Abstract
A microtiter-based assay system is described in which DNA hairpin probes with dangling ends and single-stranded, linear DNA probes were immobilized and compared based on their ability to capture single-strand target DNA. Hairpin probes consisted of a 16 bp duplex stem, linked by a T2-biotin·dT-T2 loop. The third base was a biotinylated uracil (UB) necessary for coupling to avidin coated microtiter wells. The capture region of the hairpin was a 3′ dangling end composed of either 16 or 32 bases. Fundamental parameters of the system, such as probe density and avidin adsorption capacity of the plates were characterized. The target DNA consisted of 65 bases whose 3′ end was complementary to the dangling end of the hairpin or to the linear probe sequence. The assay system was employed to measure the time dependence and thermodynamic stability of target hybridization with hairpin and linear probes. Target molecules were labeled with either a 5′-FITC, or radiolabeled with [γ-33P]ATP and captured by either linear or hairpin probes affixed to the solid support. Over the range of target concentrations from 10 to 640 pmol hybridization rates increased with increasing target concentration, but varied for the different probes examined. Hairpin probes displayed higher rates of hybridization and larger equilibrium amounts of captured targets than linear probes. At 25 and 45°C, rates of hybridization were better than twice as great for the hairpin compared with the linear capture probes. Hairpin–target complexes were also more thermodynamically stable. Binding free energies were evaluated from the observed equilibrium constants for complex formation. Results showed the order of stability of the probes to be: hairpins with 32 base dangling ends > hairpin probes with l6 base dangling ends > 16 base linear probes > 32 base linear probes. The physical characteristics of hairpins could offer substantial advantages as nucleic acid capture moieties in solid support based hybridization systems.
INTRODUCTION
Large-scale combinatorial approaches to nucleic acid analysis are emerging as powerful tools for widespread applications in detecting, discriminating and analyzing large numbers of DNA sequences via multiplex hybridization schemes (1). The use of miniaturized solid-phase surfaces for hybridization analysis has become increasingly more attractive for nucleic acid detection and analysis (1–5). Many platforms utilize solid-support bound deoxyoligonucleotide probes to hybridize, and thereby capture, single-strand targets. Hybridization of nucleic acid targets with tethered deoxyoligonucleotide probes is the central event in the detection of nucleic acids on microarrays or other high-throughput solid-phase-based assays. Attempts to model the kinetic (6–8) and thermodynamic (9) behavior of immobilized DNA probes and determine factors important in modulating hybridization efficiency have been reported (10–12). The majority of formats commonly employ linear single-stranded deoxyoligonucleotide probes for capturing targets in hybridization reactions (1,5,13). Departing from this common practice, we employ hairpins with dangling ends as capture probes (14).
Solution studies have shown that nicked duplexes comprised of dangling-ended hairpins and single-strands are thermodynamically more stable than gapped duplexes (15). At least part of the observed advantages of dangling-ended hairpins for target hybridization can be attributed to formation of stacking interactions between the 5′ terminal base(s) of the hairpin stem and the 3′ terminal base(s) of the annealed single-stranded target (15). This co-axial stacking along the helical axis of the duplex provides a thermodynamic advantage for annealing of a linear DNA strand. Another study of the hybridization of dangling-ended duplex probes with single-stranded deoxyoligonucleotide targets also showed that formation of the complexes was strongly dependent on the length of the duplex region (16). These results suggest that dangling-ended hairpins may be preferable to linear capture probes for applications in solid-support-based hybridization assays. Results of the present study provide further evidence for advantages of coaxial stacking offered by dangling-ended hairpin probes.
Here we describe a solid support microtiter plate-based DNA assay system. The system is utilized to demonstrate the superiority of hairpin capture probes and quantify comparisons of the kinetics and thermodynamics of target capture by hairpin and linear probes. The hairpins are designed with a 16 bp duplex stem linked by a 5 base unpaired loop and a 3′ 16 or 32 base dangling end for target capture. A biotin moiety attached to the central deoxyuridine of the loop via a C12 bond distance spacer allows for coupling of the hairpins to avidin-coated microtiter plates. Results demonstrate that dangling-ended hairpin probes attached to a solid support capture target DNA more rapidly and form more stable target–probe complexes than those formed with linear probes.
MATERIALS AND METHODS
Deoxyoligonucleotides
The sequences of the hairpin probes, linear probes and the target strand are shown in Figure 1. DNA deoxyoligonucleotide strands were purchased from Oligos Etc (Wilsonville, OR). Deoxyoligonucleotides were purified by polyacrylamide gel electrophoresis by the supplier and received as lyophilized product. Deoxyoligonucleotides were resuspended in 1× TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). Values of the molar extinction coefficients for all the DNA molecules were estimated from the published nearest neighbor parameters (17). Values of 9504, 9784, 9457, 9906 and 9319 M/cm/nt for the 32 base hairpin probe, 32 base linear probe, 16 base hairpin probe, 16 base linear probe and the 65 base target strand, respectively, were employed. Dilutions of the rehydrated DNA stock solutions were made; these were heated to 90°C and absorbance readings at 260 nm were recorded. Optical densities of the DNA stocks were converted to molar strand concentrations from estimated values of extinction coefficients and measured absorbances.
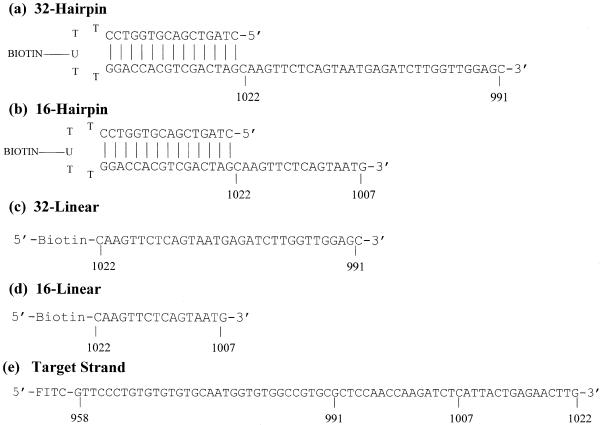
Figure 1.
