Abstract
Facile modification of oligodeoxyribonucleotides is required for efficient immobilization to a pre-activated glass surface. This report presents an oligodeoxyribonucleotide which contains a hairpin stem–loop structure with multiple phosphorothioate moieties in the loop. These moieties are used to anchor the oligo to glass slides that are pre-activated with bromoacetamidopropylsilane. The efficiency of the attachment reaction was improved by increasing the number of phosphorothioates in the loop, as shown in the remarkable enhancement of template hybridization and single base extension through catalysis by DNA polymerase. The loop and stem presumably serve as lateral spacers between neighboring oligodeoxyribonucleotides and as a linker arm between the glass surface and the single-stranded sequence of interest. The oligodeoxyribonucleotides of this hairpin stem–loop architecture with multiple phosphorothioate moieties have broad application in DNA chip-based gene analysis.
INTRODUCTION
The success of microarray technology in the analysis of genes and gene expression is highly dependent on the ability to deposit and bind DNA, DNA fragments and oligodeoxyribonucleotides to the surface of a glass microscope slide with accuracy, reproducibility and the assurance that the bound entities will survive both the attachment reaction and subsequent analysis. We have made a major effort to develop attachment chemistries that are not only compatible with the analysis of interest, but also enhance it. The efficiency of hybridization between immobilized oligonucleotide primers and a mobile DNA template depends on the length, density and topology of the primers on the surface (1). A linker arm between the glass surface and the terminus of a primer also contributes to hybridization (2,3). However, the structural requirements of DNA probes for enzyme-dependent processes have not been reported.
An array of primers with free 3′-ends that hybridize to a target of interest and are extended with single dideoxyribonucleotide triphosphates (ddNTP) through catalysis by a DNA polymerase would reveal the identities of the bases in question (4). Arrayed primer extension (APEX) on a chip is a medium to high throughput method for resequencing or for screening specific single nucleotide polymorphisms by enzymatic incorporation of a labeled dideoxyribonucleotide triphosphate on the 3′-OH of primers immobilized on a chip at their 5′-ends (4). In this study we have examined the utility of a hairpin stem structure scaffold with multiple phosphorothioates (Fig. 1) in the primer oligonucleotides for APEX. We found that the enzymatic incorporation of a cyanine dye-labeled ddNTP to a hairpin structured primer (Fig. 2) was eight times more efficient than the corresponding linear structured primer.
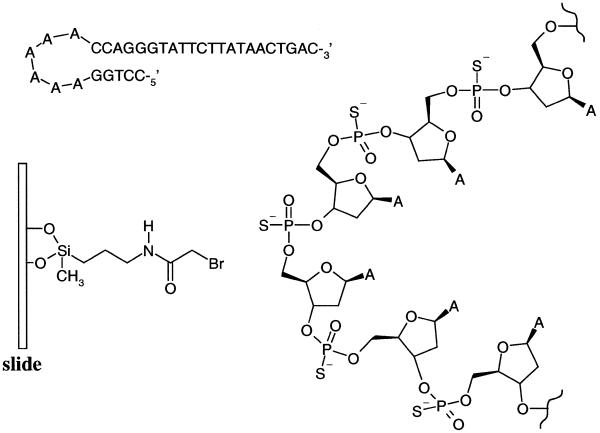
Figure 1.
The hairpin structure primer has five phosphorothioates in the loop comprising six adenosines. The surface of the glass slide is coated with bromoacetamidopropylsilane.
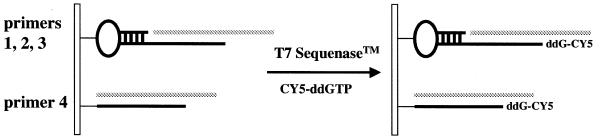
Figure 2.
Arrayed primer extension on a glass slide.
