Abstract
Previously we found that the amino-terminal region of the NS1 protein of influenza A virus plays a key role in preventing the induction of beta interferon (IFN-β) in virus-infected cells. This region is characterized by its ability to bind to different RNA species, including double-stranded RNA (dsRNA), a known potent inducer of IFNs. In order to investigate whether the NS1 RNA-binding activity is required for its IFN antagonist properties, we have generated a recombinant influenza A virus which expresses a mutant NS1 protein defective in dsRNA binding. For this purpose, we substituted alanines for two basic amino acids within NS1 (R38 and K41) that were previously found to be required for RNA binding. Cells infected with the resulting recombinant virus showed increased IFN-β production, demonstrating that these two amino acids play a critical role in the inhibition of IFN production by the NS1 protein during viral infection. In addition, this virus grew to lower titers than wild-type virus in MDCK cells, and it was attenuated in mice. Interestingly, passaging in MDCK cells resulted in the selection of a mutant virus containing a third mutation at amino acid residue 42 of the NS1 protein (S42G). This mutation did not result in a gain in dsRNA-binding activity by the NS1 protein, as measured by an in vitro assay. Nevertheless, the NS1 R38AK41AS42G mutant virus was able to replicate in MDCK cells to titers close to those of wild-type virus. This mutant virus had intermediate virulence in mice, between those of the wild-type and parental NS1 R38AK41A viruses. These results suggest not only that the IFN antagonist properties of the NS1 protein depend on its ability to bind dsRNA but also that they can be modulated by amino acid residues not involved in RNA binding.
The NS1 protein of influenza A virus is a nonstructural protein expressed at high levels in virus-infected cells that has been implicated in inhibition of the host antiviral defense mediated by alpha/beta interferon (IFN-α/β) (15), in regulation of viral translation(1, 9, 12), and in inhibition of host mRNA processing mechanisms (14, 25, 34). This viral protein is also an RNA-binding protein which has been shown to bind to several different species of RNA (17, 19, 20), including double-stranded RNA (dsRNA), through its N-terminal region (23). The presence of dsRNA in host cells is a clear signal that virus infection and replication are occurring and leads to the triggering of a plethora of antiviral host defense mechanisms (8, 36). dsRNA induces the synthesis of IFN-β and certain IFN-α molecules through the activation of several transcription factors, including IRF-3, IRF-7, NF-κB, and c-Jun/ATF2. The secreted IFN-α/β induces an antiviral state in the infected and uninfected neighboring host cells by stimulating the transcription of IFN-stimulated response element (ISRE) promoter-containing genes via the JAK/STAT pathway (36).
Most viruses have developed different mechanisms to evade the host antiviral response (21). The NS1 protein of influenza A virus has been shown to act as an IFN-α/β antagonist, exerting its function at least at two distinct stages within the virus-infected cells (15). First, the NS1 protein acts at the level of inhibition of IFN-α/β synthesis by virtue of its ability to inhibit virus-induced IRF-3, NF-κB, and c-Jun/ATF2 activation (27, 37, 40). Second, the NS1 protein has been shown to inhibit the activation of at least two IFN-induced, dsRNA-activated antiviral pathways, namely the inhibition of protein kinase R (PKR) and oligoadenylate synthetase pathways (3, 18; N. Donelan and A. García-Sastre, Abstr. 19th Annu. Meet. Am. Soc. Virol., abstr. W25-4, 2000). However, the mechanism by which the NS1 protein of influenza A virus exerts its IFN antagonistic properties is not yet fully understood. Several lines of evidence suggest that the ability of NS1 to bind to and sequester dsRNA is important for its role as an IFN antagonist (37, 40). However, interactions of NS1 with host proteins may also play an important role in its IFN antagonist function (16, 30).
The amino acids within the NS1 protein that are required for binding to RNA have been well defined (39). The basic amino acids R38 and K41 within the N-terminal domain are thought to directly interact with RNA, mediating binding. A mutant RNA-binding-defective NS1 protein was generated by replacing the R38 and K41 amino acid residues with alanines. Plasmid-mediated expression of NS1 R38AK41A protein in mammalian cells revealed that this mutant protein was severely impaired in the ability to prevent the activation of IRF-3 and NF-κB compared to wild-type NS1 (37, 40). In order to investigate the effect of these two mutations in the context of an infectious virus, we have generated by reverse genetics a recombinant influenza A/WSN/33 (WSN) virus expressing the mutant NS1 R38AK41A protein. Passaging of the WSN NS1 R38AK41A virus in tissue culture resulted in the acquisition of a compensatory mutation (S42G) within the NS1 protein. The phenotypic characterization of these mutant viruses revealed that amino acid residues at positions 38, 41, and 42 within the NS1 protein play a critical role in the prevention of IFN-β synthesis during influenza A virus infection and in virus pathogenicity.
