Abstract
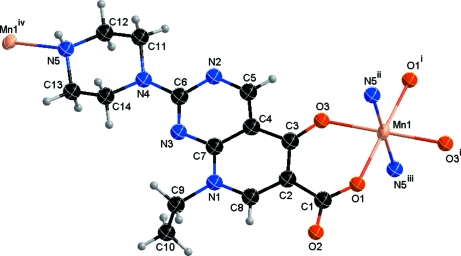
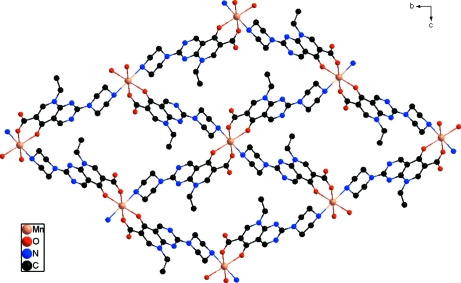
In the title compound, {[Mn(C14H16N5O3)2]·2H2O}n, the MnII atom (site symmetry  ) exhibits a distorted trans-MnN2O4 octahedral geometry defined by two monodentate N-bonded and two bidentate O,O′-bonded 8-ethyl-5-oxo-2-(piperazin-1-yl)-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxylate anions. An N—H⋯O hydrogen bond is present in the crystal structure. The extended two-dimensional structure is a square grid and the disordered uncoordinated water molecules occupy cavities within the grid.
) exhibits a distorted trans-MnN2O4 octahedral geometry defined by two monodentate N-bonded and two bidentate O,O′-bonded 8-ethyl-5-oxo-2-(piperazin-1-yl)-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxylate anions. An N—H⋯O hydrogen bond is present in the crystal structure. The extended two-dimensional structure is a square grid and the disordered uncoordinated water molecules occupy cavities within the grid.
Related literature
For background, see: Mizuki et al. (1996 ▶).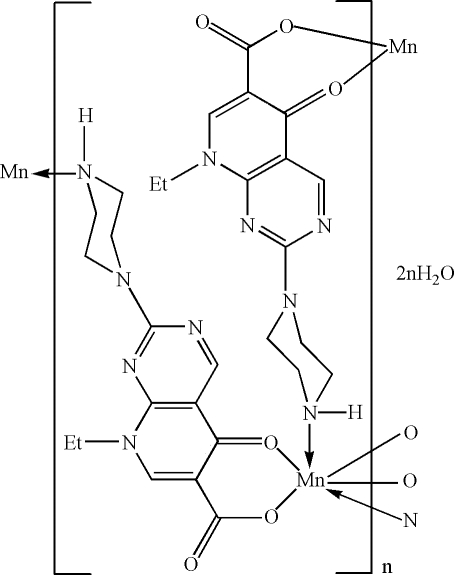
Experimental
Crystal data
[Mn(C14H16N5O3)2]·2H2O
M r = 695.58
Monoclinic,
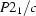
a = 6.0422 (2) Å
b = 21.5673 (8) Å
c = 12.7395 (5) Å
β = 99.617 (1)°
V = 1636.8 (1) Å3
Z = 2
Mo Kα radiation
μ = 0.47 mm−1
T = 295 (2) K
0.34 × 0.26 × 0.18 mm
Data collection
Bruker SMART CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 1998 ▶) T min = 0.861, T max = 0.910
10030 measured reflections
3938 independent reflections
3465 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.060
wR(F 2) = 0.191
S = 1.10
3938 reflections
227 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.97 e Å−3
Δρmin = −0.48 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT-Plus (Bruker, 1998 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808006533/hb2703sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808006533/hb2703Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Mn1—O1 | 2.106 (2) |
| Mn1—O3 | 2.1667 (16) |
| Mn1—N5i | 2.372 (2) |
Symmetry code: (i) 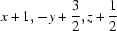 .
.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N5—H5N⋯O2ii | 0.893 (10) | 2.268 (12) | 3.149 (3) | 169 (3) |
Symmetry code: (ii) 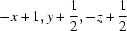 .
.
Acknowledgments
The authors thank the Innovation Science Foundation of Harbin Medical University for financial support (grant No. 060041).
supplementary crystallographic information
Comment
Pipemidic acid (Hppa, C14H17N5O3, 8-Ethyl-5,8-dihydro-5-oxo-2- (1-piperazinyl)-pyrido(2,3 - d)-pyrimidine-6-carboxylic acid) is member of a class of quinolones used to treat infections (Mizuki et al., 1996). The metal complexes of the ppa anion have not been reported; the title manganese(II) complex, (I), is reported here (Fig. 1).
