Abstract
In the title compound, C11H11NO4S, the thiazine ring adopts a distorted half-chair conformation. The enolic H atom is involved in an intramolecular O—H⋯O hydrogen bond, forming a six-membered ring. Molecules are linked through weak intermolecular C—H⋯O hydrogen bonds, resulting in chains lying along the b axis.
Related literature
For related literature, see: Bihovsky et al. (2004 ▶); Fabiola et al. (1998 ▶); Golič & Leban (1987 ▶); Zia-ur-Rehman et al. (2005 ▶, 2006 ▶, 2007 ▶); Turck et al. (1996 ▶).
Experimental
Crystal data
C11H11NO4S
M r = 253.27
Triclinic,

a = 6.8523 (1) Å
b = 8.3222 (2) Å
c = 10.4880 (2) Å
α = 72.1321 (11)°
β = 77.9619 (12)°
γ = 80.0360 (12)°
V = 552.89 (2) Å3
Z = 2
Mo Kα radiation
μ = 0.29 mm−1
T = 120 (2) K
0.40 × 0.20 × 0.14 mm
Data collection
Bruker–Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2007 ▶) T min = 0.891, T max = 0.960
12691 measured reflections
2529 independent reflections
2248 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.034
wR(F 2) = 0.092
S = 1.11
2529 reflections
157 parameters
H-atom parameters constrained
Δρmax = 0.31 e Å−3
Δρmin = −0.53 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶); data reduction: DENZO and COLLECT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: CAMERON (Pearce & Watkin, 1993 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808007721/pv2072sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808007721/pv2072Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3A⋯O4 | 0.84 | 1.78 | 2.525 (2) | 146 |
| C4—H4⋯O2i | 0.95 | 2.36 | 3.193 (2) | 146 |
Symmetry code: (i)  .
.
Acknowledgments
We gratefully acknowledge the Higher Education Commission of Pakistan and the University of the Punjab, Lahore, for financial support.
supplementary crystallographic information
Comment
In order to discover new useful therapeutic agents, many new compounds are continuously being synthesized. Benzothiazine dioxides and their derivatives are reported to possess numerous types of biological activities. Owing to their applications as non-steroidal anti-inflammatory compounds (Turck et al., 1996), considerable attention has been given to 1,2-benzothiazine 1,1-dioxides and their precursor intermediates (Golič & Leban, 1987). Various 1,2-benzothiazines derivatives are also known as potent calpain I inhibitors (Bihovsky et al., 2004), while benzothiaine-3-yl-quinazolin-4-ones showed marked activity against Bacillus subtilis (Zia-ur-Rehman et al., 2006). As part of a research program synthesizing various bioactive benzothiazines (Zia-ur-Rehman et al., 2005, 2006), we herein report the crystal structure of the title compound, (I).
In the molecule of the title compound (Fig. 1), the thiazine ring exhibits a distorted half-chair conformation with S1/C1/C6/C7 atoms lying in a plane and N1 showing significant departure from the plane due to its pyramidal geometry projecting the methyl group approximately perpendicular to the ring; the deviations of N1 and C8 from the least square plane being -0.895 (2) and -0.413 (3) Å, respectively. Like other 1,2-benzothiazine 1,1-dioxide derivatives (Fabiola et al., 1998; Zia-ur-Rehman et al., 2007), the enolic hydrogen on O3 is involved in intramolecular hydrogen bonding (Table 1). Also, C7—C8 bond length [1.378 Å] (very close to normal C?C bond; 1.36 Å) indicates a partial double-bond character indicating the dominance of enolic form in the molecule. The C1—S1 bond distance [1.7580 (14) Å] is as expected for typical C(sp2)—S bond (1.751 Å).
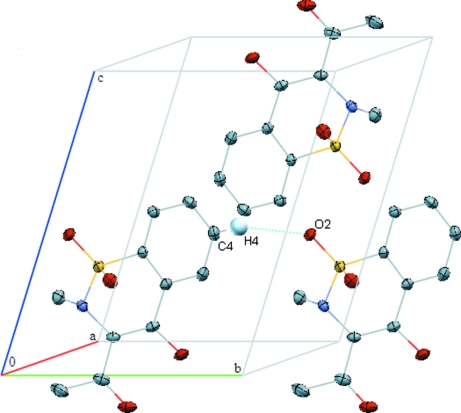
Each molecule is linked to its adjacent one through a hydrogen bond [C4—H4···O2] resulting in a chain of molecules lying along the b axis (Table 1 and Fig. 2). Each molecule in the chain is linked with its neighbour through weak slipped π-π interactions at inversion centres; the closest C–C contacts are between C3–C3i separated by 3.316 Å.
Experimental
A mixture of 1-(4-hydroxy-1,1-dioxido-2H-1,2-benzothiazin-3-yl) ethanone (239 mg, 1.0 mmol) dissolved in acetone (10 ml), aqueous NaOH (3 ml, 5%) and dimethyl sulfate (0.5 ml) was stirred for half an hour followed by careful addition of dilute HCl (5%) to maintain the pH to Congo Red. Precipitates of (I) thus obtained were filtered, washed with water and dried. Colourless crystals were grown by slow evaporation of a solution of (I) in methanol at room temperature.