MATERIALS AND METHODS
Cells and viruses.
293T, MDCK, Vero, and A549 cells were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum. Sendai virus, strain Cantell, was propagated at 37°C in 10-day-old embryonated chicken eggs. The construction and growth of Newcastle disease virus-green fluorescent protein (NDV-GFP) was previously described (32). Recombinant influenza A/WSN/33 (WSN) viruses expressing wild-type and mutant NS1 proteins were generated by transfecting 12 plasmids as previously described (6). The viral NS segments from the rescued viruses were analyzed by reverse transcriptase (RT)-PCR and sequencing.
Plasmids and GST fusion protein expression.
Glutathione S-transferase (GST) expression plasmids for the NS1 proteins were constructed as follows. The plasmid pGEX-NS1 encodes a protein consisting of GST fused to the NS1 protein of WSN virus. The wild-type NS1 cDNA was subcloned into pGEX-5X-3 between EcoRI and XhoI restriction sites. pGEX-NS1 R38AK41A was constructed by subcloning a mutated NS1 cDNA into pGEX-5X-3 between the EcoRI and XhoI unique sites. pGEX-NS1 R38AK41AS42G was constructed by subcloning the mutant NS1 cDNA into pGEX-5X-1 between BamHI (filled in with Klenow) and EcoRI sites. These fusion proteins were expressed in Escherichia coli BL21 and purified by use of glutathione-Sepharose beads (Pharmacia) as recommended by the manufacturer. Mammalian expression plasmids for NS1 were constructed by subcloning the NS1 cDNAs into the pCAGGS vector (29) between the EcoRI and XhoI sites.
EMSA.
Electrophoretic mobility shift assays (EMSAs) for NS1 binding to dsRNA were done as previously described (39), with slight modifications. Briefly, 32P-radiolabeled short dsRNA was generated by annealing complementary SP6 (76 nucleotides [nt]) and T7 (80 nt) transcripts, linearized with SfiI and HindIII, respectively, from the pGEM-11zf vector. Gel shift experiments to investigate RNA-binding activity were then performed. Briefly, 0.4 μM GST fusion proteins were incubated on ice for 30 min in 20 μl of binding buffer containing radiolabeled dsRNA (10,000 cpm, 1 nM). The protein-RNA complexes were resolved from free RNA by running them on nondenaturing 6% polyacrylamide gels at 4°C for 3 h at 150 V in 0.045 M Tris-borate-0.001 M EDTA running buffer. The gels were dried, and RNA-containing bands were visualized by autoradiography.
Reporter gene assays in 293T cells.
For the experiments investigating IFN-β and ISRE promoter activation, 293T cells were transfected with a mouse IFN-β promoter-driven chloramphenicol acetyltransferase (CAT) reporter plasmid, pIFN-CAT (40), or an ISRE-driven CAT reporter plasmid, pHISG-54-CAT (2, 5). In addition, an internal control plasmid, pGL2-control (Promega), encoding the firefly luciferase protein, and the appropriate pCAGGS-NS1 expression plasmid were transfected into the cells. The CAT reporter (0.5 μg) and 0.5 μg of the luciferase plasmid were transfected along with 4.0 μg of the pCAGGS-NS1 (wild type or mutant) plasmid into 106 293T cells by use of the calcium phosphate mammalian transfection kit (Stratagene). At 24 h posttransfection, cells were infected with Sendai virus at a multiplicity of infection (MOI) of approximately 10. At 24 h postinfection, cells were harvested and assayed for CAT expression as previously described (33) and luciferase protein expression was assayed with the Promega luciferase assay system according to the manufacturer's directions. Levels of CAT activity were normalized among samples according to luciferase values.
Virus growth curves.
MDCK cells were infected at an MOI of 0.001 with recombinant influenza A viruses expressing either NS1 R38AK41A, NS1 R38AK41AS42G, or wild-type NS1. At 24, 48, and 72 h postinfection, viruses in the cell supernatant were titrated by plaque assay on fresh MDCK cells.
NDV-GFP-based assay for IFN antagonist activity.
A549 cells were transfected with pCAGGS-NS1 (wild type or mutant) expression plasmids. Cells (105) were transfected in suspension by use of Lipofectamine 2000 (Invitrogen). Transfection mixtures were prepared in polystyrene tubes as follows: 3 μl of Lipofectamine 2000 diluted in 50 μl of OptiMEM was added to 50 μl of OptiMEM containing 2.0 μg of plasmid DNA, and the transfection mixtures were incubated for 20 min before being added to the cells. At 24 h posttransfection, cells were infected with NDV-GFP (MOI = 2), and at 24 h postinfection, the cells expressing GFP were visualized by fluorescence microscopy as previously described (32).
Determination of IFN secretion by a bioassay.