DNA sequences and schematics of the probe DNA molecules. Hairpin sequences (a) and (b) were designed to form intramolecular stem–loop structures with 3′ dangling ends with sequences complementary to the 3′ end of the target strand. Hairpins also contained a biotinylated uracil at the third base position in the loop for coupling to an avidin-coated microtiter plate. Hairpins have the same 16 bp duplex stems and either 32 (a) or 16 (b) base dangling ends. Linear probes were 32 (c) and 16 (d) bases in length. Biotin derived bases contained a 12-atom spacer at the 5′ end for coupling to plates. Target strand (e) corresponded to a 65 base region of the porcine malignant hyperthermia gene from positions 958 to 1022.
Schematic representations of the hairpin and linear probe structures are depicted in Figure 1. The target strand is a 65 base sequence that encompasses a portion of the porcine malignant hyperthermia gene and mutation site (numbered accordingly) (18). This region of the target gene (position 958–1022) contains known mutations in the porcine malignant hyperthermia gene (19). Target DNA was either radiolabeled with [γ-33P]ATP or a single 5′-FITC moiety was attached by the supplier during synthesis. Figure 1 shows that hairpin sequences were designed to form intramolecular stem–loop structures with a 3′ dangling end with sequence complementary to the 3′ end of the target strand. Hairpins also contained a biotinylated uracil at the third base position in the loop for coupling to an avidin-coated microtiter plate. Thus, the single-strand loop sequence is actually T-T-UB-T-T. The hairpins have the same 16 bp duplex stem and 3′ dangling ends with either 16 or 32 bases. Stem sequences were designed to include as many different types of unique nearest-neighbor doublets with stability sufficient to guarantee duplex fidelity of the hairpin stem at the temperatures where assays were performed. Dangling end sequences were designed so that the 3′ end of the target strand (position 1022) would hybridize adjacent to the duplex and in register with the dangling end to form a nicked junction between the stem and adjoining hybridized target strand resulting in coaxial stacking across the probe–target complex. Upon proper annealing a nicked duplex is formed. The analogous linear probes were 16 or 32 bases long and corresponded precisely to the sequences of the dangling ends of the hairpins. Linear probes contained a biotin moiety at the 5′ end for coupling to microtiter plates.
Coating of microtiter well surfaces with avidin
Stock solutions of avidin (Sigma-Aldrich Canada Ltd, Oakville, Canada) at 100 pmol/µl were prepared and diluted in carbonate coating buffer (50 mM Na2CO3/NaHCO3, pH 9.6) to a working concentration of 0.2 µM. For adsorption to well surfaces, a 100 µl working solution of avidin was added to each of the 96 flat-bottomed wells of a microwell plate and incubated for 1 h at room temperature. Then, using a Dynex plate washer, each plate was washed six times with PBS solution (150 mM NaCl, 10 mM phosphate pH 7.2). The quantity of avidin adsorbed to well surfaces was assessed semi-quantitatively by both direct and indirect methods. In the direct method, increasing amounts of FITC-labeled avidin (Molecular Probes, Eugene, OR) were added to each well and the amount of avidin adsorbed was proportional to readings of the fluorescence intensity at 520 nm. The indirect method employed an enzyme-linked immunodetection system, which incorporates an anti-FITC-alkaline phosphatase antibody catalyzed reaction to generate signal (Sigma Canada Ltd, Mississauga, Ontario, Canada).
Coupling of DNA probes to wells of the microtiter plate
Hairpin and linear probes were diluted in 1× BN buffer (1.0 M NaCl, 100 mM Tris, 0.08% Triton-X 100, pH 8.0) to working stock solutions with concentrations ranging from 0.01 to 0.50 µM. Coating reactions were initiated by adding 100 µl of the desired probe stock solution to each microtiter well. Coupling reactions proceeded at room temperature for 0.5 h. Then each well was washed six times with 200 µl of 1× BN buffer. Plates with DNA probes coupled to them were used immediately after preparation, and not allowed to dry.
Radiolabeling of probes and determination of coupled probe density on microtiter wells
Hairpin and linear probes were radiolabeled on the 3′ end with terminal deoxynucleotidyl transferase (BRL Life Technologies, Mississauga, Ontario, Canada) in the presence of a 10-fold molar excess of [γ-33P]ATP. Reactions were designed to label 80 µM strands of DNA in 120 µl reaction volumes. Unincorporated label was removed by gel filtration chromatography using Sephadex-G50. Labeled probes were diluted to 0.25, 0.5, 1, 2, 5, 7.5, 10, 25, 50 and 75 µM in BN buffer. At each probe concentration two sets of wells were coated in quadruplicate. Plates were incubated for 1 h at 25°C. For the first set, probe solutions were removed from the wells, which were then washed six times with 1× BN buffer and allowed to dry by evaporation. Amounts of probe bound to the surface, PB, were determined by Cerenkov counting of the wells of the plates. For the second set, the total amount of added probe, PT, was left in the well and the plate was allowed to dry by evaporation. The amounts of probe in the wells was determined by Cerenkov counting. The fraction of attached probe, denoted Fc was defined by the ratio: PB/PT. The total amount of probe attached to the well surface is then Fc × P, where P is the concentration of added probe.
Chemiluminescent detection to compare relative strength of probe target capture
FITC-labeled target strands that hybridized to probes attached to the wells of the plate surface were exposed to a monoclonal anti-FITC: alkaline phosphatase conjugate (Sigma). The antibody conjugate was diluted 2500-fold in BN buffer containing 200 mM NaCl. Unbound antibody was removed and the plate wells were washed six times with the 200 mM NaCl-BN buffer. Each well was equilibrated in 200 µl AP buffer (100 mM Tris, 100 mM NaCl pH 9.5 at 20°C), for 5 min at room temperature. This equilibration buffer was aspirated and replaced with 100 µl of AP buffer containing 250 µM CDP-Star (Tropix Applied Biosystems, MA). The enzymatic conversion of the substrate proceeded for 5 min after which (white) light emission was monitored on a chemiluminescent plate reader (Dynex, VA). The linear spectral response of the instrument was approximately six orders of magnitude. All data acquired in this work occurred within a spectral response between 50 and 5000 relative light units (RLU) or roughly three logs out of the entire dynamic range and therefore are assumed to lie within the linear response regime.