MATERIALS AND METHODS
All chemicals and solvents were purchased from Fluka unless noted otherwise. Oligodeoxyribonucleotides were purchased from Sigma Genosys. Cyanine-labeled dideoxyribonucleotide triphosphates (Cy5-ddNTP) (Fig. 3) and T7 Sequenase™ were obtained from Amersham Pharmacia Biotech. Glass microscope slides were purchased from VWR Scientific.
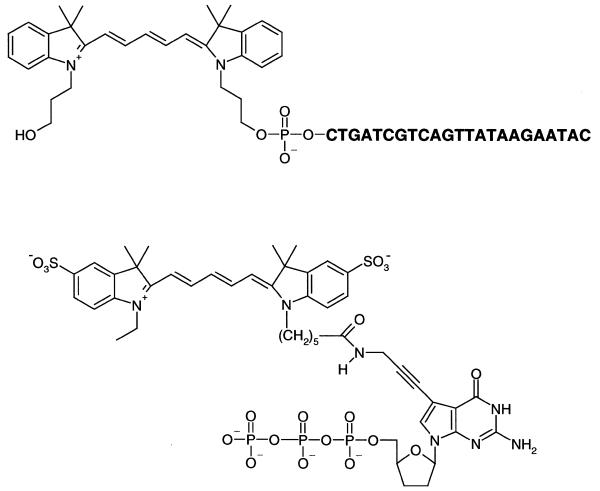
Figure 3.
Cy5-labeled template (top) and ddGTP (bottom) used in this study.
Preparation of slides
The 25 × 75 mm2 microscope slides were washed sequentially with acetone, 1 M NaOH and ethanol and immersed for 3 min with sonication in 95% ethanol containing 2% (w/v) 3-aminopropylmethyldiethoxysilane. The slides were rinsed with ethanol and cured in an oven at 75°C for 4 h. The cured slides were placed on a glass rack and immersed in 160 ml of N,N-dimethylformamide containing 0.45 g (3.3 mmol) bromoacetic acid, 0.04 g (0.3 mmol) 4-(dimethylamino)-pyridine and 0.66 g (3.2 mmol) 1,3-dicyclohexylcarbodiimide. The reaction mixture was stirred in the dark for 2 h. The slides were then washed with ethanol and dried in an oven at 75°C.
Preparation of arrayed primers on slides
The oligodeoxyribonucleotide primers were dissolved and stored in 1× TE (10 mM Tris–HCl, 1 mM EDTA, pH 7.0) and the concentrations were calculated based on spectrophotometric measurement. The primer sequences used in this study were d(CCTGGASASASASASACCAGGGTATTCTTATAACTGAC), d(CCTGGASASACCAGGGTATTCTTATAACTGAC), d(CCTGGASACCAGGGTATTCTTATAACTGAC) and d(pSGTATTCTTATAACTGAC). The superscript S denotes the position of the phosphorothioate in the sequence of oligonucleotides. The different concentrations of oligonucleotides were dispensed onto bromoacetamidosilane-coated slides by a Gen III spotter (Amersham Pharmacia Biotech–Molecular Dynamics). The volume of the spots was ∼0.7 nl and the size ∼150 µm diameter. The slides were washed with boiling MilliQ-filtered water before hybridization and primer extension.
Hybridization
A 5′-labeled complementary strand Cy5-d(CTGATCGTCAGTTATAAGAATAC) (Fig. 3) was dissolved in 5× SSC buffer (750 mM NaCl, 75 mM sodium citrate, pH 7.0) to make a 1 µM solution (50 µl), which was applied to the slide containing the immobilized primers. After incubation at 25°C for 60 min the slide was washed with 2× SSC buffer (300 mM NaCl, 30 mM sodium citrate) and 10 mM triethanolamine hydrochloride, pH 7.0. Images of template-labeled spots were obtained on an Avalanche microscanner (Amersham Pharmacia Biotech–Molecular Dynamics) by measuring emission at 670 nm with excitation at 633 nm.
