Abstract
The title compound, C18H16BrN3O4S, is a Schiff base ligand of 5-bromosalicylaldehyde and sulfisoxazole [or N-(3,4-dimethyl-5-isoxazol)sulfanilamide]. The present structure is a zwitterion and is a more precise reinterpretation of the structure which was originally reported by Hämäläinen, Lehtinen & Turpeinen [Arch. Pharm. (1986), 319, 415–420]. The two aromatic rings which make π–π interactions [centroid–centroid distance 3.7538 (18) Å] through intermolecular interactions. There is also a C—Br⋯π interaction [3.6333 (15) Å] with the heterocyclic ring. An intramolecular N—H⋯O hydrogen bond also exists. Dimers are formed due to intermolecular N—H⋯O hydrogen bonding. Intermolecular C—H⋯O hydrogen bonding links a methyl C atom and the phenolate O atom. The dimers are linked by C—H⋯N hydrogen bonds, where the C atom is from the Schiff base group and the N atom is of five-membered heterocyclic ring.
Related literature
For related literature, see: Chohan et al. (2008 ▶); Hämäläinen et al. (1986 ▶); Shad et al. (2008 ▶).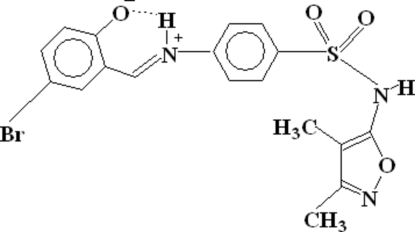
Experimental
Crystal data
C18H16BrN3O4S
M r = 450.31
Monoclinic,
a = 15.3846 (10) Å
b = 7.2235 (5) Å
c = 16.5520 (11) Å
β = 93.201 (4)°
V = 1836.6 (2) Å3
Z = 4
Mo Kα radiation radiation
μ = 2.38 mm−1
T = 296 (2) K
0.18 × 0.14 × 0.10 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.685, T max = 0.793
18874 measured reflections
3955 independent reflections
2704 reflections with I > 2σ(I)
R int = 0.058
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.115
S = 1.00
3955 reflections
252 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.46 e Å−3
Δρmin = −0.62 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: APEX2; data reduction: SAINT (Bruker, 2007 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2003 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680800682X/bq2069sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800682X/bq2069Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1 | 0.80 (3) | 1.91 (3) | 2.577 (4) | 141 (3) |
| N2—H2⋯O1i | 0.75 (3) | 2.09 | 2.828 (4) | 171 (4) |
| C17—H17C⋯O1i | 0.96 | 2.58 | 3.248 (5) | 126 |
| C7—H7⋯N3ii | 0.93 | 2.53 | 3.420 (4) | 161 |
Symmetry codes: (i) 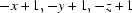 ; (ii)
; (ii) 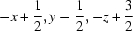 .
.
Acknowledgments
The authors acknowledge the Higher Education Commission, Islamabad, Pakistan, for funding the purchase of the diffractometer.
supplementary crystallographic information
Comment
Sulfonamides are a class of compounds, which have been found to possess a wide range of medicinal properties. In continuation of synthesizing Schiff base ligands of substituted halogen salicylaldehyde and various sulfonamides (Chohan et al., 2008, Shad et al., 2008), We report the refinement of (5-Bromosalicy1idene)-N-(3,4-dimethyl-5-isoxazolyl)sulfanilamide (Hämäläinen et al., 1986).