The MnII atom in (I) with site symmetry 1 is coordinated by four oxygen atoms and two N atoms from four ppa ligands (two monodentate-N and two O,O-bidentate) (Table 1) to form a square grid propagating in (Fig. 2). An N—H···O hydrogen bond (Table 2) helps to stabilize this arrangement.
The disordered, uncoordinated, water molecules occupy cavities within the grid. In the present study, their attached H atoms could not be located.
Experimental
A mixture of Mn(CH3COO)2.4H2O (0.061 g, 0.25 mmol), Hppa (0.15 g, 0.5 mmol), sodium hydroxide (0.04 g, 1 mmol) and water (12 ml) was stirred for 30 min in air. The mixture was then transferred to a 23 ml Teflon-lined hydrothermal bomb. The bomb was kept at 433 K for 72 h under autogenous pressure. Upon cooling, colourless prisms of (I) were obtained from the reaction mixture.
Refinement
The carbon-bound H atoms were positioned geometrically (C—H = 0.93–0.97 Å) and refined as riding with Uiso(H) = 1.2Ueq(C). The N-bound H atom was located in a difference map and its position was freely refined with Uiso(H) = 1.2Ueq(N).
The water H atoms could not be placed due to disorder.
Figures
Fig. 1.
The asymmetric unit of (I) showing the showing 50% displacement ellipsoids (water molecule O atoms have been omitted for clarity).
Fig. 2.
A view of part of a two-dimensional polymeric sheet in (I) showing the square-grid connectivity (H atoms and water molecule O atoms omitted for clarity).
Crystal data
| [Mn(C14H16N5O3)2]·2H2O | F000 = 718.0 |
| Mr = 695.58 | Dx = 1.399 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4747 reflections |
| a = 6.0422 (2) Å | θ = 2.5–28.3º |
| b = 21.5673 (8) Å | µ = 0.47 mm−1 |
| c = 12.7395 (5) Å | T = 295 (2) K |
| β = 99.617 (1)º | Prism, colorless |
| V = 1636.8 (1) Å3 | 0.34 × 0.26 × 0.18 mm |
| Z = 2 |
Data collection
| Bruker SMART CCD diffractometer | 3938 independent reflections |
| Radiation source: fine-focus sealed tube | 3465 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.019 |
| T = 295(2) K | θmax = 28.3º |
| ω scans | θmin = 2.5º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −6→8 |
| Tmin = 0.861, Tmax = 0.910 | k = −27→28 |
| 10030 measured reflections | l = −15→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.061 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.191 | w = 1/[σ2(Fo2) + (0.1066P)2 + 1.0983P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.11 | (Δ/σ)max < 0.001 |
| 3938 reflections | Δρmax = 0.98 e Å−3 |
| 227 parameters | Δρmin = −0.48 e Å−3 |
