molecular-size species correspond to incomplete glycosylation products. In contrast, plasmid-based expression of E1-E2 generated E1 proteins of 27 and 20 kDa apparent molecular sizes (Fig. 2d). The 20-kDa E1 species was not the result of hypoglycosylation, because two E1 protein species were still present after endo H treatment of cell lysates (Fig. 2d) (see below).
FIG. 2.
Characterization of E1 and E2 proteins. Unmodified HCV envelope glycoproteins (E1, E2, and E1-E2) were expressed in HeLa cells with a vaccinia virus-based (a and c) or a plasmid-based system (b and d). For immunoblotting analyses, cells were lysed in a solution of 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM EDTA buffer containing a protease inhibitor cocktail (Roche). A fraction of the cell lysates was treated with 0.25 U of endoglycosidase H (Boehringer)/ml overnight at 37°C. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 10% or 12% acrylamide (Bio-Rad) followed by transfer to Trans-Blot nitrocellulose membranes (Bio-Rad). Membranes were probed either with anti-E2 MAb A11 (1:1,000) (a and b) or anti-E1 MAb A4 (1:1000) (c and d) followed by horseradish peroxidase-labeled goat anti-mouse immunoglobulin G (1:10,000) (Amersham) and incubation with a chemifluorescent substrate (Vistra ECF; Amersham). Arrowheads indicate the positions of E1 and E2 proteins on the blots. M, molecular size marker.
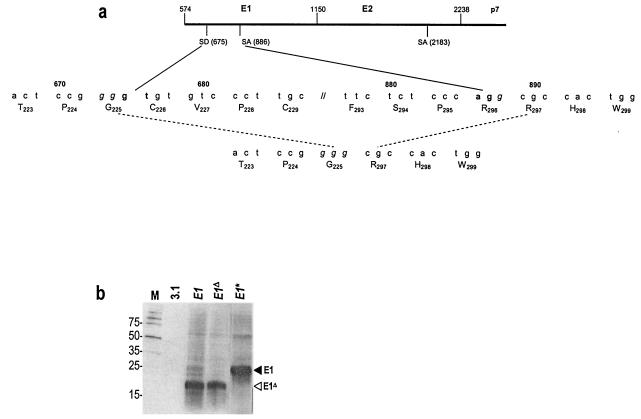
Vaccinia virus-based expression of heterologous genes results in transcription in the cytoplasm. In contrast, transient and stable expression by DNA vectors results in nuclear transcription followed by mRNA maturation and transport to the cytoplasm. We hypothesized that mRNA modifications may account for the different E1 species expressed by plasmid vectors. We therefore performed reverse transcription-PCR on RNA extracts of HeLa cells stably expressing E1. Sequence analyses of the PCR products detected a deletion between nucleotides 675 and 887 (inclusive) which preserves the E1 reading frame and would encode a protein with a 71-amino-acid deletion (Fig. 3a and data not shown). This deletion was not present in PCR products derived by amplification of genomic DNA containing the integrated E1 construct (data not shown).
FIG. 3.
Excision of a putative intron in E1 mRNA generates a protein with a deletion. (a) The HCV genome was analyzed with a splice site prediction neural network (http://www.fruitfly.org/seq_tools/splice.html). Sequences in the E1-E2-p7 coding region having >80% probability of being functional splice donor (SD) and acceptor (SA) sites are indicated. Splicing occurs between nucleotide positions 675 and 887, generating an E1 protein with a deletion spanning amino acids 230 to 292. (b) Unmodified E1 (E1), E1 with a mutated splice acceptor site (E1*), or E1 comprising a deletion of the putative intron (E1ΔHCV) was transiently expressed in HeLa cells by lipofection and analyzed by immunoblotting with anti-E1 MAb A4. The black arrowhead indicates the position of full-length E1 and the white arrowhead indicates the position of the protein species with partially deleted E1. M, molecular size marker.
Analysis of the entire HCV sequence by a splice site prediction neural network (http://www.fruitfly.org/seq_tools/splice.html) revealed the presence of a putative splice donor site in position 675 of E1, whereas putative splice acceptor sites were found in position 887 of E1 and 2183 of E2 (Fig. 3a). Based on these findings, we assumed that putative intron splicing between positions 675 and 887 resulted in expression of a partially deleted E1 protein, corresponding to the 20-kDa species observed after transient expression of E1 and E1-E2 constructs. The splice acceptor at position 2183 does not appear to be functional.