During the refinement of 4-Chloro-2-[(E)-({4-[(3,4-dimethylisoxazol-5-yl) sulfamoyl]phenyl}iminio)methyl]phenolate (Shad et al., 2008) prepared from 5-chlorosalicylaldehyde and sulfisoxazole: [N-(3,4-dimethyl-5-isoxazol) sulfanilamide], it was observed that H-atom of hydroxy group shifts to N-atom of Schiff base moiety. To see the effect of sulfisoxazole on 5-Bromosalicylaldehyde, the title compound (I) has been prepared. The X-ray structure analysis clearly shows that again a zwitterion is formed due to the reaction of 5-Bromosalicylaldehyde and sulfisoxazole. The values of bond lengths and bond angles are similar to the chloro isomer and also similar to the reported by Hämäläinen et al., 1986. There exist π interaction at a distance of 3.6333 (15) Å C5—Br1···CgAi [symmetry code i = x - 1/2, -y + 1/2, z - 1/2], where CgA is the center of gravity of the (A) five-membered heterocyclic ring containing O4. The π - π interaction also exists between the aromatic rings (B) and (C), containing C10 and C3 respectively. The distance between the centroids of neighbouring rings have value of 3.7538 (18) Å, CgB···CgCii [symmetry code ii = x, y - 1, z], here CgB and CgC represent the center of gravity of ring (B) and ring (C) respectively. The dihedral angles between the rings A/B, A/C, B/C have values of 43.30 (18)°, 44.30 (18)° and 5.56 (15)° respectively. The molecules form dimers through H-bonding between N2—H2···O1iii [symmetry code iii = -x + 1, -y + 1, -z + 1]. These dimers are linked to each other by C7—H7···N3iv [symmetry code iv = -x + 1/2, y - 1/2, -z + 3/2] and form a three-dimensional polymeric network. The intermolecular H-bond [C—H···O] between methyl C-atom and O-atom of aromatic ring with Bromom group. The detail of H-bonding is given in Table 1.
Experimental
Sulfisoxazole (0.5346 g, 2 mmol) in ethanol (20 ml) was mixed with ethanolic solution (10 ml) of 5-Bromosalicylaldehyde (0.402 g, 2 mmol). After refluxing for 3 h, the solution turned orange red. The solution was cooled to room temperature, filtered and volume reduced to about one-third using rotary evaporator. It was then allowed to stand for 10 days and obtained crystals of title compound. m.p; 481 K.
Refinement
The coordinates of H-atoms attached to N-atoms were refined. H-atoms were positioned geometrically, with C—H = 0.93 Å for aromatic like, 0.96 Å for methyl and constrained to ride on their parent atoms. The Uiso(H) = xUeq(C, N), where x = 1.5 for methyl H, and x = 1.2 for all other H atoms.
Figures
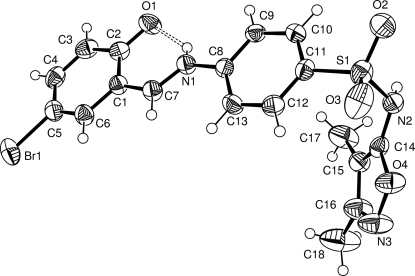
Fig. 1.
ORTEP-3 for Windows (Farrugia, 1997) drawing of the title compound, C18H16BrN3O4S, with the atom numbering scheme. The thermal ellipsoids are drawn at the 50% probability level. H-atoms are shown by small circles of arbitrary radii. The intramolecular H-bonding is shown by dashed lines.
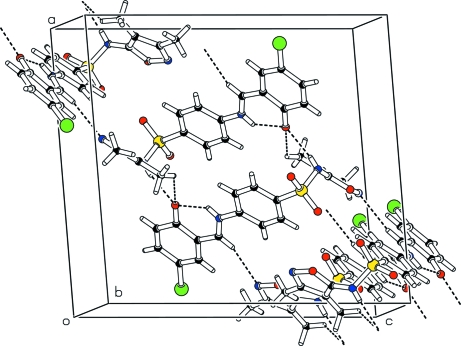
Fig. 2.
The interaction diagram of (I) (Spek, 2003) showing the intramolecular and intermolecular hydrogen bonding which leads to dimers and a three-dimensional polymeric network.