Radiolabeling of target deoxyoligonucleotides
Target strands with 5′-OH termini were radiolabeled by incubation in the presence of a 10-fold molar excess of [γ-33P]ATP and T4 polynucleotide kinase (New England Biolabs, Beverly, MA) as described above. Following the kinase reaction, unincorporated [γ-33P]ATP radiolabel was removed by gel filtration chromatography (Sephadex G-50). Resulting labeled target strands were diluted to the desired working concentrations in 1× BN buffer.
Characterization of the target strand
Radiolabeled target strands were diluted in 1× BN buffer. Target concentrations for kinetic studies were 0.001, 0.004, 0.016 and 0.064 × 10–8 M strands in 100 µl total reaction volume for each well. Hybridization rates were measured at two temperatures (25 and 45°C) over a period of 24 h. At regular time intervals, 50 µl aliquots were removed from the reaction mixtures and diluted in 5 ml of scintillation fluid. Samples were counted, yielding the amount of remaining free target strand, TFREE. As a result of the labeling procedure, each strand is presumed to contain a label, thus counts correspond directly to amounts of DNA strands. For the concentrations of target strand studied, total counts corresponding to these amounts were determined as follows. Target solutions at bulk concentrations of 0.001, 0.004, 0.016 and 0.064 × 10–8 M were added in tandem to plate wells with and without DNA probes coupled to them. Reactions were incubated for 3 h. For wells without probes attached, the liquid volumes in each well were removed and the wells were washed six times with 1× BN buffer, followed by a final wash with 6 N NaOH solution. Wells were neutralized by addition of an equal volume of 6 N HCl. The solution was removed from each well and counted. Counts of these solutions were taken to correspond to non-specifically-bound target strands, TNSB. Presumably, this non-specific binding results from interactions of the target strand with the avidin surface, or uncoated portions of the plate in the reaction volume. For wells with probes attached, solutions were removed and counted. These counts corresponded to the amount of free target strands, TFREE. The total number of counts, TTOT, is the sum of TFREE, the counts corresponding to non-specific binding, TNSB, and the counts corresponding to the target strands specifically bound to the attached probes in the well, TWELL. Thus:
TWELL = TTOT – TFREE – TNSB 1
The fraction of target strands specifically bound to probe strands on the surface is:
FSB = TWELL/TTOT 2
For hairpin and linear probes attached to the avidin-coated plate, concentrations of target strands specifically bound to probe strands, [T]BOUND, were found from the product FSB × [TTOT] as a function of time.
Analysis of hybridization kinetics
Hybridization of free target DNA strands (T) with probe strands (linear or hairpin) (P) coupled to the surface of avidin-coated microtiter wells was modeled by the simple irreversible second order process:
k
T + P → P*T 3
P is the concentration of free (unbound) DNA probe on the surface, T is the concentration of free DNA target strand in solution and P*T is the concentration of probe–target complexes that form on the surface. The total concentrations of probe and target strands are designated P0 and T0, respectively. The observed second order rate constant for P*T complex formation is given by k. Initially at time t = 0, P0 = P, T0 = T and P*T = 0. When t > 0, P = (P0 – X), T = (T0 – X) where P*T = X. The rate equation for the formation of target–probe complexes, X, is:
dX/dt = k(P0 – X)(T0 – X) 4
Integration of this equation and solving for the constant of integration yields the expression:
kt = [1/(T0 – P0)]{ln[P0(T0 – X)/T0(P0 – X)]} 5
which can be solved for X to yield:
X = (T0{exp[(T0 – P0)kt]–1})/{T0/P0exp[(T0 – P0)kt]–1} 6
Plots of the concentration of X = P*T versus time were fit with equation 6 using the non-linear least squares curve fitting routines in SigmaPlot version 4.0. In these fits, the second order rate constant k and the effective total target strand concentration T0 were the adjustable fitting parameters. The effective molar strand concentration of probe P0 was constant and determined from a standard curve of the amount of probe coupled to the surface versus the amount of probe added. Of course, the overall reaction rate is the product (k × T0 × P0).