Levels of IFN secreted by virus-infected cells were determined as previously described (31). A549 cells were infected at an MOI of 2 with WSN (wild type), WSN-NS1 R38AK41A, or WSN-NS1 R38AK41AS42G virus. Following infection, cells were incubated with minimum essential medium containing 0.3% bovine serum albumin, and at 24 h postinfection, supernatants were harvested. Viruses present in the supernatant were UV inactivated by placing samples on ice 6 in. below an 8-W UV lamp for 15 min with constant stirring. Vero cells were seeded in 24-well plates the day before and incubated with the UV-inactivated supernatants for 24 h. The preincubated Vero cells were then infected with NDV-GFP (MOI = 2), and at 24 h postinfection, the cells expressing GFP were visualized by fluorescence microscopy. Green fluorescence intensity was quantified by fluorescence-activated cell-sorting analysis with a Beckman-Coulter Epics XL-MCL fluorescence-activated cell sorter.
Analysis of IFN-β mRNA by RT-PCR.
A549 cells were infected at an MOI of 2, and at 24 h posttransfection, total RNA was extracted by using Trizol reagent (Invitrogen). RT-PCR was done by using the following primer pairs specific for human IFN-β and β-actin mRNAs: IFN-β 5′+, GGCCATGACCAACAAGTGTCTCCTCC, and IFN-β 3′−, GCGCTCAGTTTCGGAGGTAACCTGT, resulting in a product of approximately 550 bp; and β-actin forward, TGGGTCAGAAGGACTCCTATG, and β-actin reverse, CAGGCAGCTCATAGCTCTTCT, also resulting in a 550-bp product.
Animal infections.
Six-week-old female BALB/c mice were anesthetized and infected intranasally with 50 μl of phosphate-buffered saline containing the indicated amounts of influenza A viruses. For viral lung titrations, mice were sacrificed at either day 2, 4, or 6 postinfection. Lungs were homogenized, resuspended in sterile phosphate-buffered saline, and titrated by plaque assay on MDCK cells. For monitoring of viral disease, animals were weighed daily and were euthanized when observed in extremis. All procedures were done in accordance with the National Institutes of Health guidelines on care and use of laboratory animals.
RESULTS
Generation of a recombinant influenza A virus expressing a mutant NS1 protein (NS1 R38AK41A) defective in dsRNA binding.
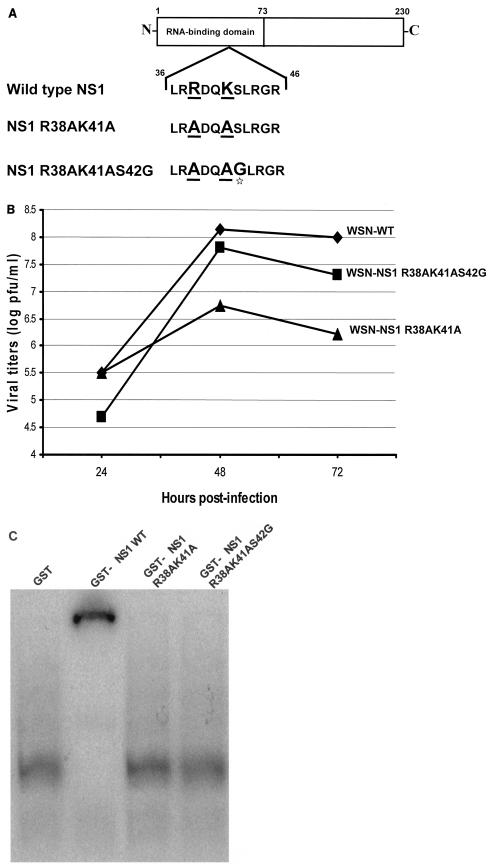
In order to investigate the role of the dsRNA-binding activity of the NS1 protein during influenza A virus infection, we generated WSN-NS1 R38AK41A virus by plasmid transfection. This recombinant virus is isogenic with wild-type WSN virus, except for the NS1 gene, which encodes a mutant NS1 protein with two alanine residues at positions 38 and 41 instead of arginine and lysine, respectively (Fig. 1A). These two amino acid changes were previously found to result in attenuation of dsRNA binding without affecting protein dimerization (39). WSN-NS1 R38AK41A virus was rescued upon transfection in 293T-MDCK cocultures of eight plasmids encoding the influenza virus RNA segments from a human polymerase I promoter together with four viral protein expression plasmids, as described previously (13, 35). Rescued viruses were passaged three times in MDCK cells. Interestingly, viral titers as measured by plaque assay dramatically increased ∼2 log between the first passage and the last passage. The NS gene of the virus that was passaged three times was amplified by RT-PCR using specific primers, and the obtained cDNA was cloned into pGEM-T. Sequence analysis of several clones revealed that the majority of these clones (seven of eight) differed from the original NS1 R38AK41A sequence by a single nucleotide substitution, resulting in an amino acid change (S42G) immediately after the original R38AK41A mutations (Fig. 1A). The mutant NS1 R38AK41A and NS1 R38AK41AS42G genes were reintroduced into recombinant viruses by plasmid transfection. Viruses present in the supernatants of transfected cells were plaque purified in MDCK cells, and seed viruses derived from one plaque were prepared in MDCK cells. RT-PCR and sequence analysis of the NS genes in the new WSN-NS1 R38AK41A and WSN-NS1 R38AK41AS42G virus seeds confirmed the presence of the introduced mutations and the absence of additional changes. These virus seeds were used for all other experiments involving recombinant viruses.