Crystal data
| C18H16BrN3O4S | F000 = 912 |
| Mr = 450.31 | Dx = 1.629 Mg m−3 |
| Monoclinic, P21/n | Melting point: 481 K |
| Hall symbol: -P 2yn | Mo Kα radiation radiation λ = 0.71073 Å |
| a = 15.3846 (10) Å | Cell parameters from 2704 reflections |
| b = 7.2235 (5) Å | θ = 1.8–27.0º |
| c = 16.5520 (11) Å | µ = 2.38 mm−1 |
| β = 93.201 (4)º | T = 296 (2) K |
| V = 1836.6 (2) Å3 | Prismatic, red |
| Z = 4 | 0.18 × 0.14 × 0.10 mm |
Data collection
| Bruker KappaAPEXII CCD diffractometer | 3955 independent reflections |
| Radiation source: fine-focus sealed tube | 2704 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.058 |
| Detector resolution: 7.40 pixels mm-1 | θmax = 27.0º |
| T = 296(2) K | θmin = 1.8º |
| ω scans | h = −19→19 |
| Absorption correction: multi-scan(SADABS; Bruker, 2005) | k = −9→9 |
| Tmin = 0.685, Tmax = 0.793 | l = −21→18 |
| 18874 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.115 | w = 1/[σ2(Fo2) + (0.00.057P)2 + 0.7324P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 3955 reflections | Δρmax = 0.46 e Å−3 |
| 252 parameters | Δρmin = −0.62 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.11987 (2) | −0.44715 (5) | 0.41651 (3) | 0.05783 (16) | |
| S1 | 0.39184 (6) | 0.99341 (12) | 0.67050 (5) | 0.0440 (2) | |
| O1 | 0.38204 (15) | 0.1394 (3) | 0.34949 (14) | 0.0491 (6) | |
| O2 | 0.4329 (2) | 1.1298 (3) | 0.62307 (15) | 0.0647 (7) | |
| O3 | 0.31683 (17) | 1.0380 (4) | 0.71321 (17) | 0.0645 (8) | |
| O4 | 0.40342 (15) | 0.8167 (3) | 0.85228 (14) | 0.0514 (6) | |
| N1 | 0.32829 (17) | 0.3238 (4) | 0.47030 (16) | 0.0365 (6) | |
| H1 | 0.364 (2) | 0.300 (5) | 0.439 (2) | 0.044* | |
| N2 | 0.46806 (18) | 0.9255 (4) | 0.73775 (17) | 0.0380 (6) | |
| H2 | 0.510 (2) | 0.918 (5) | 0.717 (2) | 0.046* | |
| N3 | 0.3987 (2) | 0.6529 (5) | 0.89824 (19) | 0.0669 (10) | |
| C1 | 0.26586 (19) | 0.0371 (4) | 0.42687 (18) | 0.0352 (7) | |
| C2 | 0.3239 (2) | 0.0154 (4) | 0.36301 (19) | 0.0382 (7) | |
| C3 | 0.3146 (2) | −0.1448 (5) | 0.3150 (2) | 0.0466 (8) | |
| H3 | 0.3501 | −0.1604 | 0.2718 | 0.056* | |
| C4 | 0.2547 (2) | −0.2782 (5) | 0.33041 (19) | 0.0449 (8) | |
| H4 | 0.2503 | −0.3839 | 0.2984 | 0.054* | |
| C5 | 0.1997 (2) | −0.2558 (5) | 0.3945 (2) | 0.0416 (7) | |