Refinement
The hydrogen atoms were included in the refinements in a riding mode with the following constraints: aryl, methyl and hydroxyl C/O—H distances 0.95, 0.98 and 0.84 Å, respectively, and Uiso(H) = 1.2 Ueq(aryl C) and 1.5 Ueq(methyl C and O).
Figures
Fig. 1.
The molecular structure of (I), showing displacement ellipsoids at the 50% probability level for non-H atoms; dashed lines denote hydrogen bonds.
Fig. 2.
Unit cell packing of (I), showing intermolecular H-bonds resulting in the chains of molecules lying along the b axis; H atoms not involved in H-bonding have been omitted.
Crystal data
| C11H11NO4S | Z = 2 |
| Mr = 253.27 | F000 = 264 |
| Triclinic, P1 | Dx = 1.521 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 6.8523 (1) Å | Cell parameters from 15360 reflections |
| b = 8.3222 (2) Å | θ = 2.9–27.5º |
| c = 10.4880 (2) Å | µ = 0.30 mm−1 |
| α = 72.1321 (11)º | T = 120 (2) K |
| β = 77.9619 (12)º | Shard, colourless |
| γ = 80.0360 (12)º | 0.40 × 0.20 × 0.14 mm |
| V = 552.892 (19) Å3 |
Data collection
| Bruker–Nonius CCD camera on κ-goniostat diffractometer | 2529 independent reflections |
| Radiation source: Bruker Nonius FR591 Rotating Anode | 2248 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.033 |
| Detector resolution: 9.091 pixels mm-1 | θmax = 27.5º |
| T = 120(2) K | θmin = 3.1º |
| φ & ω scans to fill the asymmetric unit | h = −8→8 |
| Absorption correction: multi-scan(SADABS; Sheldrick, 2007) | k = −10→10 |
| Tmin = 0.891, Tmax = 0.960 | l = −13→13 |
| 12691 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.034 | H-atom parameters constrained |
| wR(F2) = 0.092 | w = 1/[σ2(Fo2) + (0.041P)2 + 0.3066P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.11 | (Δ/σ)max = 0.030 |
| 2529 reflections | Δρmax = 0.31 e Å−3 |
| 157 parameters | Δρmin = −0.53 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Experimental. SADABS was used to perform the Absorption correction Estimated minimum and maximum transmission: 0.6696 0.7456 The given Tmin and Tmax were generated using the SHELX SIZE command |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3007 (2) | 0.32223 (18) | 0.37041 (14) | 0.0135 (3) | |
| C2 | 0.2992 (2) | 0.31686 (19) | 0.50412 (15) | 0.0164 (3) | |
| H2 | 0.3246 | 0.2117 | 0.5702 | 0.020* | |
| C3 | 0.2595 (2) | 0.4692 (2) | 0.53933 (16) | 0.0191 (3) | |
| H3 | 0.2575 | 0.4684 | 0.6304 | 0.023* | |
| C4 | 0.2228 (2) | 0.6220 (2) | 0.44168 (16) | 0.0199 (3) | |
| H4 | 0.1973 | 0.7252 | 0.4665 | 0.024* | |
| C5 | 0.2229 (2) | 0.62640 (19) | 0.30802 (15) | 0.0184 (3) | |
| H5 | 0.1983 | 0.7320 | 0.2422 | 0.022* | |
| C6 | 0.2593 (2) | 0.47505 (18) | 0.27093 (14) | 0.0148 (3) | |
| C7 | 0.2558 (2) | 0.47385 (19) | 0.13150 (14) | 0.0153 (3) | |
| C8 | 0.2396 (2) | 0.32881 (19) | 0.09905 (14) | 0.0157 (3) | |
| C9 | 0.2507 (2) | 0.3316 (2) | −0.04098 (16) | 0.0204 (3) | |
| C10 | 0.2518 (3) | 0.1701 (2) | −0.07671 (18) | 0.0332 (4) | |
| H10A | 0.1136 | 0.1532 | −0.0764 | 0.050* | |
| H10B | 0.3113 | 0.0740 | −0.0098 | 0.050* | |
| H10C | 0.3310 | 0.1775 | −0.1672 | 0.050* | |
| C11 | 0.0024 (2) | 0.1379 (2) | 0.25625 (16) | 0.0210 (3) | |