We therefore generated new E1 expression constructs wherein the splice acceptor site in E1 was removed by conservative replacement of A886GG with C886GT (E1*), or the sequence encoding the putative intron between positions 675 and 887 was deleted (E1Δ). Transient, plasmid-based expression of E1 as well as E1Δ generated a single 20-kDa protein species (Fig. 3b). A single 27-kDa protein species was generated by E1*, wherein the splice acceptor site was mutated (Fig. 3b). The 20-kDa protein species generated by unmodified E1 is therefore the result of E1 mRNA splicing, whereas the 27-kDa protein species corresponds to full-length E1. Secondary structure may partially obstruct splice sites in E1-E2 mRNA, leading to expression of full-length E1 (27 kDa) as well as those with partially deleted E1 (20 kDa).
The putative splice acceptor site in position 887 was eliminated by conservative mutagenesis in all constructs to ensure that splicing would not occur. We also modified the splice acceptor site in position 2183 by a conservative A2183->T2183 substitution in case it became functional in the absence of the upstream splice acceptor. Constructs expressing modified E1, E2, or E1-E2 (indicated by asterisks) were stably transfected into HeLa cells. Stable clones were also generated with constructs expressing E1/E2 in conjunction with p7 (including nucleotides 511 to 2427), an HCV structural protein of unknown function. Reverse transcription-PCR analyses of RNA extracts showed that the length of transcripts matched the full length of the coding sequences, indicating that putative intron splicing was no longer occurring (data not shown). E2* and E1*-E2* expression generated a major 62-kDa protein corresponding to E2 (Fig. 4a). Immunoblotting demonstrated that E1 was now expressed as a single 27-kDa species by E1* and E1*-E2* constructs (Fig. 4b).
FIG. 4.
Stable expression of E1 and E2 lacking putative splice acceptor sites. In order to generate cell lines stably expressing the HCV envelope glycoproteins, HeLa cells were lipofected with different constructs and were placed in medium containing 1 mg of G418 (Sigma)/ml. G418-resistant cells were pooled and labeled with anti-E2 MAb H53. The 10% most strongly labeled cells were sorted by using the fluorescence-activated cell sorterVantage SE (Becton Dickinson) and were subcloned by limiting dilution in order to generate clonal populations. For E1-expressing stable cell lines, cells were subcloned directly after G418 selection and individual clones were tested for E1 expression by immunoblotting. Proteins from whole-cell lysates were analyzed by immunoblotting with anti-E2 MAb A11 (a) or anti-E1 MAb A4 (b). Arrowheads indicate the positions of full-length E1 and E2 proteins. M, molecular size markers.
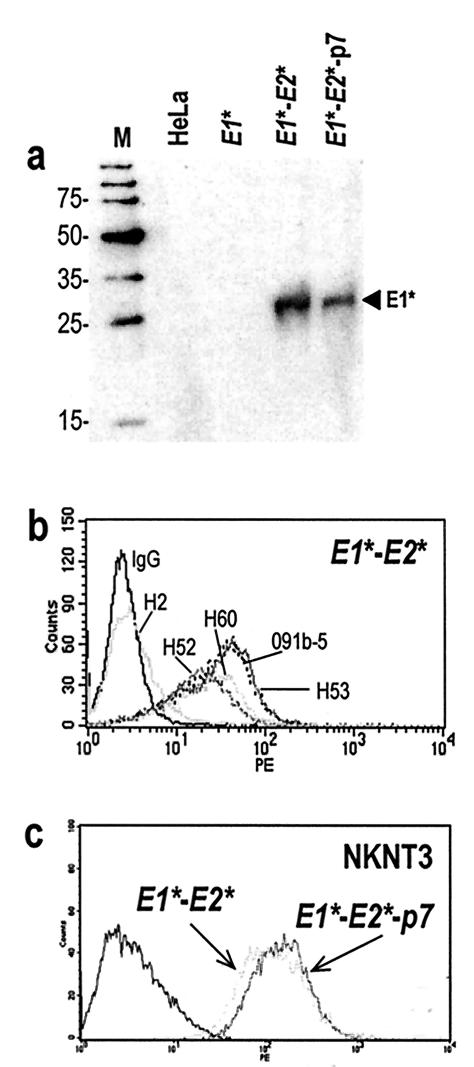
Cell surface-associated E1 protein could not be detected in any of the stable HeLa clones by flow cytometry using two different anti-E1 monoclonal antibodies (MAbs), A4 (15) and 081-5 (Austral Biologicals) (data not shown). We reason that their epitopes may not be accessible in the full-length protein. Cell surface-associated E1 was readily detected, however, by cell surface biotinylation followed by streptavidin capture and immunoblotting of E1*-E2* and E1*-E2*-p7-expressing cells with an anti-E1 MAb (Fig. 5a). When E1 was expressed alone it was not detectable on the cell surface, suggesting that coexpression of E2 is required for efficient transport of E1 to the plasma membrane (Fig. 5a).