| C6 | 0.2043 (2) | −0.1014 (5) | 0.4411 (2) | 0.0407 (8) | |
| H6 | 0.1666 | −0.0871 | 0.4826 | 0.049* | |
| C7 | 0.2714 (2) | 0.1925 (4) | 0.47838 (19) | 0.0383 (7) | |
| H7 | 0.2330 | 0.2017 | 0.5197 | 0.046* | |
| C8 | 0.34005 (19) | 0.4812 (4) | 0.52001 (18) | 0.0329 (7) | |
| C9 | 0.39958 (18) | 0.6121 (4) | 0.49609 (18) | 0.0349 (7) | |
| H9 | 0.4295 | 0.5926 | 0.4495 | 0.042* | |
| C10 | 0.41460 (19) | 0.7704 (4) | 0.54089 (18) | 0.0375 (7) | |
| H10 | 0.4549 | 0.8577 | 0.5255 | 0.045* | |
| C11 | 0.3682 (2) | 0.7976 (4) | 0.60991 (18) | 0.0361 (7) | |
| C12 | 0.3092 (2) | 0.6689 (5) | 0.63357 (19) | 0.0414 (8) | |
| H12 | 0.2786 | 0.6893 | 0.6797 | 0.050* | |
| C13 | 0.2952 (2) | 0.5096 (5) | 0.5892 (2) | 0.0411 (8) | |
| H13 | 0.2558 | 0.4214 | 0.6056 | 0.049* | |
| C14 | 0.45153 (18) | 0.7779 (4) | 0.78862 (17) | 0.0346 (7) | |
| C15 | 0.4783 (2) | 0.6014 (5) | 0.7904 (2) | 0.0410 (8) | |
| C16 | 0.4434 (3) | 0.5296 (5) | 0.8609 (2) | 0.0572 (10) | |
| C17 | 0.5315 (3) | 0.5017 (5) | 0.7328 (3) | 0.0692 (13) | |
| H17A | 0.5086 | 0.3792 | 0.7242 | 0.104* | |
| H17B | 0.5906 | 0.4938 | 0.7546 | 0.104* | |
| H17C | 0.5300 | 0.5672 | 0.6823 | 0.104* | |
| C18 | 0.4540 (4) | 0.3344 (6) | 0.8916 (3) | 0.0978 (18) | |
| H18A | 0.3996 | 0.2914 | 0.9103 | 0.147* | |
| H18B | 0.4976 | 0.3314 | 0.9354 | 0.147* | |
| H18C | 0.4714 | 0.2559 | 0.8486 | 0.147* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0474 (2) | 0.0454 (2) | 0.0799 (3) | −0.01217 (17) | −0.00429 (19) | −0.00371 (19) |
| S1 | 0.0541 (5) | 0.0327 (4) | 0.0448 (5) | 0.0070 (4) | −0.0016 (4) | −0.0070 (4) |
| O1 | 0.0517 (13) | 0.0490 (14) | 0.0487 (13) | −0.0079 (12) | 0.0226 (11) | −0.0059 (11) |
| O2 | 0.105 (2) | 0.0305 (13) | 0.0578 (16) | −0.0076 (14) | −0.0076 (15) | 0.0067 (12) |
| O3 | 0.0561 (15) | 0.0677 (18) | 0.0693 (17) | 0.0268 (13) | −0.0017 (13) | −0.0284 (14) |
| O4 | 0.0569 (14) | 0.0549 (15) | 0.0449 (13) | 0.0158 (12) | 0.0248 (12) | −0.0003 (12) |
| N1 | 0.0342 (14) | 0.0397 (15) | 0.0362 (15) | −0.0020 (12) | 0.0080 (11) | −0.0048 (12) |
| N2 | 0.0384 (14) | 0.0388 (15) | 0.0372 (15) | −0.0013 (12) | 0.0068 (12) | −0.0050 (12) |
| N3 | 0.083 (2) | 0.062 (2) | 0.059 (2) | 0.0189 (19) | 0.0412 (18) | 0.0139 (17) |
| C1 | 0.0343 (15) | 0.0365 (17) | 0.0350 (17) | −0.0005 (13) | 0.0044 (13) | −0.0045 (14) |