| 1 restraint | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Mn1 | 0.5000 | 0.5000 | 0.5000 | 0.02831 (19) | |
| O1W | 0.388 (4) | 0.5235 (7) | 0.9613 (10) | 0.244 (9) | 0.50 |
| O1 | 0.7068 (3) | 0.49893 (7) | 0.38228 (16) | 0.0341 (4) | |
| O2W | −0.081 (2) | 0.4426 (8) | 0.9246 (7) | 0.194 (6) | 0.50 |
| O2 | 0.8721 (5) | 0.51863 (13) | 0.2451 (2) | 0.0717 (8) | |
| O3 | 0.3574 (3) | 0.58106 (8) | 0.41354 (14) | 0.0378 (4) | |
| N1 | 0.5078 (5) | 0.67158 (12) | 0.1504 (2) | 0.0533 (7) | |
| N2 | −0.0007 (4) | 0.73723 (11) | 0.2985 (2) | 0.0464 (6) | |
| N3 | 0.2395 (4) | 0.74681 (10) | 0.16596 (18) | 0.0432 (5) | |
| N4 | −0.0102 (4) | 0.82368 (10) | 0.19027 (19) | 0.0384 (5) | |
| N5 | −0.2339 (3) | 0.93726 (9) | 0.11005 (17) | 0.0322 (4) | |
| H5N | −0.140 (4) | 0.9649 (12) | 0.146 (2) | 0.048* | |
| C1 | 0.7282 (4) | 0.52991 (11) | 0.3018 (2) | 0.0358 (5) | |
| C2 | 0.5782 (4) | 0.58537 (11) | 0.2731 (2) | 0.0354 (5) | |
| C3 | 0.4044 (4) | 0.60559 (10) | 0.33118 (19) | 0.0313 (5) | |
| C4 | 0.2852 (4) | 0.66087 (11) | 0.28824 (19) | 0.0336 (5) | |
| C5 | 0.1042 (5) | 0.68542 (12) | 0.3304 (2) | 0.0425 (6) | |
| H5A | 0.0546 | 0.6635 | 0.3848 | 0.051* | |
| C6 | 0.0805 (4) | 0.76813 (12) | 0.2189 (2) | 0.0360 (5) | |
| C7 | 0.3383 (5) | 0.69362 (12) | 0.2010 (2) | 0.0395 (6) | |
| C8 | 0.6170 (5) | 0.61896 (14) | 0.1875 (2) | 0.0496 (7) | |
| H8A | 0.7282 | 0.6046 | 0.1512 | 0.059* | |
| C9 | 0.5717 (8) | 0.7043 (2) | 0.0566 (3) | 0.0725 (12) | |
| H9A | 0.5531 | 0.7486 | 0.0645 | 0.087* | |
| H9B | 0.7285 | 0.6963 | 0.0537 | 0.087* | |
| C10 | 0.4373 (10) | 0.6840 (4) | −0.0400 (5) | 0.116 (2) | |
| H10A | 0.4583 | 0.6403 | −0.0488 | 0.174* | |
| H10B | 0.4811 | 0.7059 | −0.0989 | 0.174* | |
| H10C | 0.2822 | 0.6923 | −0.0375 | 0.174* | |
| C11 | −0.1446 (6) | 0.85820 (14) | 0.2560 (2) | 0.0499 (7) | |
| H11A | −0.0477 | 0.8852 | 0.3045 | 0.060* | |
| H11B | −0.2170 | 0.8295 | 0.2980 | 0.060* | |
| C12 | −0.3226 (5) | 0.89702 (13) | 0.1855 (2) | 0.0433 (6) | |
| H12A | −0.4326 | 0.8693 | 0.1460 | 0.052* | |
| H12B | −0.3996 | 0.9225 | 0.2308 | 0.052* | |
| C13 | −0.1018 (4) | 0.89946 (12) | 0.0466 (2) | 0.0372 (5) | |
| H13A | −0.0347 | 0.9264 | −0.0003 | 0.045* | |
| H13B | −0.2015 | 0.8710 | 0.0025 | 0.045* | |
| C14 | 0.0812 (4) | 0.86281 (12) | 0.1146 (2) | 0.0382 (6) | |
| H14A | 0.1573 | 0.8371 | 0.0693 | 0.046* | |
| H14B | 0.1906 | 0.8911 | 0.1530 | 0.046* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0350 (3) | 0.0187 (3) | 0.0304 (3) | −0.00041 (16) | 0.0030 (2) | 0.00255 (16) |