FIG. 5.
Cell surface expression of E1 and E2 lacking putative splice acceptor sites. Cell surface proteins of HeLa cells stably expressing E1*, E1*-E2*, and E1*-E2*-p7 were tagged with EZ-Link Sulfo-NHS-LC-Biotin (Pierce) before lysis, as described previously (31). (a) Biotinylated proteins were recovered by incubation of lysates with streptavidin-coupled agarose beads for 1 h at 4°C (Molecular Probes) followed by three washes with the lysis buffer. Recovered proteins were immunoblotted with anti-E1 MAb A4. The arrowhead indicates the position of E1 proteins. M, molecular size marker. (b) Cell surface-associated E2 proteins generated by stable expression of E1*-E2* and E1*-E2*-p7 were detected by flow cytometry analyses after labeling of cells with five different anti-E2 MAbs, H2, H52, H53, H60, and 091b-5 or a control mouse immunoglobulin G (IgG). (c) Cell surface-associated E2 proteins generated by stable expression of E1*-E2* and E1*-E2*-p7 in NKNT3 cells were detected by anti-E2 MAb H53.
Cell surface-associated E2 was detected in stable HeLa clones by flow cytometry after labeling with four different anti-E2 MAbs of E2*-, E1*-E2*-, and E1*-E2*-p7-expressing cells (Fig. 5b and data not shown). MAb H2, which has been reported to recognize E1/E2 heterodimers, did not recognize cell surface E2, but this antibody is also not reactive with HCV particles in patient sera (13). E1*-E2* and E1*-E2*-p7 also were stably expressed in hepatic NKNT3 cells, which display morphological characteristics of liver parenchyma cells, express key genes of liver metabolism, and are not tumorigenic in SCID mice (24, 25). E2 was readily detected on the surface of NKNT3 cells, suggesting that plasma membrane localization is an inherent property of HCV envelope glycoproteins rather than of the cell line that they are expressed in (Fig. 5c). Coexpression of E1/E2 with p7 does not appear to influence the processing and cell surface localization of the envelope glycoproteins.
Finally, cell surface-associated E1 and E2 were analyzed for their ability to form noncovalent heterodimers. HeLa cells stably expressing different combination of E1, E2, and p7 were preincubated with an anti-E2 MAb, and protein-antibody complexes were recovered by immunoprecipitation of cell lysates with G protein-coupled agarose beads. In this manner, only cell surface-associated envelope glycoproteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with an anti-E1 MAb. E1 readily coimmunoprecipitated with E2 only in cells expressing E1*-E2*, and only if the cells were preincubated with an anti-E2 MAb (Fig. 6a). Similarly, E2 was detected in cells expressing E2*, E1*-E2*, or E1*-E2*-p7 only if the cells were preincubated with an anti-E2 MAb (Fig. 6b). Therefore, E1 and E2 proteins associated with the plasma membrane also form noncovalent heterodimers.
FIG. 6.
Heterodimerization of E1 and E2 on the cell surface. Intact HeLa cells stably expressing modified HCV envelope glycoproteins were incubated with the anti-E2 MAb H53 (1:100), lysed, and incubated with protein G-coupled agarose beads overnight at 4°C (Oncogene Research Products) followed by three washes with the lysis buffer. As a control, lysates from cells that had not been treated with H53 were used. The presence of E1 was detected by immunoblotting with anti-E1 MAb A4 (a), whereas the presence of E2 was detected by immunoblotting with anti-E2 MAb A11 (b). Arrowheads indicate the positions of full-length E1* and E2* proteins. M, molecular size markers; IP, immunoprecipitation; IgG, immunoglobulin G.