Primer extension
Primer extension was carried out in 50 µl of T7 Sequenase buffer (10 mM Tris–HCl pH 7.5, 2 mM dithiothreitol, 1 mM ATP) with 1 µM dye terminator Cy5-ddGTP, 1 µM templates, and 10 U T7 Sequenase™ for 10 min at 25°C. The template is the 23mer d(CTGATCGTCAGTTATAAGAATAC). The reaction mixture was then washed with boiling water. Images of the dye terminator-labeled spots were obtained on an Avalanche Gen III microscanner by measuring emission at 670 nm with excitation at 633 nm.
RESULTS AND DISCUSSION
Thiophosphorylation of oligonucleotides
Covalent attachment of oligonucleotides to the surface of glass slides has the advantage of conferring substantial resistance to washing in pursuit of background reduction. Oligonucleotides have been directly synthesized on a planar glass surface in a 3′→5′ direction (5,6). Since oligonucleotide synthesis in the reverse direction is more problematical (7–9), the covalent immobilization of pre-synthesized oligonucleotides to a surface in the 5′→3′ direction is commonly used.
To make DNA chips using the spotting technique, the attachment kinetics must be extremely fast because the sub-nanoliter solution at each spot evaporates quickly during spotting. Pirrung et al. (10) have reported an elegant method of attaching oligonucleotides with a 5′-terminal thiophosphate to bromoacetamidosilane-coated glass slides by adapting the work of Letsinger et al. (11,12) used for chemical ligation of thiophosphorylated and bromoacetylated oligonucleotides. Phosphorothioates do not form dimers via formation of an intermolecular –S–S– bond (13) and function as nucleophiles without pre-reduction, as in the case of disulfides.
The automated synthesis of oligonucleotides having phosphorothioate moieties in the backbone is simpler than phosphorothioate modification at the 5′-terminus because the incorporation of internucleotide phosphorothioates involves replacing an oxidation reagent with a sulfurization reagent, e.g. Beaucage reagent (14), after each coupling of phosphoramidite in solid phase synthesis. Although precedent exists for the reaction of backbone phosphorothioates with azidophenylacylbromide in the preparation of photocrosslinkers to study DNA–protein interactions (15), such phosphorothioates have not yet been used as anchors for attaching oligonucleotides to a glass surface. Herein we demonstrate that an oligonucleotide modified with multiple phosphorothioates in its backbone can undergo efficient anchorage to a glass surface by reaction with a bromoacetyl functionality.
Primer of hairpin structure
Primers 1, 2 and 3 were spotted onto the surface of glass slides pre-activated by bromoacetamidopropylmethyldiethoxysilane at concentrations ranging from 0.25 to 32 µM. A Cy5-labeled template was hybridized to the primers and the fluorescent signals were compared (Fig. 4). By increasing the number of phosphorothioate moieties in the loop, the attachment efficiency increased, as measured by the hybridization signal. The hybridization signal on primer 1 (which has five phosphorothioates) is ∼4-fold stronger than that of primer 2 (which has two phosphorothioates) and 7-fold stronger than that of primer 3 (which has a single phosphorothioate in the loop). This may be explained by an increased reaction probability of phosphorothioates during the attachment process with an increasing number of reactive sites. A clean silicon dioxide surface is believed to have ∼5 × 1012 silanols/mm2 (16). If most of the silanols are converted to bromoacetyl groups, then the number of bromoacetyl groups would greatly exceed the maximum number of oligonucleotides that can be covalently attached to the surface (∼1 × 1011 molecules/mm2) (17). With the assumption that phosphorothioates in the loop have equal reactivity towards bromoacetyl on the surface, the oligonucleotide would have multiple opportunities for attachment.
Figure 4.