FIG. 1.
Characterization of recombinant influenza A viruses and their NS1 mutations. (A) Schematic diagram of wild-type and mutant NS1 proteins. Underlined amino acid residues were mutated to alanine. The star highlights the serine-to-glycine mutation in the NS1 R38AK41AS42G protein. (B) Multicycle growth curves of recombinant influenza A/WSN/33 viruses expressing wild-type and mutant NS1 proteins. MDCK cells were infected at an MOI of 0.001 with the different recombinant viruses. Viruses released to the supernatant were titrated at 24, 48, and 72 h postinfection by plaque assay on fresh MDCK cells. (C) Binding of GST-NS1 proteins to dsRNA in vitro. Each GST-NS1 fusion protein (0.4 μM) was incubated with 32P-radiolabeled dsRNA (10,000 cpm, 1 nM). The protein-RNA complexes were then resolved from free RNA by running them on a 6% nondenaturing polyacrylamide gel at 4°C for 3 h at 150 V in 0.045 M Tris-borate-0.001 M EDTA running buffer.
Decreased replication of WSN-NS1 R38AK41A, but not of WSN-NS1 R38AK41AS42G, virus in MDCK cells.
The multicycle growth kinetics of the mutant NS1 viruses were investigated in MDCK cells. These viruses were compared to the isogenic wild-type WSN virus, also generated by plasmid transfection. Infections were done at a low MOI (0.001), and viruses in supernatants from infected cells were titrated at 24, 48, and 72 h postinfection by plaque assay. The wild-type and WSN-NS1 R38AK41AS42G viruses grew to approximately 108 PFU/ml after 48 h, whereas WSN-NS1 R38AK41A virus grew to titers between 1.0 and 1.5 log lower (Fig. 1B). These experiments indicate that the R38AK41A mutations in the NS1 protein result in viral growth attenuation in MDCK cells and that this phenotype can be compensated for by a third amino acid substitution (S42G) within the NS1 N-terminal region.
Both the NS1 R38AK41A and NS1 R38AK41AS42G mutant proteins are defective in binding to dsRNA in vitro.
In order to investigate whether the amino acid change S42G in the NS1 R38K41 protein results in a gain of dsRNA-binding activity, we determined by EMSA the ability of the wild-type NS1 and mutant NS1 R38K41 and NS1 R38K41S42G proteins to bind to dsRNA. These proteins were expressed in bacteria as GST fusion proteins and used in an in vitro dsRNA-binding assay. GST (Fig. 1C, lane 1) does not bind to dsRNA, while dsRNA incubated with GST-NS1 wild type (Fig. 1C, lane 2) shows a typical gel shift consistent with dsRNA binding. The GST-NS1 R38AK41A mutant protein (Fig. 1C, lane 3) was unable to bind to dsRNA, as previously reported (39). Interestingly, the GST-NS1 R38AK41AS42G mutant protein was also defective in binding to dsRNA (Fig. 1C, lane 4).
Expression of wild-type NS1 and NS1 R38AK41AS42G, but not NS1 R38AK41A, proteins prevents IFN-β and ISRE promoter activation.
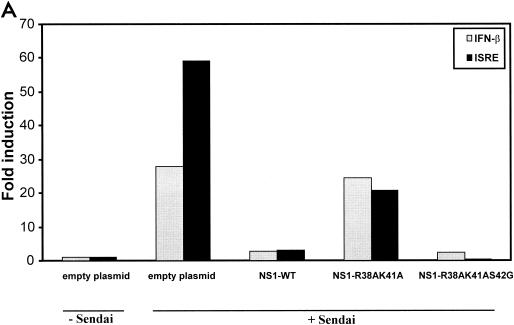
The NS1 protein of influenza A virus has been shown to prevent the virus-induced activation of ISRE and IFN-β promoters (37, 40). We next investigated, by using a transfection-based assay, whether the NS1 R38AK41A and R38AK41AS42G mutant proteins, which are defective in binding to dsRNA in vitro, are also defective in preventing the activation of these promoters. 293T cells were transfected with expression plasmids for the NS1 wild-type and R38AK41A and R38AK41AS42G mutant proteins together with a reporter gene under the control of the ISG54-ISRE or IFN-β promoter. Activation of these promoters was induced 24 h later by infection with Sendai virus. Interestingly, the NS1 R38AK41AS42G mutant protein was able to inhibit Sendai virus-induced IFN-β and ISRE promoter activation to levels similar to those for wild-type NS1 (Fig. 2A). However, the NS1 R38AK41A mutant was severely impaired in the ability to inhibit activation of these promoters, as was previously found (40). All NS1 proteins were expressed in 293T cells at similar levels, as measured by Western blotting (data not shown).