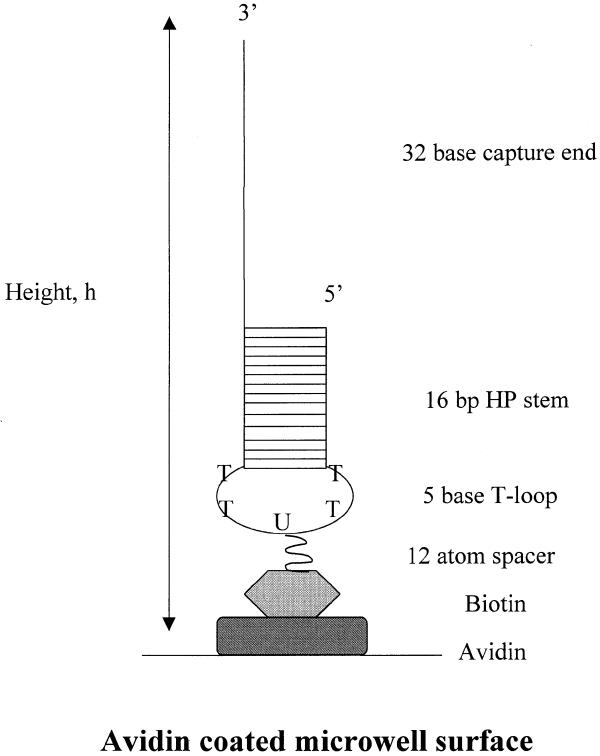
In the fitting procedure, the effective target and probe strand concentrations T0 and P0, respectively, correspond to concentrations in the effective reaction volume (ERV). The ERV is defined by the reactive surface area of the plate well and estimated height of DNA probes coupled to the surface. The ERV and appropriate dimensions are schematically represented in Figure 2 and shown specifically for the 32 base hairpin probe. Obviously, defined in this way, the ERV is actually much smaller than the volume of the target strand solution added to each well (100 µl). The probe concentration in the ERV is constant, and because the ERV is much smaller than the volume defined by the added target solution, the actual target concentrations in the ERV (where hybridization to probes occurs) are much higher than the concentration of target added at t = 0 in the bulk reaction volume. Because the target strands in solution and the probe strands affixed to the surface do not possess the same degrees of freedom, this analysis assumes that target–probe hybridization is restricted to occur only within the ERV, and thus the observed rates evaluated from fitting the data correspond to hybridization in the ERV. We assume that both types of probes reside on the well surface in an extended configuration, as depicted in Figure 2. This assumption seems reasonable based on recent investigations of effects of DNA length on immobilization on gold surfaces (20). The ERV was estimated using the reaction surface area of the well covered by probe in a 100 µl reaction volume and the height of the immobilized probe DNAs. Since linear probes differ from hairpins by a 16 bp duplex and a 5 base loop, linear probes are estimated to be ~60 Å shorter. However, because differences in the actual hydrated and dynamic lengths of the strands in solvent were not determined quantitatively, we used an estimation as an upper limit on the probe height (that of the 32 base hairpin) to determine the upper limit on the ERV. The hairpin stem was assumed to be average B-form with 3.4 Å/bp. Thus, the reaction surface area of the plate well covered by 100 µl was estimated to be 0.9482 cm2, and the height of the biotinylated 32 base hairpin probe strand was estimated to be 250 Å. These values result in an estimated ERV of 2.371 × 10–3 µl. This value should be considered as an upper limit on the actual ERV. Thus, all target, probe and target–probe complex concentrations determined using this value should be considered as lower limits on the actual concentrations.
Figure 2.
Schematic of the ERV. Dimensions of the ERV are shown. The upper limit of the ERV is defined by the reactive surface area of the plate well covered by 100 µl and estimated height (h) of the 32 base hairpin capture probe coupled to the surface. For our system the surface area of the plate well covered by 100 µl was estimated to be 0.9482 cm2, and the height of the biotinylated 32 base hairpin probe was estimated to be 250 Å. These values result in an estimated ERV of 2.371 × 10–3 µl.
The observed equilibrium association constant, KA, was determined as:
KA = [P*T]asm/([P0] –[P*T]asm)([T0] – [P*T]asm) 7
where [P*T]asm is the value of [P*T] at the asymptote determined by extrapolating the best fit line through the asymptotic region of plots of [P*T] versus time, back to t = 0. P0 is fixed with values determined as described above. The binding free energy for hybridization between the target and probe is given by:
–ΔGbind = –RTlnKA 8
Where T is the temperature (K) and R the gas constant.
RESULTS
Coating of plates with avidin
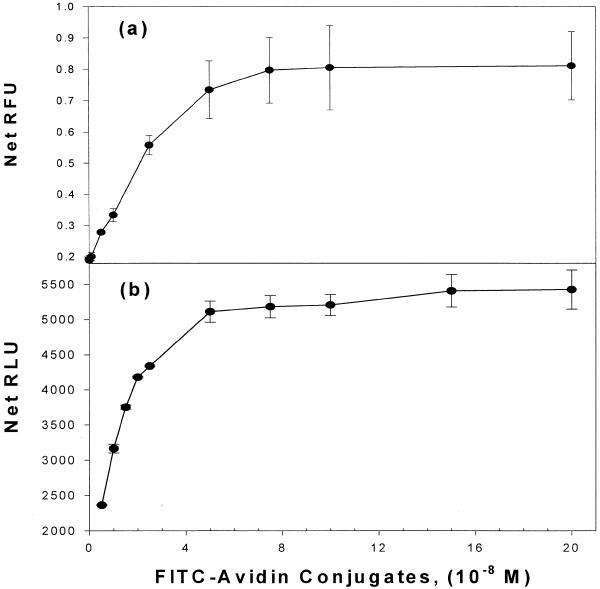
The amount of avidin attached to the plate surface was estimated directly and indirectly using FITC-labeled avidin in place of native avidin. The FITC–avidin does not exhibit dramatically different behavior from native avidin and its biotin-binding ability is not affected (21). The amount of avidin required to saturate a microwell was determined by coating wells with increasing concentrations of FITC–avidin until saturation was observed. Results of such coating experiments are shown in Figure 3 for two different types of plates as determined by the direct method (Fig. 3a) and the indirect method (Fig. 3b). Because DNA probes could potentially have different coupling properties depending on the type of plate surface used, two different types of plates were examined by two different methods. The curve in Figure 3a corresponds to Black Flourotiter B plates (Dynex) and direct fluorometric detection of bound FITC–avidin using a Tecan Fluorometer. Error bars denote the standard deviation (SD) of at least four independent coating experiments. In Figure 3b, indirect measurement of bound avidin involved determination of the amount of bound FITC–avidin by immunodetection using an anti-FITC-alkaline phosphatase antibody conjugate. The saturation curves measured directly on white high binding plates are shown in Figure 3b. These curves were generated by indirect measurement of the chemiluminescence (RLU). The saturation profiles determined indirectly by immunodetection (Fig. 3b) or by direct measurement of the flourescent signal of the attached FITC–avidin (Fig. 3a) are qualitatively quite comparable, and both types of plates display very similar saturation points. As comparison of the curves in Figure 3a and b indicate, the normalized curves (if shown) would be nearly identical. Binding profiles were essentially the same whether the wells were coated for 1 h (as in Fig. 3) or 24 h (data not shown).