HCV envelope glycoproteins E1 and E2 have been described to form membrane-anchored, noncovalent heterodimers that are retained in the ER, where HCV budding is believed to occur (37). The signals for heterodimerization and ER retention colocalize to residues in the TM domains of E1 and E2, hence it seems the two functions cannot be dissociated (37). This has made it difficult to generate cell surface-associated variants of E1/E2 heterodimers, which would be invaluable for the development of cell fusion assays and virus pseudotypes. Attempts to create such variants have thus far focused on fusing E1 and E2 ectodomains to the TM domains of vesicular stomatitis virus (VSV) G or influenza hemagglutinin envelope glycoproteins, which have no known dimerization function (20, 27, 55). Additionally, in all of these studies, chimeric E1 and E2 proteins were translated from separate mRNAs, which may have further minimized their potential to form native heterodimers. Even though E2-HA chimeras underwent pH-dependent conformational changes and were incorporated into influenza virus particles, they did not induce fusion with target cells (20). HCV-VSV chimeric envelope glycoproteins also did not appear to reproducibly model HCV fusion and entry (4, 27, 28, 33, 35, 55).
The initial goal of our study was to create chimeric HCV envelope glycoproteins that would be expressed on the cell surface as E1/E2 heterodimers. We therefore chose a strategy wherein the ectodomains of HCV E1 and E2 would be fused to the TM domains of E1 and E2 from a related alphavirus, SFV. Chimeric HCV-SFV envelope glycoproteins were expressed on the cell surface and resembled unmodified HCV envelope glycoproteins in size and processing. The most surprising finding, and one that changed the focus of our studies, was the expression of unmodified HCV E1 and E2 on the cell surface. E2 was detected on the cell surface by flow cytometry with four different anti-E2 MAbs. Cell surface associated E2 expression was also detected in a hepatic cell line and was not influenced by the presence of p7. By biotin-tagging cell surface proteins we demonstrated that full-length E1 was also associated with the plasma membrane. Most importantly, we were able to specifically coimmunoprecipitate E1 protein with an anti-E2 MAb, thus demonstrating that cell surface-associated E1 and E2 form noncovalent heterodimers.
One of the complicating factors in identifying properly folded and functional E1 and E2 has been the host of expression systems used to study these proteins, and a careful survey of the literature reveals significant diversity in the number and size of protein species corresponding to E1 and E2. In this study, unmodified and chimeric envelope glycoproteins were generated by using two different expression systems. The use of vaccinia virus-based expression is justifiable on the premise that it circumvents the nucleus, just as HCV replication does. In this expression system, E1 and E2 remain intracellular. Vaccinia replication, however, modifies internal cellular membranes as well as the translation machinery (41, 44, 46, 48-50), and the apparent trapping of HCV envelope glycoproteins inside the cell may be an artifact. Indeed, vaccinia virus-based expression has been shown to cause ER retention of other viral envelope glycoproteins (50, 54). The observation that vaccinia virus-based expression generates hypoglycosylated E1 proteins led us to explore plasmid delivery of HCV envelope glycoproteins. Plasmid-based expression of proteins typically does not adversely affect cellular protein synthesis but does involve nuclear transcription, which is not a natural part of HCV replication. Indeed, we clearly show that plasmid-based expression of HCV envelope glycoproteins results in putative intron excision in E1 mRNA that translates into a protein with a deletion. Other modifications such as mRNA editing were not observed (unpublished results). Our finding highlights an inherent complication in expressing RNA virus proteins by DNA-based expression systems.
Recently it was reported that HCV envelope glycoproteins are able to pseudotype retroviral particles and mediate their entry into target cells (3, 22). Both studies used plasmid vectors to express unmodified E1/E2, meaning that pseudoviral envelopes contained both full-length proteins and those with E1 deleted. We have confirmed that unmodified HCV envelope glycoproteins are able to mediate entry of retroviral pseudotypes into several hepatic and nonhepatic cell lines as well as primary hepatocytes (unpublished results). We are presently determining how the presence of partially deleted E1 species in pseudoviral envelopes affects entry into different target cells. These studies will allow us to further optimize pesudovirion entry mediated by HCV envelope glycoproteins, which will facilitate structure/function studies of HCV envelope glycoproteins as well as the identification of HCV receptors and target cells.