| C2 | 0.0399 (17) | 0.0392 (18) | 0.0357 (17) | 0.0051 (14) | 0.0052 (14) | −0.0004 (14) |
| C3 | 0.058 (2) | 0.046 (2) | 0.0358 (18) | 0.0073 (17) | 0.0095 (16) | −0.0042 (16) |
| C4 | 0.056 (2) | 0.0372 (18) | 0.0403 (19) | 0.0041 (16) | −0.0072 (16) | −0.0082 (15) |
| C5 | 0.0401 (17) | 0.0379 (18) | 0.0461 (19) | −0.0026 (14) | −0.0036 (15) | 0.0010 (15) |
| C6 | 0.0347 (16) | 0.0437 (19) | 0.0440 (19) | −0.0029 (14) | 0.0059 (14) | −0.0044 (15) |
| C7 | 0.0359 (16) | 0.0411 (18) | 0.0384 (17) | −0.0021 (14) | 0.0066 (14) | −0.0043 (15) |
| C8 | 0.0318 (15) | 0.0353 (16) | 0.0315 (16) | −0.0006 (12) | 0.0007 (12) | −0.0019 (13) |
| C9 | 0.0348 (16) | 0.0408 (17) | 0.0294 (16) | −0.0005 (13) | 0.0057 (13) | 0.0017 (14) |
| C10 | 0.0394 (16) | 0.0372 (17) | 0.0363 (17) | −0.0053 (14) | 0.0058 (14) | 0.0014 (14) |
| C11 | 0.0398 (16) | 0.0340 (17) | 0.0338 (17) | 0.0032 (13) | −0.0032 (13) | −0.0038 (13) |
| C12 | 0.0407 (17) | 0.048 (2) | 0.0365 (17) | −0.0022 (15) | 0.0117 (14) | −0.0057 (15) |
| C13 | 0.0414 (17) | 0.0431 (18) | 0.0399 (18) | −0.0120 (14) | 0.0126 (15) | −0.0035 (15) |
| C14 | 0.0329 (15) | 0.0420 (18) | 0.0294 (15) | 0.0027 (13) | 0.0068 (13) | −0.0095 (14) |
| C15 | 0.0448 (17) | 0.0390 (18) | 0.0405 (18) | −0.0009 (14) | 0.0132 (15) | −0.0056 (14) |
| C16 | 0.067 (2) | 0.049 (2) | 0.058 (2) | 0.0105 (19) | 0.025 (2) | 0.0077 (18) |
| C17 | 0.096 (3) | 0.041 (2) | 0.076 (3) | 0.005 (2) | 0.048 (3) | −0.008 (2) |
| C18 | 0.140 (5) | 0.066 (3) | 0.094 (3) | 0.028 (3) | 0.065 (3) | 0.029 (3) |
Geometric parameters (Å, °)
| Br1—C5 | 1.898 (3) | C6—H6 | 0.9300 |
| S1—O3 | 1.424 (3) | C7—H7 | 0.9300 |
| S1—O2 | 1.429 (3) | C8—C13 | 1.385 (4) |
| S1—N2 | 1.646 (3) | C8—C9 | 1.389 (4) |
| S1—C11 | 1.760 (3) | C9—C10 | 1.375 (4) |
| O1—C2 | 1.294 (4) | C9—H9 | 0.9300 |
| O4—C14 | 1.350 (3) | C10—C11 | 1.395 (4) |
| O4—N3 | 1.411 (4) | C10—H10 | 0.9300 |
| N1—C7 | 1.302 (4) | C11—C12 | 1.371 (4) |
| N1—C8 | 1.409 (4) | C12—C13 | 1.376 (5) |
| N1—H1 | 0.80 (3) | C12—H12 | 0.9300 |
| N2—C14 | 1.391 (4) | C13—H13 | 0.9300 |
| N2—H2 | 0.75 (4) | C14—C15 | 1.339 (4) |
| N3—C16 | 1.302 (5) | C15—C16 | 1.410 (5) |
| C1—C6 | 1.406 (4) | C15—C17 | 1.478 (5) |
| C1—C7 | 1.409 (4) | C16—C18 | 1.504 (6) |