Figure 3.
Coating of microtiter plates with avidin. Results of avidin-coating experiments are shown for two different detection methods. The amount of avidin required to saturate a microwell is plotted versus the amount of FITC–avidin added to the wells with increasing concentrations of FITC–avidin until saturation as shown. (a) Relative fluorescence units (RFU) determined by fluorometry of bound avidin. Error bars denote the SD of at least four independent coating experiments. (b) FITC–avidin binding curves determined indirectly by an anti-FITC alkaline phosphatase antibody and chemiluminescence detection.
Coupling of hairpin and linear probes to the plate surface
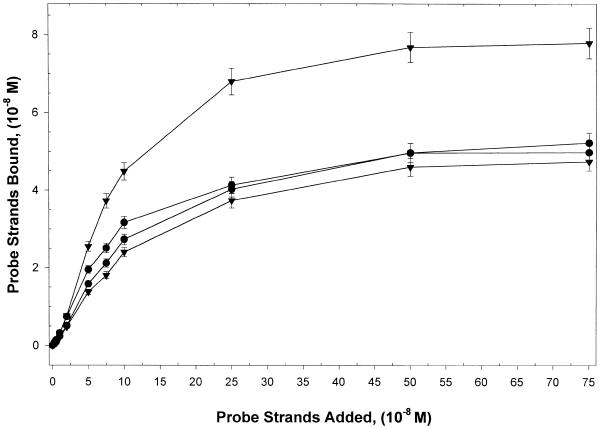
In order to properly establish standard conditions for hybridization experiments, it was essential to know precisely the amount of DNA probe (linear or hairpin) that was actually coupled to the avidin-coated well surface. For this purpose, standard curves relating the amount of probe added in 100 µl to that actually coupled to the well surfaces were constructed. To assess the binding capacity of the avidin surface, increasing concentrations of radiolabeled hairpin or linear probes (16mers and 32mers) were added to the wells and the probe density on the surface was determined. Typically, quantities of avidin at saturation were ~10 pmol/well (regardless of the type of microwell plate). The standard curves derived for each type of DNA probe are shown in Figure 4, where the amounts of biotinylated linear and hairpin probes attached to the surface are plotted versus the amounts of probe added in 100 µl. Error bars are SDs of at least four independent experiments. Clearly, for the different types of probes the amounts coupled to the avidin-coated wells increase as the amount of added probe increases. When >1 pmol of probe is added, smaller amounts of hairpin compared with linear DNA probes are coupled to the surface. As a result, densities of the attached hairpin probes on the surface are lower than for linear probes. Also, length of the capture region of the hairpin does not significantly affect coupling of the hairpin at any probe concentration. In contrast, for the linear probes, when >2 pmol of probe are added, the shorter 16mer capture strand is coupled to a much greater extent and thus is able to achieve much higher surface densities than the longer 32mer linear strand. This observation is in agreement with recently published results for linear DNA probes immobilized to gold surfaces in the presence of a blocking agent (20).
Figure 4.
Coupling of probes to the plate surface. Standard curves derived for each type of DNA probe where the amounts of biotinylated linear and hairpin probes bound to the surface are plotted against the amounts of probe added in 100 µl. Error bars are SDs of at least four independent experiments. Clearly, for the different types of probes the amounts coupled to the avidin wells increase with increasing amounts of added probe, but the amount of bound probe is not the same for all probes.
Microtiter plates that were used in the kinetic studies described below were prepared by adding 2 µM probe (hairpin or linear). Figure 4 indicates that different probes did not couple to the well surface in the same quantity. Although not clearly evident because of the scale in Figure 4, at 2 µM of probe this corresponds to 0.51, 0.74, 0.48 and 0.76 µM in the 100 µl bulk reaction volume for 32mer hairpin, 32mer linear, 16mer hairpin and 16mer linear probes, respectively. Differences in coupling densities for the 32mer and 16mer hairpins probes were <5%, and, regardless of their length, linear probes coupled at ~50% higher density than the hairpin probes.
Relative comparison between hairpin and linear probe target capture
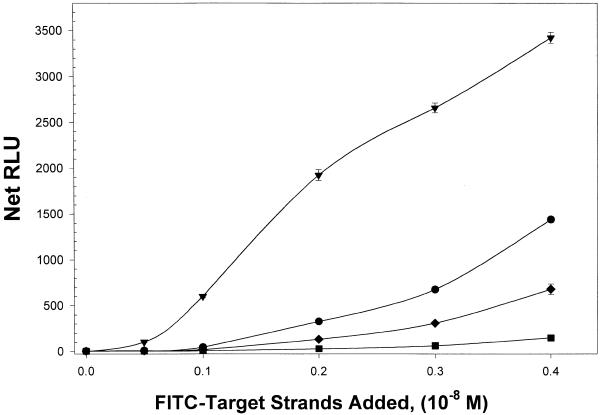
Semi-quantitative comparison of target capture by hairpin and linear probes is shown in Figure 5. There the summed RLU (net RLU), corresponding to captured FITC-labeled target strand is plotted against the amount of target strand added. For these experiments FITC-target strand was added at concentrations ranging from 0.05 to 0.4 × 10–8 M strands to microwells coated with 5 × 10–8 M probe strands. This probe-coating amount corresponded to ~2 ×10–8 M strands of probe coupled to the well surfaces for all probes. Hybridizations were performed at 25°C for 1 h. Hybridized target was detected indirectly by chemiluminescence generated by an enzyme-linked immunodetection system as described above. All RLU values were well within the linear response range of the assay and therefore the RLU values are assumed to be proportional to the amount of FITC-target strands on the well surface. The plot in Figure 5 reveals that hairpin probes capture relatively more target than linear probes. The 32 base hairpin captured the highest relative quantity of FITC-target strand at all input target strand concentrations. Appreciable amounts of target strand are captured only when target concentrations are >0.2 × 10–8 M as assessed by the chemiluminescent-based assay employed. The relative order of target capture strength is 32 base hairpin > 16 base hairpin > 32 base linear > 16 base linear probe.