FIG. 2.
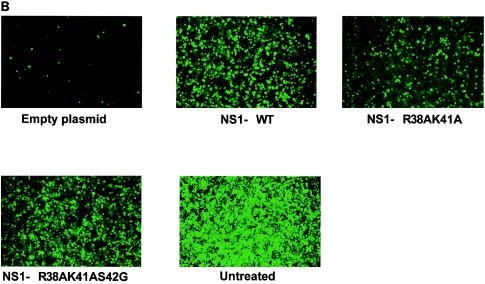
IFN inhibitory activities of wild-type and mutant NS1 proteins. (A) Inhibition of Sendai virus-induced IFN-β and ISRE promoter activation by plasmid-mediated expression of wild-type and mutant NS1 proteins. The IFN-β or ISRE promoter-driven CAT reporter plasmid (0.5 μg) and 0.5 μg of the luciferase control plasmid were transfected along with 4.0 μg of empty plasmid or the pCAGGS-NS1 (wild-type or mutant) plasmid into 106 293T cells. At 24 h posttransfection, the cells were infected with Sendai virus at an MOI of approximately 10. At 24 h postinfection, cells were harvested and assayed for CAT and luciferase expression. (B) NDV-GFP replication in A549 cells expressing wild-type and mutant NS1 proteins. A549 cells (105) were transfected with 2.0 μg of empty plasmid or pCAGGS-NS1 (wild-type or mutant) expression plasmid. At 24 h posttransfection, cells were infected with NDV-GFP (MOI ∼ 2), and at 24 h postinfection, the cells expressing GFP were monitored by fluorescence microscopy. The results shown are from one representative experiment of three total experiments.
Enhancement of replication of IFN-sensitive NDV-GFP by expression of wild-type NS1 and NS1 R38AK41AS42G, but not NS1 R38AK41A, proteins.
Transfection of plasmid DNA into A549 cells results in the induction of an IFN-mediated antiviral state that prevents replication of IFN-sensitive NDV-GFP. However, transfection of a plasmid encoding an IFN antagonist protein prevents the induction of the IFN-mediated antiviral state, allowing for replication of NDV-GFP. This can be easily monitored by GFP expression (32). In order to investigate the ability of NS1 wild-type and R38AK41A and R38AK41AS42G mutant proteins to inhibit the IFN system in this virus-based assay, we transfected A549 cells with either empty plasmid or pCAGGS-NS1 expression plasmids and at 24 h posttransfection infected them with NDV-GFP. At 24 h postinfection, cells were monitored for NDV replication by fluorescence microscopy. In the cells transfected with pCAGGS-empty plasmid, NDV-GFP replication was significantly inhibited. However, cells expressing wild-type and NS1 R38AK41AS42G proteins replicated NDV-GFP to high levels, as shown in Fig. 2B. The levels of replication of NDV-GFP in A549 cells expressing the NS1 R38AK41A mutant protein were significantly lower than those in cells expressing the wild-type or the NS1 R38AK41AS42G protein.
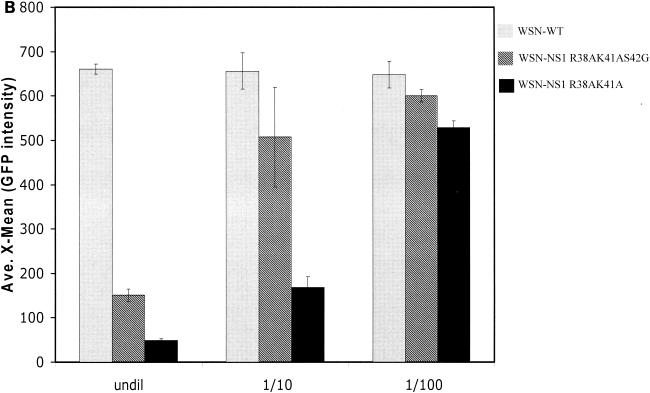
Infection of A549 cells with WSN-NS1 R38AK41A virus induces higher levels of IFN-β than infection with WSN-NS1 R38AK41AS42G and wild-type viruses.