| C1—C2 | 1.429 (4) | C17—H17A | 0.9600 |
| C2—C3 | 1.406 (5) | C17—H17B | 0.9600 |
| C3—C4 | 1.368 (5) | C17—H17C | 0.9600 |
| C3—H3 | 0.9300 | C18—H18A | 0.9600 |
| C4—C5 | 1.402 (5) | C18—H18B | 0.9600 |
| C4—H4 | 0.9300 | C18—H18C | 0.9600 |
| C5—C6 | 1.355 (5) | ||
| O3—S1—O2 | 120.84 (18) | C9—C8—N1 | 116.6 (3) |
| O3—S1—N2 | 107.36 (15) | C10—C9—C8 | 120.4 (3) |
| O2—S1—N2 | 104.87 (16) | C10—C9—H9 | 119.8 |
| O3—S1—C11 | 108.49 (16) | C8—C9—H9 | 119.8 |
| O2—S1—C11 | 108.95 (16) | C9—C10—C11 | 118.7 (3) |
| N2—S1—C11 | 105.22 (14) | C9—C10—H10 | 120.7 |
| C14—O4—N3 | 107.1 (2) | C11—C10—H10 | 120.7 |
| C7—N1—C8 | 126.3 (3) | C12—C11—C10 | 121.1 (3) |
| C7—N1—H1 | 114 (3) | C12—C11—S1 | 120.1 (2) |
| C8—N1—H1 | 119 (3) | C10—C11—S1 | 118.7 (2) |
| C14—N2—S1 | 119.4 (2) | C11—C12—C13 | 120.0 (3) |
| C14—N2—H2 | 114 (3) | C11—C12—H12 | 120.0 |
| S1—N2—H2 | 108 (3) | C13—C12—H12 | 120.0 |
| C16—N3—O4 | 105.9 (3) | C12—C13—C8 | 119.7 (3) |
| C6—C1—C7 | 119.0 (3) | C12—C13—H13 | 120.2 |
| C6—C1—C2 | 119.9 (3) | C8—C13—H13 | 120.2 |
| C7—C1—C2 | 121.0 (3) | C15—C14—O4 | 111.2 (3) |
| O1—C2—C3 | 121.4 (3) | C15—C14—N2 | 132.6 (3) |
| O1—C2—C1 | 121.2 (3) | O4—C14—N2 | 116.1 (3) |
| C3—C2—C1 | 117.4 (3) | C14—C15—C16 | 103.8 (3) |
| C4—C3—C2 | 121.5 (3) | C14—C15—C17 | 129.1 (3) |
| C4—C3—H3 | 119.3 | C16—C15—C17 | 127.2 (3) |
| C2—C3—H3 | 119.3 | N3—C16—C15 | 112.1 (3) |
| C3—C4—C5 | 120.1 (3) | N3—C16—C18 | 122.1 (3) |
| C3—C4—H4 | 120.0 | C15—C16—C18 | 125.8 (3) |
| C5—C4—H4 | 120.0 | C15—C17—H17A | 109.5 |
| C6—C5—C4 | 120.7 (3) | C15—C17—H17B | 109.5 |
| C6—C5—Br1 | 120.2 (3) | H17A—C17—H17B | 109.5 |
| C4—C5—Br1 | 119.0 (3) | C15—C17—H17C | 109.5 |
| C5—C6—C1 | 120.3 (3) | H17A—C17—H17C | 109.5 |
| C5—C6—H6 | 119.8 | H17B—C17—H17C | 109.5 |
| C1—C6—H6 | 119.8 | C16—C18—H18A | 109.5 |
| N1—C7—C1 | 122.4 (3) | C16—C18—H18B | 109.5 |
| N1—C7—H7 | 118.8 | H18A—C18—H18B | 109.5 |
| C1—C7—H7 | 118.8 | C16—C18—H18C | 109.5 |
| C13—C8—C9 | 120.1 (3) | H18A—C18—H18C | 109.5 |
| C13—C8—N1 | 123.3 (3) | H18B—C18—H18C | 109.5 |
| O3—S1—N2—C14 | 56.9 (3) | C9—C10—C11—S1 | 176.4 (2) |
| O2—S1—N2—C14 | −173.4 (2) | O3—S1—C11—C12 | −29.4 (3) |
| C11—S1—N2—C14 | −58.5 (3) | O2—S1—C11—C12 | −162.7 (3) |
| C14—O4—N3—C16 | 0.5 (4) | N2—S1—C11—C12 | 85.3 (3) |