| O1W | 0.47 (3) | 0.155 (12) | 0.111 (9) | 0.000 (15) | 0.058 (14) | −0.057 (9) |
| O1 | 0.0407 (10) | 0.0237 (9) | 0.0383 (10) | 0.0041 (6) | 0.0073 (8) | 0.0040 (6) |
| O2W | 0.203 (11) | 0.307 (17) | 0.088 (6) | −0.110 (12) | 0.076 (7) | −0.028 (8) |
| O2 | 0.0854 (18) | 0.0730 (16) | 0.0674 (16) | 0.0491 (15) | 0.0441 (14) | 0.0338 (13) |
| O3 | 0.0448 (10) | 0.0297 (9) | 0.0403 (9) | 0.0085 (7) | 0.0110 (7) | 0.0122 (7) |
| N1 | 0.0730 (17) | 0.0433 (13) | 0.0500 (14) | 0.0245 (12) | 0.0294 (12) | 0.0196 (11) |
| N2 | 0.0498 (13) | 0.0386 (12) | 0.0550 (14) | 0.0152 (10) | 0.0211 (11) | 0.0213 (11) |
| N3 | 0.0549 (13) | 0.0339 (11) | 0.0434 (12) | 0.0165 (10) | 0.0157 (10) | 0.0150 (9) |
| N4 | 0.0418 (11) | 0.0301 (10) | 0.0458 (12) | 0.0107 (9) | 0.0146 (9) | 0.0133 (9) |
| N5 | 0.0344 (10) | 0.0231 (9) | 0.0382 (10) | 0.0029 (7) | 0.0034 (8) | 0.0012 (8) |
| C1 | 0.0410 (13) | 0.0311 (12) | 0.0355 (12) | 0.0080 (10) | 0.0071 (10) | 0.0029 (9) |
| C2 | 0.0429 (13) | 0.0281 (11) | 0.0360 (12) | 0.0084 (9) | 0.0092 (10) | 0.0040 (9) |
| C3 | 0.0367 (11) | 0.0227 (10) | 0.0337 (11) | 0.0032 (9) | 0.0033 (9) | 0.0042 (8) |
| C4 | 0.0396 (12) | 0.0265 (11) | 0.0352 (12) | 0.0060 (9) | 0.0073 (9) | 0.0067 (9) |
| C5 | 0.0491 (15) | 0.0339 (13) | 0.0475 (15) | 0.0103 (11) | 0.0167 (12) | 0.0168 (11) |
| C6 | 0.0377 (12) | 0.0307 (12) | 0.0398 (13) | 0.0066 (9) | 0.0066 (10) | 0.0090 (10) |
| C7 | 0.0506 (14) | 0.0313 (12) | 0.0386 (13) | 0.0109 (11) | 0.0132 (11) | 0.0082 (10) |
| C8 | 0.0609 (17) | 0.0438 (15) | 0.0485 (16) | 0.0220 (13) | 0.0219 (13) | 0.0120 (12) |
| C9 | 0.091 (3) | 0.071 (2) | 0.065 (2) | 0.035 (2) | 0.041 (2) | 0.0293 (19) |
| C10 | 0.111 (4) | 0.154 (6) | 0.085 (4) | 0.032 (4) | 0.022 (3) | 0.021 (4) |
| C11 | 0.0604 (17) | 0.0465 (15) | 0.0478 (15) | 0.0251 (14) | 0.0234 (13) | 0.0176 (13) |
| C12 | 0.0427 (13) | 0.0368 (13) | 0.0535 (16) | 0.0129 (11) | 0.0169 (12) | 0.0133 (12) |
| C13 | 0.0394 (13) | 0.0308 (12) | 0.0414 (13) | 0.0087 (10) | 0.0071 (10) | 0.0078 (10) |
| C14 | 0.0365 (12) | 0.0304 (12) | 0.0497 (14) | 0.0075 (9) | 0.0128 (11) | 0.0136 (10) |
Geometric parameters (Å, °)
| Mn1—O1 | 2.106 (2) | C2—C8 | 1.361 (4) |
| Mn1—O1i | 2.106 (2) | C2—C3 | 1.450 (3) |
| Mn1—O3 | 2.1667 (16) | C3—C4 | 1.452 (3) |
| Mn1—O3i | 2.1667 (16) | C4—C7 | 1.399 (3) |
| Mn1—N5ii | 2.372 (2) | C4—C5 | 1.400 (4) |
| Mn1—N5iii | 2.3723 (19) | C5—H5A | 0.9300 |
| O1—C1 | 1.248 (3) | C8—H8A | 0.9300 |
| O2—C1 | 1.244 (3) | C9—C10 | 1.425 (8) |
| O3—C3 | 1.249 (3) | C9—H9A | 0.9700 |