When the NS1 R38AK41AS42G mutant protein was expressed in cells by plasmid transfection, it displayed IFN antagonistic properties similar to those of wild-type NS1 protein, while the NS1 R38AK41A protein was defective in preventing the activation of the IFN system. However, the wild-type and mutant NS1 proteins are overexpressed from a mammalian promoter in these assays, and therefore subtle differences in activity between these proteins might not be detectable. We therefore investigated the IFN-inducing properties of recombinant influenza virus expressing the wild-type and mutant NS1 proteins. Human epithelial lung A549 cells were infected at an MOI of 2 with either wild-type recombinant WSN, mutant WSN-NS1 R38AK41A, or mutant WSN-NS1 R38AK41AS42G virus, and at 24 h postinfection, supernatants from these infected cells were tested for the presence of IFN by a bioassay based on inhibition of NDV-GFP replication in Vero cells (31). Figure 3A and B show that supernatants from WSN-NS1 R38AK41A virus-infected A549 cells caused the highest level of inhibition of NDV-GFP replication, as measured by GFP expression in pretreated Vero cells. In contrast, levels of NDV-GFP replication were similar between mock-pretreated Vero cells (data not shown) and Vero cells pretreated with supernatants from wild-type WSN virus-infected A549 cells, indicating that levels of IFN induced by wild-type influenza A/WSN virus infection in A549 cells were not detectable by this bioassay. Infection of A549 cells by WSN-NS1 R38AK41AS42G resulted in levels of secreted IFN that were intermediate between those present in supernatants from cells infected with wild-type and WSN-NS1 R38AK41A viruses. The levels of NS1 expression in the virus-infected A549 cells were comparable among the three viruses, as monitored by Western blotting (data not shown). These results indicate that while the NS1 R38AK41AS42G protein is a better inhibitor of IFN production than the NS1 R38AK41A protein in the context of an infectious influenza A virus, the S42G mutation did not completely revert the phenotype of the recombinant WSN-NS1 R38AK41AS42G virus to that of the wild-type virus.
FIG. 3.
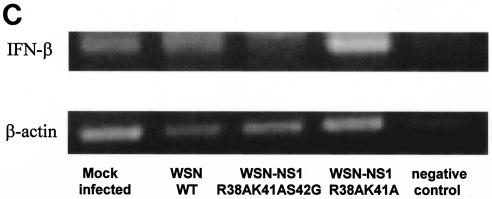
Induction of IFN-α/β synthesis in A549 cells infected with recombinant influenza A viruses expressing wild-type and mutant NS1 proteins. (A) Vero cells were pretreated for 24 h with UV-inactivated supernatant from A549 cells infected with the indicated influenza viruses. The pretreated Vero cells were then infected with NDV-GFP, and at 24 h postinfection, the cells expressing GFP were monitored by fluorescence microscopy. (B) Quantification by fluorescence-activated cell sorting of GFP expression in NDV-GFP-infected Vero cells pretreated with UV-inactivated supernatant from WSN or WSN-NS1 mutant virus-infected A549 cells. undil, not diluted. (C) RT-PCR analysis of IFN-β mRNA levels in virus-infected A549 cells. Cells were infected at an MOI of 2, and at 24 h posttransfection, total RNA was extracted and RT-PCR was done using primer pairs specific for human IFN-β and β-actin mRNA.
RT-PCR analysis of the IFN-β mRNA present in virus-infected A549 cells confirmed that infection with WSN-NS1 R38AK41A virus induces higher levels of IFN-β than infection with wild-type WSN or WSN-NS1 R38AK41AS42G virus (Fig. 3C, compare lane 4 with lanes 2 and 3). It should be noted that this analysis does not allow a precise quantitative comparison of IFN-β mRNA levels in virus-infected cells.
WSN-NS1 R38AK41AS42G virus is attenuated in mice but shows increased replication and virulence when compared to WSN-NS1 R38AK41A virus.
In order to study the phenotypes of the recombinant WSN viruses bearing mutations in the NS1 protein during host infections, we next conducted infection experiments in mice. Six-week-old BALB/c mice were intranasally infected with either wild-type or mutant NS1-expressing virus. We monitored body weight loss (Fig. 4A) and viral lung titers (Fig. 4B) as indicators of virus pathogenicity and replication, respectively. Infection with 105 PFU of WSN-NS1 R38AK41A did not result in any loss of body weight and the mice consistently gained weight over the course of the experiment. However, infection with WSN-NS1 R38AK41AS42G virus caused a transient reduction (between 5 and 10%) in the body weight of the mice, which recovered to their original weight at 7 days postinfection. Mice infected with wild-type WSN virus lost approximately 25% of their total body weight by day 5 postinfection and all eventually succumbed to the infection (Fig. 4A). Virus replication in lungs, as determined by plaque assay in MDCK cells, was monitored at days 2, 4, and 6 in mice intranasally infected with 104 PFU of the different viruses (Fig. 4B). The wild-type WSN virus was consistently found to replicate to titers of ∼106 PFU/ml at all days postinfection. WSN-NS1 R38AK41A virus replicated very poorly in mouse lungs and could only be detected, at a very low titer of ∼102 PFU/ml, in one of three mice at day 2. WSN-NS1 R38AK41AS42G virus showed an intermediate profile in the ability to replicate in mice, consistent with its virulence phenotype. At day 2, titers of approximately 104 PFU/ml were detected, but by day 4 this mutant virus could only be detected, at a low titer (5 × 102 PFU/ml), in one of three mice, and by day 6 there was no detectable virus in the lungs from three infected mice. It should be noted that the limit of detection of viral lung titers was 102 PFU/ml.