It remains to be determined whether cell surface-associated E1/E2 heterodimers have any physiological relevance in the viral replication cycle. Our observation that HCV envelope glycoproteins are expressed on the surface of cells that closely resemble primary hepatocytes implies that there is no specific retention mechanism for HCV envelope glycoproteins in liver cells. The postulated HCV replication cycle is based on analogies to the closely related flavi- and pestiviruses and it is generally assumed that flaviviruses bud into the ER and mature by passage into cytoplasmic vesicles (42). Thus far the cellular localization of HCV envelope glycoproteins and particles has mostly been studied in cells transfected or infected in vitro. Virus-like particles mostly occurred in cytoplasmic vesicles, suggesting vesicle-based morphogenesis of HCV (12, 18, 21, 23, 43, 52, 53). No study, however, has clearly documented the HCV budding and maturation process, perhaps because it does not occur in these experimental systems or perhaps because it is an extremely rare event that is difficult to detect by standard methods. We are presently addressing these questions by expressing E1/E2 envelope glycoproteins in human primary hepatocytes.
Acknowledgments
We are especially thankful to Jean Dubuisson (Institut Pasteur, Lilles, France) for repeatedly and generously providing MAbs A4, A11, H2, H52, H53, and H60. We also thank Charles Rice (Rockefeller University, New York, N.Y.) for providing the p90/HCV FL-long clone and Margaret Kielian (Albert Einstein College of Medicine, Bronx, N.Y.) for the pSP6SFV-4 clone.
This work was supported by The Speaker's Fund for Biomedical Research Award to Young Investigators to T.D., by grant AI051134 to J.P.G., and by Progenics Pharmaceuticals, Inc. J.D. was financed in part by the Schuller-Bettencourt Foundation (France).
REFERENCES
- 1.Alter, H. J., and L. B. Seef. 1993. Transfusion-associated hepatitis. In Z. A. Thomas (ed.), Viral hepatitis. Churchill Livingstone, Edinburgh, United Kingdom.
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonocore, L., K. J. Blight, C. M. Rice, and J. K. Rose. 2002. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 76:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charloteaux, B., L. Lins, H. Moereels, and R. Brasseur. 2002. Analysis of the C-terminal membrane anchor domains of hepatitis C virus glycoproteins E1 and E2: toward a topological model. J. Virol. 76:1944-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocquerel, L., J. C. Meunier, A. Op de Beeck, D. Bonte, C. Wychowski, and J. Dubuisson. 2001. Coexpression of hepatitis C virus envelope proteins E1 and E2 in cis improves the stability of membrane insertion of E2. J. Gen. Virol. 82:1629-1635. [DOI] [PubMed] [Google Scholar]
- 8.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., A. Op de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 12.Dash, S., A. B. Halim, H. Tsuji, N. Hiramatsu, and M. A. Gerber. 1997. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am. J. Pathol. 151:363-373. [PMC free article] [PubMed] [Google Scholar]
- 13.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 15.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 17.Earl, P. L., and B. Moss. 1991. Generation of recombinant vaccinia viruses, p. 16.17.1-16.17.16. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York, N.Y.
- 18.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 20.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greive, S. J., R. I. Webb, J. M. Mackenzie, and E. J. Gowans. 2002. Expression of the hepatitis C virus structural proteins in mammalian cells induces morphology similar to that in natural infection. J. Viral Hepatol. 9:9-17. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iacovacci, S., A. Manzin, S. Barca, M. Sargiacomo, A. Serafino, M. B. Valli, G. Macioce, H. J. Hassan, A. Ponzetto, M. Clementi, C. Peschle, and G. Carloni. 1997. Molecular characterization and dynamics of hepatitis C virus replication in human fetal hepatocytes infected in vitro. Hepatology 26:1328-1337. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, N., T. Fujiwara, K. A. Westerman, Y. Inoue, M. Sakaguchi, H. Noguchi, M. Miyazaki, J. Cai, N. Tanaka, I. J. Fox, and P. Leboulch. 2000. Prevention of acute liver failure in rats with reversibly immortalized human hepatocytes. Science 287:1258-1262. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, N., H. Noguchi, K. A. Westerman, T. Watanabe, T. Matsumura, T. Totsugawa, T. Fujiwara, P. Leboulch, and N. Tanaka. 2001. Cre/loxP-based reversible immortalization of human hepatocytes. Cell Transplant. 10:383-386. [PubMed] [Google Scholar]