| N1—C8 | 1.357 (3) | C9—H9B | 0.9700 |
| N1—C7 | 1.382 (4) | C10—H10A | 0.9600 |
| N1—C9 | 1.493 (4) | C10—H10B | 0.9600 |
| N2—C5 | 1.315 (3) | C10—H10C | 0.9600 |
| N2—C6 | 1.371 (4) | C11—C12 | 1.531 (4) |
| N3—C7 | 1.335 (3) | C11—H11A | 0.9700 |
| N3—C6 | 1.344 (3) | C11—H11B | 0.9700 |
| N4—C6 | 1.343 (3) | C12—H12A | 0.9700 |
| N4—C14 | 1.457 (3) | C12—H12B | 0.9700 |
| N4—C11 | 1.463 (3) | C13—C14 | 1.510 (3) |
| N5—C12 | 1.461 (3) | C13—H13A | 0.9700 |
| N5—C13 | 1.474 (3) | C13—H13B | 0.9700 |
| N5—Mn1iv | 2.3723 (19) | C14—H14A | 0.9700 |
| N5—H5N | 0.90 (3) | C14—H14B | 0.9700 |
| C1—C2 | 1.508 (3) | ||
| O1—Mn1—O1i | 180.0 | N4—C6—N3 | 117.5 (2) |
| O1—Mn1—O3 | 83.09 (6) | N4—C6—N2 | 117.0 (2) |
| O1i—Mn1—O3 | 96.91 (6) | N3—C6—N2 | 125.4 (2) |
| O1—Mn1—O3i | 96.91 (6) | N3—C7—N1 | 117.7 (2) |
| O1i—Mn1—O3i | 83.09 (6) | N3—C7—C4 | 123.4 (2) |
| O3—Mn1—O3i | 180.0 | N1—C7—C4 | 118.9 (2) |
| O1—Mn1—N5ii | 90.17 (7) | N1—C8—C2 | 125.8 (3) |
| O1i—Mn1—N5ii | 89.83 (7) | N1—C8—H8A | 117.1 |
| O3—Mn1—N5ii | 90.74 (7) | C2—C8—H8A | 117.1 |
| O3i—Mn1—N5ii | 89.26 (7) | C10—C9—N1 | 111.2 (5) |
| O1—Mn1—N5iii | 89.83 (7) | C10—C9—H9A | 109.4 |
| O1i—Mn1—N5iii | 90.17 (7) | N1—C9—H9A | 109.4 |
| O3—Mn1—N5iii | 89.26 (7) | C10—C9—H9B | 109.4 |
| O3i—Mn1—N5iii | 90.73 (7) | N1—C9—H9B | 109.4 |
| N5ii—Mn1—N5iii | 180.0 | H9A—C9—H9B | 108.0 |
| C1—O1—Mn1 | 137.22 (16) | C9—C10—H10A | 109.5 |
| C3—O3—Mn1 | 129.93 (16) | C9—C10—H10B | 109.5 |
| C8—N1—C7 | 118.7 (2) | H10A—C10—H10B | 109.5 |
| C8—N1—C9 | 119.8 (3) | C9—C10—H10C | 109.5 |
| C7—N1—C9 | 121.4 (2) | H10A—C10—H10C | 109.5 |
| C5—N2—C6 | 115.3 (2) | H10B—C10—H10C | 109.5 |
| C7—N3—C6 | 116.3 (2) | N4—C11—C12 | 110.2 (2) |
| C6—N4—C14 | 120.9 (2) | N4—C11—H11A | 109.6 |
| C6—N4—C11 | 122.6 (2) | C12—C11—H11A | 109.6 |
| C14—N4—C11 | 113.1 (2) | N4—C11—H11B | 109.6 |
| C12—N5—C13 | 108.90 (19) | C12—C11—H11B | 109.6 |
| C12—N5—Mn1iv | 116.15 (15) | H11A—C11—H11B | 108.1 |
| C13—N5—Mn1iv | 111.54 (14) | N5—C12—C11 | 114.3 (2) |
| C12—N5—H5N | 110 (2) | N5—C12—H12A | 108.7 |
| C13—N5—H5N | 107 (2) | C11—C12—H12A | 108.7 |
| Mn1iv—N5—H5N | 103 (2) | N5—C12—H12B | 108.7 |
| O2—C1—O1 | 123.4 (2) | C11—C12—H12B | 108.7 |
| O2—C1—C2 | 117.6 (2) | H12A—C12—H12B | 107.6 |
| O1—C1—C2 | 118.9 (2) | N5—C13—C14 | 112.8 (2) |
| C8—C2—C3 | 119.0 (2) | N5—C13—H13A | 109.0 |
| C8—C2—C1 | 116.1 (2) | C14—C13—H13A | 109.0 |
| C3—C2—C1 | 124.9 (2) | N5—C13—H13B | 109.0 |
| O3—C3—C2 | 126.0 (2) | C14—C13—H13B | 109.0 |
| O3—C3—C4 | 119.8 (2) | H13A—C13—H13B | 107.8 |