FIG. 4.
Pathogenicity in mice of recombinant influenza A viruses expressing wild-type and mutant NS1 proteins. (A) Weight loss of BALB/c mice was monitored after intranasal inoculation with 105 PFU of WSN-WT, WSN-NS1 R38AK41A, or WSN-NS1 R38AK41AS42G virus. Five mice per group were infected and the average percentages of original body weight were monitored for 15 days postinfection. (B) Viral lung titers in mice on days 2, 4, and 6 postinfection with 104 PFU of intranasally administered virus. Three mice per group were infected and viral lung titers were determined by plaque assay on MDCK cells.
DISCUSSION
We have shown in this report that amino acid residues in the NS1 protein of influenza A virus that play a critical role in the ability of this protein to bind to dsRNA (R38 and K41) are also critical for the optimal IFN antagonist activity of NS1. Infection with influenza A viruses with mutated NS1 proteins at these amino acid residues results in increased IFN secretion. There is a good correlation between the levels of IFN induced by viral infection in lung epithelial A549 cells and the attenuation of the viruses in mice. Of the two studied mutant viruses, WSN-NS1 R38AK41A virus induced the highest levels of IFN secretion in A549 lung epithelial cells and was the most attenuated virus in mice. WSN-NS1 R38AK41AS42G mutant virus induced lower levels of IFN in A549 cells than WSN-NS1 R38AK41A, but the levels were still higher than in the wild-type WSN virus, and this virus had an attenuated phenotype in mice that was between those of the other two viruses. Neither the NS1 R38AK41A nor the NS1 R38AK41AS42G mutant protein was able to bind in vitro to dsRNA. These results suggest that although the ability of the NS1 protein of influenza A virus to bind to RNA is needed to efficiently inhibit IFN-α/β production during viral infection, other mechanisms also play a role in this inhibition. Our results also demonstrate a definitive role for the N-terminal RNA-binding domain of the NS1 protein in the inhibition of the IFN response. This inhibition is independent of the NS1-mediated effects on host mRNA splicing and export (7, 14, 25, 28, 30) that appear not to be affected by the R38AK41A mutations (22).
Our first attempt to generate a recombinant WSN-NS1 R38AK41A virus expressing an NS1 protein defective in RNA-binding activity resulted in a virus that upon a few passages in MDCK cells grew to higher titers and acquired a third amino acid mutation in its NS1 protein, S42G. In order to demonstrate that this new amino acid substitution was sufficient to confer increased viral growth properties in MDCK cells, we generated a new recombinant WSN virus from plasmids encoding wild-type virus RNAs, except for the NS viral RNA that codes for an NS1 R38AK41AS42G mutant protein. When the growth properties of clonal populations of recombinant wild-type WSN and mutant WSN-NS1 R38AK41 and WSN-NS1 R38AK41S42G viruses were compared, it was evident that the S42G mutation in the background of an NS1 R38AK41A protein was sufficient to increase the replication levels of the mutant virus in MDCK cells to wild-type levels. Due to the quasispecies nature of RNA viruses, we cannot conclude that the S42G mutation is the only difference between WSN-NS1 R38AK41A and WSN-NS1 R38AK41S42G. However, experiments involving cells transfected with plasmids expressing the NS1 mutant proteins clearly support a role for the S42G mutation in promoting viral replication. The crystal structure of the first 73 amino acids of the NS1 protein has been solved and consists of three α-helices interconnected by loops (23). Amino acid residues 39, 41, and 42 are all located in the second α-helix, which is comprised of amino acids 30 to 50, and therefore our results highlight a critical role for this α-helix structure in the IFN antagonistic activity of the NS1 protein of influenza A virus.