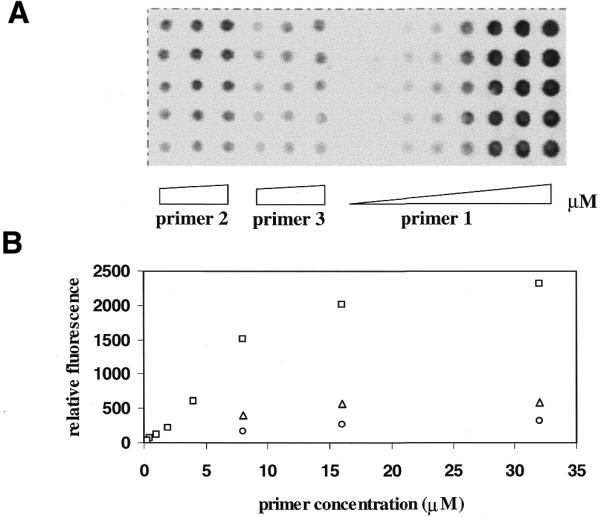
(A) An image of hybridization of primer 1 at increasing concentrations of 0.25, 0.5, 1, 2, 4, 8, 16 and 32 µM and primers 2 and 3 at increasing concentrations of 8, 16 and 32 µM to excess Cy5-labeled complementary strand. (B) A plot of relative fluorescence intensity of the hybridized template versus concentration of primers. Each data point is the average of the integral of five spots. Open squares, primer 1; open triangles, primer 2; open circles, primer 3.
The primers, which were attached to slides at their loops, were extended with a single dye-labeled terminator, Cy5-ddGTP, by T7 DNA Sequenase™. The incorporation of Cy5-ddGTP on the primer increased with primer concentration until a plateau in signal intensity was reached. Interestingly, we observed that the number of phosphorothioate moieties also correlated with the signal strength of primer extension (Fig. 5), with five phosphorothioates performing best (primer 1 > primer 2 > primer 3). In addition, the signal intensity of primer extension was not found to change with the location of a single phosphorothioate in the loop (data not shown), suggesting that every phosphorothioate in the loop might be equally reactive.
Figure 5.
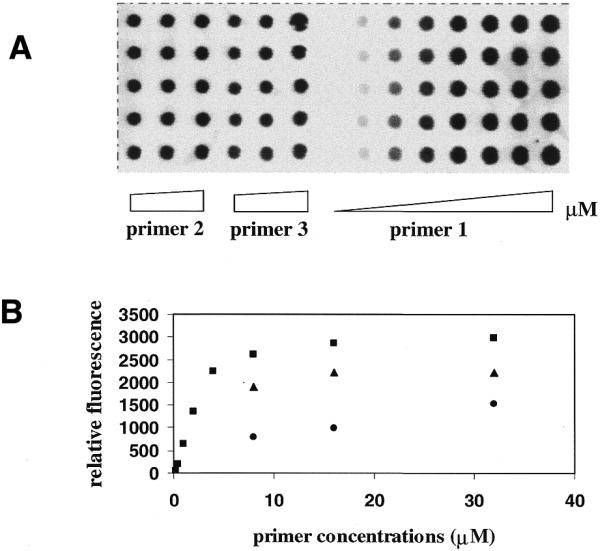
(A) An image of primer extension on primer 1 at increasing concentrations of 0.25, 0.5, 1, 2, 4, 8, 16 and 32 µM and on primers 2 and 3 at increasing concentrations of 8, 16 and 32 µM. (B) A plot of relative fluorescence intensity of the Cy5-ddGTP-labeled primers versus concentration. Each data point is the average of the integral of five spots. Filled squares, primer 1; filled triangles, primer 2; filled circles, primer 3.
Hairpin versus linear structure
We compared hybridization and primer extension of the hairpin oligonucleotide 1 with multiple phosphorothioates in the loop and the 5′-thiophosphorylated oligonucleotide 4. At each dispensing concentration, a comparable amount of the Cy5-labeled complementary strand was revealed on the immobilized primers (Fig. 6), suggesting that approximately the same amount of primers and presumably similar attachment yields were achieved for primers 1 and 4. Cy5, labeled at the 5′-terminus of the template, was expected to overhang from the double-stranded region between primer and template and not to affect hybridization. The hybridization profile reached saturation and did not increase above 5 µM concentration, suggesting that the density of primer approached maximum capacity.
Figure 6.