- 26.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 27.Lagging, L. M., K. Meyer, R. J. Owens, and R. Ray. 1998. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J. Virol. 72:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagging, L. M., K. Meyer, J. Westin, R. Wejstal, G. Norkrans, M. Lindh, and R. Ray. 2002. Neutralization of pseudotyped vesicular stomatitis virus expressing hepatitis C virus envelope glycoprotein 1 or 2 by serum from patients. J. Infect. Dis. 185:1165-1169. [DOI] [PubMed] [Google Scholar]
- 29.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 30.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, Y. E., and M. Kielian. 2000. Semliki forest virus budding: assay, mechanisms, and cholesterol requirement. J. Virol. 74:7708-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martire, G., A. Viola, L. Iodice, L. V. Lotti, R. Gradini, and S. Bonatti. 2001. Hepatitis C virus structural proteins reside in the endoplasmic reticulum as well as in the intermediate compartment/cis-Golgi complex region of stably transfected cells. Virology 280:176-182. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 34.Meunier, J. C., A. Fournillier, A. Choukhi, A. Cahour, L. Cocquerel, J. Dubuisson, and C. Wychowski. 1999. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J. Gen. Virol. 80:887-896. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, K., A. Basu, and R. Ray. 2000. Functional features of hepatitis C virus glycoproteins for pseudotype virus entry into mammalian cells. Virology 276:214-226. [DOI] [PubMed] [Google Scholar]
- 36.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 37.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 38.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 39.Patel, J., A. H. Patel, and J. McLauchlan. 1999. Covalent interactions are not required to permit or stabilize the non-covalent association of hepatitis C virus glycoproteins E1 and E2. J. Gen. Virol. 80:1681-1690. [DOI] [PubMed] [Google Scholar]
- 40.Patel, J., A. H. Patel, and J. McLauchlan. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58-68. [DOI] [PubMed] [Google Scholar]
- 41.Person-Fernandez, A., and G. Beaud. 1986. Purification and characterization of a protein synthesis inhibitor associated with vaccinia virus. J. Biol. Chem. 261:8283-8289. [PubMed] [Google Scholar]
- 42.Pettersson, R. F. 1991. Protein localization and virus assembly at intracellular membranes. Curr. Top. Microbiol. Immunol. 170:67-106. [DOI] [PubMed] [Google Scholar]
- 43.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. Gonzalez, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ralston, R., K. Thudium, K. Berger, C. Kuo, B. Gervase, J. Hall, M. Selby, G. Kuo, M. Houghton, and Q. L. Choo. 1993. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J. Virol. 67:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice, A. P., and B. E. Roberts. 1983. Vaccinia virus induces cellular mRNA degradation. J. Virol. 47:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice, C. M. 1996. Flaviviridiae: The viruses and their replication, p. 931-1034. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 48.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanger, C., E. Muhlberger, H. D. Klenk, and S. Becker. 2001. Adverse effects of MVA-T7 on the transport of Marburg virus glycoprotein. J. Virol. Methods 91:29-35. [DOI] [PubMed] [Google Scholar]
- 51.Selby, M. J., E. Glazer, F. Masiarz, and M. Houghton. 1994. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology 204:114-122. [DOI] [PubMed] [Google Scholar]
- 52.Serafino, A., M. B. Valli, A. Alessandrini, A. Ponzetto, G. Carloni, and L. Bertolini. 1997. Ultrastructural observations of viral particles within hepatitis C virus-infected human B lymphoblastoid cell line. Res. Virol. 148:153-159. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu, Y. K., S. M. Feinstone, M. Kohara, R. H. Purcell, and H. Yoshikura. 1996. Hepatitis C virus: detection of intracellular virus particles by electron microscopy. Hepatology 23:205-209. [DOI] [PubMed] [Google Scholar]
- 54.Szepanski, S., M. Veit, S. Pleschka, H. D. Klenk, M. F. Schmidt, and G. Herrler. 1994. Post-translational folding of the influenza C virus glycoprotein HEF: defective processing in cells expressing the cloned gene. J. Gen. Virol. 75:1023-1030. [DOI] [PubMed] [Google Scholar]
- 55.Takikawa, S., K. Ishii, H. Aizaki, T. Suzuki, H. Asakura, Y. Matsuura, and T. Miyamura. 2000. Cell fusion activity of hepatitis C virus envelope proteins. J. Virol. 74:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]