Figure 5.
Relative binding as a function of input target concentration. The summed RLU (net RLU), corresponding to captured FITC-labeled target strand, is plotted against the amount of target strand added for each type of probe. The 32 base hairpin (triangles) captured the highest quantity of FITC-target strand. At somewhat lesser amounts are the 16mer hairpins (circles), 32 base linear (diamonds) and 16 base linear (squares) probes. The avidin-coated wells were coated with 5 × 10–8 M probe strands.
Target–probe hybridization kinetics for 16 and 32 base hairpin and linear probes
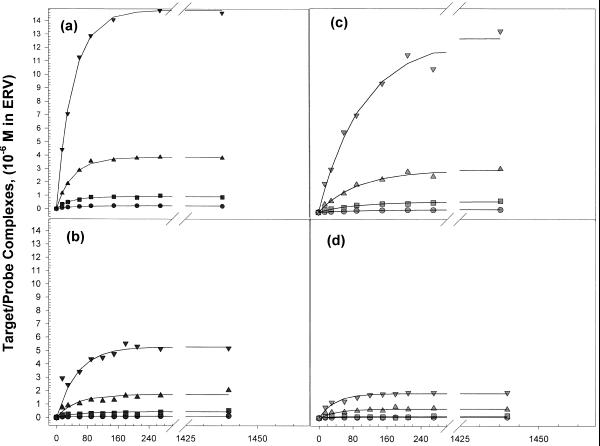
Hybridization of single-stranded target DNA to the hairpin and linear DNA probes immobilized onto avidin-coated microtiter plate wells was measured as a function of time, at four different target concentrations and two temperatures, 25 and 45°C. Figure 6 shows plots of the concentration of target–probe complexes, [P*T], versus time for four target concentrations at 45°C. Solid lines drawn through the data in Figure 6 are best fits obtained from non-linear least squares regression fits of the data with equation 6. The fits in Figure 6 are typical in that the same quality of data and fits were also obtained at 25°C. For each probe, the average regression coefficient was >0.92 in all cases. At the highest target concentration, a substantial increase in P*T complex formation is seen for the 32mer hairpin (Fig. 6a) and linear (Fig. 6c) probes versus time. Hairpin and linear probes with 32 base capture regions display the highest rates of capture and largest amounts of hybridized complexes at equilibrium (when d[P*T]/dt = 0). At 45°C the hairpin probes (Fig. 6a and c) capture more target at a faster rate than their linear counterparts. Probes with the 16 base capture regions display at least a 2-fold lower amount of product formed at equilibrium than their 32mer counterparts. At the lowest target concentrations, the amounts of hybridized product formed with either linear or hairpin probes is about 10 times less than that found for the same type of probe, at higher input target concentrations. For instance, Figure 6 reveals that for any of the probes, no additional complexes are formed after ~5 h. Results for these experiments at 45 and 25°C at four target concentrations are summarized in Table 1. For the most part, the analysis revealed that the measured rates of hybridization, k × T0 × P0 at both temperatures, increased with increasing T0. At both temperatures hairpin probes displayed significantly higher rates of hybridization compared with their linear counterparts. Ratios of the rates of hybridization for linear and hairpin probes of the same length (normalized for differences in surface concentrations for the hairpin and linear probes) are summarized in Table 2. At 25°C the ratio of the rates for the hairpin versus linear probes with 16 base capture sequences is >2 at all target concentrations. At 45°C the ratio of the rates for the hairpin versus linear 16mers ranges from 1.2 to 2.0 over the target concentration range examined, while the ratio of rates for the 32mer hairpin and linear probes is nearly 3. These comparisons show that the overall rates of hybridization are higher for the hairpin than linear probes of the same length. This is notable since the probe density on well surfaces was ~40% higher for the linear probes. These observations reveal substantial differences in the hybridization behaviors for the hairpin versus linear probes on the surface.
Figure 6.
Probe–target complex formation as a function of time. Plots of the concentration of target–probe complexes, [P*T], versus time for four target concentrations at 45°C are shown for each probe. (a) 32mer hairpin, (b) 16mer hairpin, (c) 32mer linear, (d) 16mer linear. Solid lines drawn through the data are best fits obtained from non-linear least square regression fits of the data with equation 6.
Table 1. Results of kinetic measurements and analysis.