| C6—C1—C2—O1 | −178.9 (3) | O3—S1—C11—C10 | 154.8 (3) |
| C7—C1—C2—O1 | −1.4 (5) | O2—S1—C11—C10 | 21.5 (3) |
| C6—C1—C2—C3 | 1.8 (5) | N2—S1—C11—C10 | −90.5 (3) |
| C7—C1—C2—C3 | 179.3 (3) | C10—C11—C12—C13 | 0.2 (5) |
| O1—C2—C3—C4 | 178.3 (3) | S1—C11—C12—C13 | −175.5 (3) |
| C1—C2—C3—C4 | −2.3 (5) | C11—C12—C13—C8 | −0.9 (5) |
| C2—C3—C4—C5 | 1.0 (5) | C9—C8—C13—C12 | 0.7 (5) |
| C3—C4—C5—C6 | 0.9 (5) | N1—C8—C13—C12 | −178.7 (3) |
| C3—C4—C5—Br1 | −177.9 (3) | N3—O4—C14—C15 | −0.2 (4) |
| C4—C5—C6—C1 | −1.4 (5) | N3—O4—C14—N2 | −176.6 (3) |
| Br1—C5—C6—C1 | 177.4 (2) | S1—N2—C14—C15 | 105.7 (4) |
| C7—C1—C6—C5 | −177.5 (3) | S1—N2—C14—O4 | −78.9 (3) |
| C2—C1—C6—C5 | 0.0 (5) | O4—C14—C15—C16 | −0.1 (4) |
| C8—N1—C7—C1 | −178.3 (3) | N2—C14—C15—C16 | 175.5 (4) |
| C6—C1—C7—N1 | 178.1 (3) | O4—C14—C15—C17 | 179.7 (4) |
| C2—C1—C7—N1 | 0.6 (5) | N2—C14—C15—C17 | −4.7 (7) |
| C7—N1—C8—C13 | 4.9 (5) | O4—N3—C16—C15 | −0.6 (5) |
| C7—N1—C8—C9 | −174.5 (3) | O4—N3—C16—C18 | −180.0 (4) |
| C13—C8—C9—C10 | 0.1 (5) | C14—C15—C16—N3 | 0.5 (5) |
| N1—C8—C9—C10 | 179.6 (3) | C17—C15—C16—N3 | −179.4 (4) |
| C8—C9—C10—C11 | −0.8 (5) | C14—C15—C16—C18 | 179.8 (5) |
| C9—C10—C11—C12 | 0.6 (5) | C17—C15—C16—C18 | 0.0 (7) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1 | 0.80 (3) | 1.91 (3) | 2.577 (4) | 141 (3) |
| N2—H2···O1i | 0.75 (3) | 2.09 | 2.828 (4) | 171 (4) |
| C17—H17C···O1i | 0.96 | 2.58 | 3.248 (5) | 126 |
| C7—H7···N3ii | 0.93 | 2.53 | 3.420 (4) | 161 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1/2, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BQ2069).
References
- Bruker (2005). SADABS Bruker AXS Inc. Madison, Wisconsion, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc. Madison, Wisconsion, USA.
- Chohan, Z. H., Tahir, M. N., Shad, H. A. & Khan, I. U. (2008). Acta Cryst. E64, o648. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Hämäläinen, R., Lehtinen, M. & Turpeinen, U. (1986). Arch. Pharm.319, 415–420.
- Shad, H. A., Chohan, Z. H., Tahir, M. N. & Khan, I. U. (2008). Acta Cryst. E64, o635. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680800682X/bq2069sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800682X/bq2069Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report