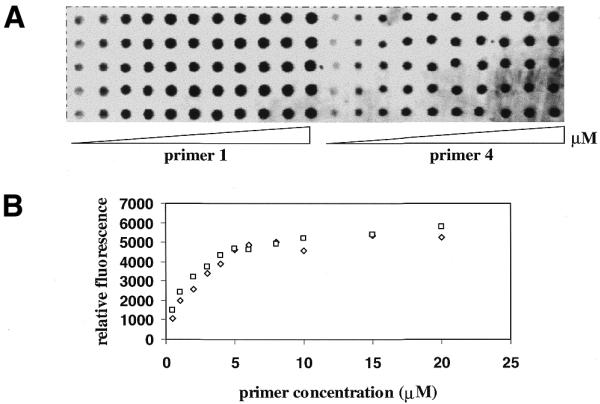
(A) An image of hybridization of primers 1 and 4 at increasing concentrations of 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 15 and 20 µM to excess of Cy5-labeled complementary strand. (B) A plot of relative fluorescence intensity of the hybridized template versus concentration of primers. Each data point is the average of the integral of five spots. Open squares, primer 1; open diamonds, primer 4.
For primer extension incorporation of Cy5-ddGTP on primer 1 increased with increasing primer concentration and reached a plateau at >4 µM (Fig. 7). In contrast, the signal of labeled primer 4 increased with increasing concentration from 0.5 to 4 µM. After peaking the signal intensity started to decrease with increasing concentration from 5 to 20 µM. At the maximum intensity the signal of primer 1 was found to be eight times stronger than that of primer 4.
Figure 7.
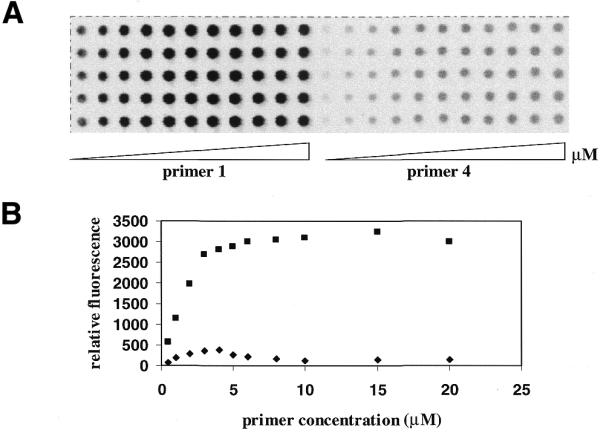
(A) An image of primer extension on primers 1 and 4 at increasing concentrations of 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 15 and 20 µM. The quantum yields of fluorescence emission of Cy5-labeled template is 3.3 times that of Cy5-ddGTP in aqueous solution. (B) A plot of relative fluorescence intensity of the Cy5-ddGTP-labeled primers versus concentration. Each data point is the average of the integral of five spots. Filled squares, primer 1; filled diamonds, primer 4.
The difference in hybridization signal for primers 1 and 4 is negligible at each concentration. This suggests that comparable amounts of primers were covalently attached for spots of equivalent concentration. The higher attachment yield of primer 4 over 2 and 3 might be related to its less congested single-stranded form. However, the incorporation of Cy5-ddGTP on primer 1 is superior to primer 4 in spite of the similar hybridization signal. This could be due to the weak electrostatic repulsion of single-stranded DNA and possibly a tethered coil orientation of the linear primer on the surface (1). A densely populated substrate may not hinder hybridization, but may encumber the participation of enzymes (18). The hairpin stem of primer 1 could function as a lateral spacer to accommodate primer extension following hybridization, whereas the randomly coiled primer 4 may limit the accessibility of enzymes. The loop and stem region of primer 1 could also serve as a linker arm so that the 3′-terminus of the primer, namely the enzyme-binding site on the primer, is further distant from the glass. This may well be favorable for enzymatic reactions.