The increased replication properties of WSN-NS1 R38AK41AS42G virus in MDCK cells correlated with a gain of function of the NS1 protein with respect to the ability to inhibit IFN-β promoter activation and to promote replication of an IFN-sensitive virus when expressed in trans by plasmid transfection. However, experiments using recombinant WSN viruses demonstrated that the S42G mutation, although resulting in an increased inhibition of IFN production during viral infection, did not restore the potency of the NS1 protein to wild-type levels. Interestingly, this partial gain of IFN inhibitory function conferred by the S42G mutation was not accompanied by a gain of dsRNA-binding activity. These results suggest that the NS1-dependent inhibition of IFN synthesis is not completely mediated by binding to and sequestering dsRNA generated during virus infection. It is possible that interactions of the N-terminal domain of the NS1 protein with host proteins involved in induction of IFN synthesis may also contribute to the IFN antagonist activity of this protein. The S42G change would then be responsible for restoring these protein-protein interactions. On the other hand, we cannot exclude the possibility that the NS1 R38AK41AS42G protein, while not binding to the in vitro-synthesized dsRNA used in our experiments, may have acquired specific binding to influenza virus-generated dsRNA in vivo. However, given the important role that basic residues usually play on RNA-protein interactions, we do not favor this hypothesis.
In addition to attenuating IFN synthesis during viral infection, the NS1 protein of influenza A virus inhibits an important IFN-induced antiviral pathway, namely, the dsRNA-activated PKR (3, 18, 26). The dsRNA-binding activity of NS1 appears to be responsible for PKR inhibition (26), although direct protein-protein interactions between NS1 and PKR have also been suggested (38). We are currently investigating the ability of NS1 R38AK41A and NS1 R38AK41AS42G proteins to inhibit PKR activation.
In summary, our results demonstrate a critical role for amino acid residues located within the second α-helix of the NS1 protein of influenza A virus in preventing IFN-α/β secretion in virus-infected cells and in viral replication and pathogenicity in the host. IFN-α/β secretion is not only a critical component of the innate antiviral immune response but also appears to modulate the specific antiviral cellular immune response (4). Further studies are required in order to determine whether the NS1 protein of influenza A virus has an impact in the induction of specific immune responses. In this respect, it is interesting that induction of IFN-α/β secretion by influenza virus infection in dendritic cells is also attenuated by the NS1 protein (10, 11, 24). Finally, our studies indicate that the N-terminal domain of the NS1 protein might be an attractive target for antiviral drug design.
Acknowledgments
We acknowledge members of the A.G.-S. and P. Palese laboratories for critical discussions, especially Man-Seong Park for his invaluable assistance. We gratefully acknowledge Estanislao Nistal-Villán and Richard Cadagan for excellent technical assistance.
This work was supported by NIH grants to C.F.B. and A.G.-S. C.F.B. is an Ellison Medical Foundation New Scholar in Infectious Diseases.
REFERENCES
- 1.Aragón, T., S. de La Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. García-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 5.Bluyssen, H. A., R. J. Vlietstra, A. van der Made, and J. Trapman. 1994. The interferon-stimulated gene 54 K promoter contains two adjacent functional interferon-stimulated response elements of different strength, which act synergistically for maximal interferon-alpha inducibility. Eur. J. Biochem. 220:395-402. [DOI] [PubMed] [Google Scholar]
- 6.Bourmakina, S. V., and A. García-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly, C., and N. C. Reich. 1993. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol. Cell. Biol. 13:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. R. Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 11.Efferson, C. L., J. Schickli, B. K. Ko, K. Kawano, S. Mouzi, P. Palese, A. García-Sastre, and C. G. Ioannides. 2003. Activation of tumor antigen-specific cytotoxic T lymphocytes (CTLs) by human dendritic cells infected with an attenuated influenza A virus expressing a CTL epitope derived from the HER-2/neu proto-oncogene. J. Virol. 77:7411-7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enami, M., and K. Enami. 2000. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J. Virol. 74:5556-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes, P., A. Beloso, and J. Ortín. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. García-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 18.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatada, E., S. Saito, N. Okishio, and R. Fukuda. 1997. Binding of the influenza virus NS1 protein to model genome RNAs. Virology 78:1059-1063. [DOI] [PubMed] [Google Scholar]
- 20.Hatada, E., T. Takizawa, and R. Fukuda. 1992. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 73:17-25. [DOI] [PubMed] [Google Scholar]
- 21.Levy, D. E., and A. García-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 24.López, C. B., A. García-Sastre, B. R. Williams, and T. M. Moran. 2003. Type I interferon induction pathway, but not released interferon, participates in the maturation of dendritic cells induced by negative-strand RNA viruses. J. Infect. Dis. 187:1126-1136. [DOI] [PubMed] [Google Scholar]
- 25.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 26.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. García-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 29.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 30.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 31.Park, M. S., A. García-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, M. S., M. L. Shaw, J. Muñoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. García-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percy, N., W. S. Barclay, A. García-Sastre, and P. Palese. 1994. Expression of a foreign protein by influenza A virus. J. Virol. 68:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schickli, J. H., A. Flandorfer, T. Nakaya, L. Martínez-Sobrido, A. García-Sastre, and P. Palese. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B 356:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 37.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 39.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]