| 32 base hairpin capture probe | 16 base hairpin capture probe | ||||||
| Temperature |
[Target]a |
k × [T0]b |
Ratec |
R2 |
k × [T0]b |
Ratec |
R2 |
| 25°C | 0.001 | 1.1 ± 0.2 | 0.23 ± 0.02 | 0.9250 | 0.53 ± 0.08 | 0.11 ± 0.02 | 0.9175 |
| 0.004 | 4.9 ± 1.0 | 1.1 ± 0.3 | 0.8882 | 2.2 ± 0.1 | 0.44 ± 0.07 | 0.8874 | |
| 0.016 | 19 ± 3 | 4.2 ± 1.2 | 0.9441 | 8.6 ± 1.3 | 1.7 ± 0.2 | 0.9414 | |
| 0.064 | 64 ± 9 | 14 ± 3 | 0.9633 | 53 ± 6 | 11 ± 1 | 0.9850 | |
| 45°C | 0.001 | 4.2 ± 1.0 | 0.90 ± 0.09 | 0.9820 | 1.7 ± 0.2 | 0.33 ± 0.05 | 0.8547 |
| 0.004 | 19 ± 4 | 4.0 ± 0.5 | 0.9796 | 6.1 ± 1.0 | 1.2 ± 0.2 | 0.8367 | |
| 0.016 | 70 ± 3 | 15 ± 1 | 0.9970 | 28 ± 4 | 5.6 0.5 | 0.9097 | |
| 0.064 | 270 ± 10 | 58 ± 3 | 0.9986 | 82 ± 8 | 17 ± 2 | 0.9854 | |
| 32 base linear capture probe | 16 base linear capture probe | ||||||
| 25°C | 0.001 | ND | ND | ND | 0.15 ± 0.03 | 0.048 ± 0.009 | 0.8764 |
| 0.004 | 1.8 ± 0.5 | 0.58 ± 0.03 | 0.8983 | 0.62 ± 0.08 | 0.20 ± 0.02 | 0.8983 | |
| 0.016 | 9.0 ± 3.0 | 2.8 ± 0.4 | 0.8158 | 2.6 ± 0.4 | 0.82 ± 0.08 | 0.9665 | |
| 0.064 | 23 ± 2 | 7.3 ± 0.8 | 0.9826 | 8.8 ± 0.9 | 2.8 ± 0.3 | 0.9690 | |
| 45°C | 0.001 | 1.0 ± 0.2 | 0.31 ± 0.05 | 0.9935 | 0.84 ± 0.2 | 0.27 0.05 | 0.7405 |
| 0.004 | 4.9 ± 1.1 | 1.5 ± 0.8 | 0.9743 | 2.1 ± 0.2 | 0.69 ± 0.1 | 0.8491 | |
| 0.016 | 18 ± 2 | 5.6 ± 2.9 | 0.9828 | 8.6 ± 0.9 | 2.8 ± 0.4 | 0.8856 | |
| 0.064 | 64 ± 11 | 20 ± 3 | 0.9854 | 26 ± 3 | 8.4 ± 0.8 | 0.9615 | |
a10–8 M strands.
b10–9/s (k and [T0] were determined from fits to equation 6).
cEvaluated from k × [T0] × [P0], 10–12 M/s. See text for details.
ND, not determined.
Table 2. Ratios of the rates of hybridization for the hairpin and linear probes.
| Probe |
[T]Bulka |
25°C |
45°C |
| 32 base hairpin:32 base linear | 0.001 | ND | 2.9 |
| 0.004 | 2.4 | 2.6 | |
| 0.016 | 2.4 | 2.7 | |
| 0.064 | 2.3 | 2.9 | |
| 16 base hairpin:16 base linear | 0.001 | 2.2 | 1.2 |
| 0.004 | 2.2 | 1.8 | |
| 0.016 | 2.1 | 2.0 | |
| 0.064 | 3.7 | 2.0 |
a10–8 M strands.
ND, not determined
Analysis of the temperature dependence of the rates
Attempts were made to systematically analyze the temperature dependence of the hybridization rates of the probes via the Arrhenius method. Plots of the natural log of the rate constant, determined by fitting curves like those shown in Figure 6 with equation 6, versus the inverse of the hybridization temperature, were constructed at each target concentration (data not shown) and the slopes connecting the two temperature data points were determined. Unfortunately, because rates were only measured at two temperatures, 25 and 45°C, the aforementioned plots were difficult to analyze quantitatively. Only qualitative results were obtained (data not shown), which suggest, consistent with observations of the measured rates, that single-stranded targets have relatively lower activation barriers to overcome in order to form specific hybridized products with hairpin probes than with linear probes. Target capture by linear probes is affected by length of the capture region to a greater extent for hairpins, although in both cases the effect is minimal.
Thermodynamics of hybridization
For each probe type, values of the probe–target binding free energies were estimated from equilibrium constants evaluated from the asymptotic regions of plots like those shown in Figure 6. These values evaluated at 25 and 45°C are summarized in Table 3. Stated errors are SDs for binding free-energy values determined at four target concentrations. Plots of complex formation versus time (as in Fig. 6) revealed that at the higher temperatures considerably larger amounts of probe–target complexes are formed with hairpin than with linear probes (higher values of [P*T] when d[P*T]/dt = 0). Comparison of the values in Table 3 shows that probe–target complexes comprised of hairpins are more stable than linear complexes by as much as 1.6 kcal/mol. Thus, not only do hairpins form hybridized products faster than linear probes but once formed, probe–target complexes with hairpins are thermodynamically more stable.
Table 3. Equilibrium binding parameters.
| Probe | 25°C | 45°C | ||||
| |
KAa,
(105/M) |
ΔGb kcal/mol |
ΔΔGc |
KAa,(105/M) |
ΔGb kcal/mol |
ΔΔGc |
| 32 base hairpin | 1.44 | –6.91 | 1.61 | 0.940 | –6.67 | 0.95 |
| 32 base linear | 0.0899 | –5.30 | 0.184 | –5.72 | ||
| 16 base hairpin | 0.318 | –6.04 | 0.32 | 1.30 | –6.86 | 0.85 |
| 16 base linear | 0.184 | –5.72 | 0.302 | –6.01 |
a±15% error.
bAssociation free energies calculated using the expression –RTlnKA, with T = 293.15 K.
cΔΔG values are for comparison between hairpin and linear probe–target complex formation.
DISCUSSION
Capture via hybridization of nucleic acid targets by deoxyoligonucleotide probes attached to a solid support surface is crucial for target detection on DNA microarrays and other high-throughput solid-phase-based assays. In this study the capture of linear target DNA by hairpin and linear probes coupled to a solid support surface were compared. There were two inter-related components of the study. The first was to define standard experimental parameters for performing hybridization assays on microtiter plates. This required precise determination of the coating capacity of microtiter plate surfaces with DNA probes and evaluation of the saturation (binding) capacity of the coated plates. The second component of the study was to employ the developed and characterized assay system to measure hybridization rates and stabilities of hairpin and linear capture probes.
Our results show that target capture by hairpin probes is faster than with linear probes and that, once formed, hairpin complexes are thermodynamically more stable. This is despite the fact that the coupling density on the surface for linear probes was nearly twice that for hairpin probes. At 25 and 45°C and all target concentration examined, rates of hybridization for hairpins are at least twice those of their linear probe counterparts. Length of the single-stranded capture region was also found to affect target capture and in general longer capture regions were favored.