| C2—C3—C4 | 114.3 (2) | N4—C14—C13 | 111.1 (2) |
| C7—C4—C5 | 114.3 (2) | N4—C14—H14A | 109.4 |
| C7—C4—C3 | 123.3 (2) | C13—C14—H14A | 109.4 |
| C5—C4—C3 | 122.4 (2) | N4—C14—H14B | 109.4 |
| N2—C5—C4 | 124.8 (2) | C13—C14—H14B | 109.4 |
| N2—C5—H5A | 117.6 | H14A—C14—H14B | 108.0 |
| C4—C5—H5A | 117.6 | ||
| O3—Mn1—O1—C1 | −1.4 (3) | C7—N3—C6—N4 | 175.3 (3) |
| O3i—Mn1—O1—C1 | 178.6 (3) | C7—N3—C6—N2 | −6.2 (4) |
| N5ii—Mn1—O1—C1 | −92.1 (3) | C5—N2—C6—N4 | −174.7 (3) |
| N5iii—Mn1—O1—C1 | 87.9 (3) | C5—N2—C6—N3 | 6.8 (4) |
| O1—Mn1—O3—C3 | 1.4 (2) | C6—N3—C7—N1 | −177.9 (3) |
| O1i—Mn1—O3—C3 | −178.6 (2) | C6—N3—C7—C4 | −0.5 (4) |
| N5ii—Mn1—O3—C3 | 91.5 (2) | C8—N1—C7—N3 | 177.3 (3) |
| N5iii—Mn1—O3—C3 | −88.5 (2) | C9—N1—C7—N3 | −2.9 (5) |
| Mn1—O1—C1—O2 | 179.3 (2) | C8—N1—C7—C4 | −0.1 (5) |
| Mn1—O1—C1—C2 | 1.5 (4) | C9—N1—C7—C4 | 179.7 (3) |
| O2—C1—C2—C8 | −1.0 (4) | C5—C4—C7—N3 | 5.6 (4) |
| O1—C1—C2—C8 | 177.0 (3) | C3—C4—C7—N3 | −175.2 (3) |
| O2—C1—C2—C3 | −179.1 (3) | C5—C4—C7—N1 | −177.1 (3) |
| O1—C1—C2—C3 | −1.2 (4) | C3—C4—C7—N1 | 2.1 (4) |
| Mn1—O3—C3—C2 | −1.8 (4) | C7—N1—C8—C2 | −1.3 (5) |
| Mn1—O3—C3—C4 | −179.39 (16) | C9—N1—C8—C2 | 178.8 (4) |
| C8—C2—C3—O3 | −176.6 (3) | C3—C2—C8—N1 | 0.8 (5) |
| C1—C2—C3—O3 | 1.5 (4) | C1—C2—C8—N1 | −177.5 (3) |
| C8—C2—C3—C4 | 1.1 (4) | C8—N1—C9—C10 | 92.7 (5) |
| C1—C2—C3—C4 | 179.2 (2) | C7—N1—C9—C10 | −87.1 (5) |
| O3—C3—C4—C7 | 175.3 (2) | C6—N4—C11—C12 | −148.9 (3) |
| C2—C3—C4—C7 | −2.5 (4) | C14—N4—C11—C12 | 51.9 (3) |
| O3—C3—C4—C5 | −5.6 (4) | C13—N5—C12—C11 | 54.0 (3) |
| C2—C3—C4—C5 | 176.6 (3) | Mn1iv—N5—C12—C11 | −179.1 (2) |
| C6—N2—C5—C4 | −0.7 (5) | N4—C11—C12—N5 | −52.9 (4) |
| C7—C4—C5—N2 | −5.0 (4) | C12—N5—C13—C14 | −55.0 (3) |
| C3—C4—C5—N2 | 175.8 (3) | Mn1iv—N5—C13—C14 | 175.47 (16) |
| C14—N4—C6—N3 | −8.0 (4) | C6—N4—C14—C13 | 146.2 (3) |
| C11—N4—C6—N3 | −165.6 (3) | C11—N4—C14—C13 | −54.2 (3) |
| C14—N4—C6—N2 | 173.4 (3) | N5—C13—C14—N4 | 55.9 (3) |
| C11—N4—C6—N2 | 15.8 (4) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x+1, −y+3/2, z+1/2; (iii) −x, y−1/2, −z+1/2; (iv) −x, y+1/2, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5N···O2v | 0.893 (10) | 2.268 (12) | 3.149 (3) | 169 (3) |
Symmetry codes: (v) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB2703).
References
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808006533/hb2703sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808006533/hb2703Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report