In summary, the attachment method described is an efficient way to immobilize oligonucleotides through covalent bonds at a desirable density and orientation. Since both the 3′- and 5′-ends are not modified, they can be labeled with various reporters, for such purposes as preparing molecular beacons (19). The attachment of hairpin oligonucleotides may offer the advantage of immobilizing cDNA or any PCR products to microchips by ligation following hybridization. The versatility of using such oligonucleotides is also complemented by their low cost of synthesis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Anthony Murray and Carl Fuller for discussions and critical reading of the manuscript. We are also grateful for financial support from a NIST/ATP award (70NANBSH1112) and Amersham Pharmacia Biotech Inc.
References
- 1.Levicky R., Herne,T.M., Tarlov,M.J. and Satija,S.K. (1998) Using self-assembly to control the structure of DNA monolayers on gold: a neutron reflectivity study. J. Am. Chem. Soc., 120, 9787–9792. [Google Scholar]
- 2.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Z., Guilfoyle,R.A., Thiel,A.J, Wang,R. and Smith,L.M. (1994) Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res., 22, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastinen T., Kurg,A., Metsapalu,A., Peltonen,L. and Syvanen,A.C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 5.Fodor S.P.A., Read,J.L., Pirrung,M.C., Stryer,L., Lu,A.T. and Solas,D. (1991) Light-directed, spatially addressable parallel chemical synthesis. Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 6.Southern E.M., Maskos,U. and Elder,J.K. (1992) Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 7.Pirrung M.C., Fallon,L., Lever,D.C. and Shuey,S.W. (1996) Inverse phosphotriester DNA synthesis using photochemically-removable dimethoxybenzoin phosphate protecting groups. J. Org. Chem., 61, 2129–2136. [Google Scholar]
- 8.Pirrung M.C., Fallon,L. and McGall,G. (1998) Proofing of photolithographic DNA synthesis with 3′,5′-dimethoxybenzoinyl-oxycarbonyl-protected deoxynucleoside phosphoramidites. J. Org. Chem., 63, 241–246. [Google Scholar]
- 9.Kwiatkowski M., Fredriksson,S., Lsaksson,A., Nilsson,M. and Landegren,U. (1999) Inversion of in situ synthesized oligonucleotides: improved reagents for hybridization and primer extension in DNA microarrays. Nucleic Acids Res., 24, 4710–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirrung M.C., Davis,J.D. and Odenbaugh,A.L. (2000) Novel reagents and procedures for immobilization of DNA on glass microchips for primer extension. Langmuir, 16, 2185–2191. [Google Scholar]
- 11.Gryaznov S.M. and Letsinger,R.L. (1993) Chemical ligation of oligonucleotides in the presence and absence of a template. J. Am. Chem. Soc., 115, 3808–3809. [Google Scholar]
- 12.Herrlein M.K. and Letsinger,R.L. (1994) Selective chemical autoligation on a double-stranded DNA template. Nucleic Acids Res., 22, 5076–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassignol M. and Thoung,N.T. (1998) Phosphodisulfide bond: a new linker for the oligonucleotide conjugation. Tetrahedron Lett., 39, 8271–8274. [Google Scholar]
- 14.Iyer R.P., Egan,W., Regan,J.B. and Beaucage,S.L. (1990) 3H-1,2-benzodithiole-3-one 1,1-dioxide as an improved sulfurizing reagent in the solid-phase synthesis of oligodeoxyribonucleoside phosphorothioates. J. Am. Chem. Soc., 112, 1253–1254. [Google Scholar]
- 15.Yang S.W. and Nash,H.A. (1994) Specific photocrosslinking of DNA–protein complexes: identification of contacts between integration host factor and its target DNA. Proc. Natl Acad. Sci. USA, 91, 12183–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuravlev L.T. (1987) Concentration of hydroxyl groups on the surface of amorphous silicas. Langmuir, 3, 316–318. [Google Scholar]
- 17.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 18.Mrksich M. and Houseman,B.T. (1999) The role of ligand density in the enzymatic glycosylation of carbohydrates presented on self-assembled monolayers of alkanethiolates on gold. Angew. Chem. Int. Ed. Engl., 38, 782–785. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]