Recent studies have suggested that increasing the distance of capture probes from the surface improves target hybridization (11). We also investigated whether the capture performance of the hairpins could be enhanced by increasing the distance of the hairpin from the surface. For this purpose, hairpins with a standard 12-atom spacer were prepared and their hybridization properties were compared with hairpins with an ~28-atom spacer. By definition, the ERV increases with increasing spacer length. Results of the comparison (data not shown) indicated that increasing atomic spacer length only marginally improved hybridization performance at the highest target concentrations. At low target concentrations both probes exhibited similar behavior. Effects of increasing the spacer for linear capture probes above the surface were also investigated (data not shown). Target capture by linear probes, wherein the spacer region was increased from 12 to 60 atoms, a distance that approximates the height of the hairpin duplex stem and linker region, was also measured. For the linear molecules, a significant advantage was observed for the longer 60-atom spacer compared with the 12-atom spacer, but the hairpin with a 12-atom spacer still maintained an advantage in hybridization over the linear probe with a 60-atom spacer. Thus, independent of the spacer size, hairpins display better capture capabilities.
Improvements in the rates and efficiencies of nucleic acid hybridizations by tethered DNA probes on solid support surfaces (leading to increased sensitivity) are necessary to validate the use of high-throughput arrays for various types of multiplex sequence analysis. In our application, DNA capture probes on the surface have been designed as dangling-ended hairpin structures and found to improve assay characteristics. Put simply, because of their faster kinetics and higher thermodynamic stabilities, hairpin probes attached to an avidin-coated microtiter plate offer distinct advantages as nucleic capture moieties. It is anticipated that these advantages of hairpins will also be realized on other solid-phase formats including microarrays, micro-particle beads or standard high-throughput microwell plates. Practically, it is expected that with their superior physical characteristics hairpin probes will significantly enhance the performance of nucleic acid assays for a variety of applications including high-throughput diagnostics, single nucleotide polymorphism detection and gene expression profiling, where the current challenge is the development of rapid and sensitive detection methods.
References
- 1.Duggan D.J., Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Expression profiling using cDNA microarrays. Nature Genet., 21 (Suppl.), 10–14. [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz R.J., Morris,D., Chee,M., Hubbell,E., Kozal,M.J., Shah,N., Shen,N., Yang,R. and Fodor,S.P.A. (1995) Using oligonucleotide probe arrays to access genetic diversity. Biotechniques, 19, 442–447. [PubMed] [Google Scholar]
- 3.O’Donnell-Maloney M.J., Smith,C.L. and Cantor,C.R. (1996) The development of microfabricated arrays for DNA sequencing and analysis. Trends Biochem. Technol., 14, 401–407. [DOI] [PubMed] [Google Scholar]
- 4.de Saizieu A., Certa,U., Warrington,J., Gray,C., Keck,W. and Mous,J. (1998) Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nature Biotechnol., 16, 45–48. [DOI] [PubMed] [Google Scholar]
- 5.Southern E.M, Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21 (Suppl.), 5–9. [DOI] [PubMed] [Google Scholar]
- 6.Day P.J.R., Flora,P.S., Fox,J.E. and Walker,M.R. (1991) Immobilization of polynucleotides on magnetic particles—factors influencing hybridization efficiency. Biochem. J., 278, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan V., Graves,D.J. and McKenzie,S.E. (1995) The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys. J., 69, 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkins-Stevens P., Henry,M.R. and Kelso,D.M. (1999) DNA hybridization on microparticles: determining capture-probe density and equilibrium dissociation constants. Nucleic Acids Res., 27, 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maskos U. and Southern,E.M. (1992) Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. 1. analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res., 20, 1675–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams J.C., Case-Green,S.C., Mir,K.U. and Southern,E.M. (1994) Studies of oligonucleotide interactions by hybridization to arrays—the influence of dangling ends on duplex yield. Nucleic Acids Res., 22, 1365–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall A. and Hodgson,J. (1998) DNA chips: an array of possibilities. Nature Biotechnol., 16, 27–31. [DOI] [PubMed] [Google Scholar]
- 14.Lane M.J., Benight,A.S. and Faldasz,B.D. (1998) US Patent No. 5,770,365. June 23, 1998.
- 15.Lane M.J., Paner,T., Kashin,I., Faldasz,B.D., Li,B., Gallo,F.J. and Benight,A.S. (1997) The thermodynamic advantage of DNA oligonucleotide ‘stacking hybridization’ reactions: energetics of a DNA nick. Nucleic Acids Res., 25, 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Meara D., Nilsson,P., Nygren,P., Uhlen,M. and Lundeberg,J. (1998) Capture of single-stranded DNA assisted by oligonucleotide modules. Anal. Biochem., 255, 195–203. [DOI] [PubMed] [Google Scholar]
- 17.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide Interactions. III. Circulardichroism Studies of the Conformation of Deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 18.Otsu K., Khanna,V.K., Archibald,A.L. and MacLennan,D.H. (1991) Cosegregation of porcine malignant hyperthermia and a probable causal mutation in the skeletal-muscle ryanodine receptor gene in backcross families. Genomics, 11, 744–750. [DOI] [PubMed] [Google Scholar]
- 19.Otsu K., Phillips,M.S., Khanna,V.K., Deleon,S. and MacLennon,D.H. (1992) Refinement of diagnostic assays for a probable causal mutation for porcine and human-malignant hyperthermia. Genomics, 13, 835–837. [DOI] [PubMed] [Google Scholar]
- 20.Steel A.B., Levicky,R.L., Herne,T.M. and Tarlov,M.J. (2000) Immobilization of nucleic acids at solid surfaces: effect of oligonucleotide length on layer assembly. Biophys. J., 79, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emans N., Biwersi,J. and Verkman,A.S. (1995) Imaging of endosome fusion in bhk fibroblasts based on a novel fluorometric avidin-biotin binding assay. Biophys. J., 69, 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]