| H11A | −0.0710 | 0.2301 | 0.2933 | 0.031* | |
| H11B | −0.0022 | 0.0298 | 0.3279 | 0.031* | |
| H11C | −0.0598 | 0.1321 | 0.1819 | 0.031* | |
| N1 | 0.21494 (18) | 0.17080 (16) | 0.20411 (12) | 0.0152 (3) | |
| O1 | 0.56456 (16) | 0.12980 (14) | 0.24794 (11) | 0.0202 (2) | |
| O2 | 0.30103 (17) | −0.00587 (13) | 0.42939 (11) | 0.0212 (3) | |
| O3 | 0.27313 (18) | 0.62312 (14) | 0.03808 (11) | 0.0220 (3) | |
| H3A | 0.2785 | 0.6110 | −0.0392 | 0.033* | |
| O4 | 0.26152 (18) | 0.46897 (16) | −0.13289 (11) | 0.0264 (3) | |
| S1 | 0.36145 (5) | 0.13574 (4) | 0.31790 (3) | 0.01424 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0111 (6) | 0.0140 (7) | 0.0157 (7) | −0.0025 (5) | −0.0013 (5) | −0.0046 (5) |
| C2 | 0.0141 (7) | 0.0192 (7) | 0.0154 (7) | −0.0029 (6) | −0.0034 (5) | −0.0033 (6) |
| C3 | 0.0167 (7) | 0.0257 (8) | 0.0181 (7) | −0.0037 (6) | −0.0031 (6) | −0.0101 (6) |
| C4 | 0.0189 (7) | 0.0187 (8) | 0.0251 (8) | −0.0044 (6) | −0.0014 (6) | −0.0107 (6) |
| C5 | 0.0181 (7) | 0.0145 (7) | 0.0205 (7) | −0.0024 (5) | −0.0011 (6) | −0.0032 (6) |
| C6 | 0.0128 (7) | 0.0160 (7) | 0.0148 (7) | −0.0026 (5) | −0.0010 (5) | −0.0034 (5) |
| C7 | 0.0120 (7) | 0.0171 (7) | 0.0132 (7) | 0.0003 (5) | −0.0008 (5) | −0.0009 (5) |
| C8 | 0.0135 (7) | 0.0193 (7) | 0.0129 (7) | 0.0005 (5) | −0.0023 (5) | −0.0037 (5) |
| C9 | 0.0164 (7) | 0.0277 (8) | 0.0176 (7) | 0.0039 (6) | −0.0049 (6) | −0.0092 (6) |
| C10 | 0.0463 (11) | 0.0342 (10) | 0.0245 (9) | 0.0043 (8) | −0.0133 (8) | −0.0164 (7) |
| C11 | 0.0152 (7) | 0.0230 (8) | 0.0249 (8) | −0.0049 (6) | −0.0035 (6) | −0.0056 (6) |
| N1 | 0.0144 (6) | 0.0165 (6) | 0.0157 (6) | −0.0018 (5) | −0.0045 (5) | −0.0050 (5) |
| O1 | 0.0138 (5) | 0.0234 (6) | 0.0246 (6) | 0.0025 (4) | −0.0036 (4) | −0.0108 (5) |
| O2 | 0.0292 (6) | 0.0133 (5) | 0.0202 (5) | −0.0044 (4) | −0.0086 (5) | 0.0005 (4) |
| O3 | 0.0287 (6) | 0.0182 (6) | 0.0143 (5) | −0.0019 (5) | −0.0022 (5) | 0.0009 (4) |
| O4 | 0.0300 (6) | 0.0322 (7) | 0.0140 (5) | 0.0020 (5) | −0.0058 (5) | −0.0038 (5) |
| S1 | 0.01453 (19) | 0.01270 (19) | 0.01608 (19) | 0.00015 (13) | −0.00467 (13) | −0.00442 (13) |
Geometric parameters (Å, °)
| C1—C2 | 1.387 (2) | C8—C9 | 1.448 (2) |
| C1—C6 | 1.404 (2) | C9—O4 | 1.251 (2) |
| C1—S1 | 1.7580 (14) | C9—C10 | 1.501 (2) |
| C2—C3 | 1.394 (2) | C10—H10A | 0.9800 |
| C2—H2 | 0.9500 | C10—H10B | 0.9800 |
| C3—C4 | 1.388 (2) | C10—H10C | 0.9800 |
| C3—H3 | 0.9500 | C11—N1 | 1.4864 (19) |
| C4—C5 | 1.391 (2) | C11—H11A | 0.9800 |
| C4—H4 | 0.9500 | C11—H11B | 0.9800 |
| C5—C6 | 1.397 (2) | C11—H11C | 0.9800 |
| C5—H5 | 0.9500 | N1—S1 | 1.6438 (12) |
| C6—C7 | 1.471 (2) | O1—S1 | 1.4319 (11) |
| C7—O3 | 1.3316 (17) | O2—S1 | 1.4314 (11) |
| C7—C8 | 1.378 (2) | O3—H3A | 0.8400 |
| C8—N1 | 1.4430 (18) | ||
| C2—C1—C6 | 122.15 (13) | O4—C9—C10 | 119.76 (14) |
| C2—C1—S1 | 120.70 (11) | C8—C9—C10 | 120.31 (15) |
| C6—C1—S1 | 117.13 (11) | C9—C10—H10A | 109.5 |
| C1—C2—C3 | 118.54 (14) | C9—C10—H10B | 109.5 |
| C1—C2—H2 | 120.7 | H10A—C10—H10B | 109.5 |
| C3—C2—H2 | 120.7 | C9—C10—H10C | 109.5 |
| C4—C3—C2 | 120.14 (14) | H10A—C10—H10C | 109.5 |
| C4—C3—H3 | 119.9 | H10B—C10—H10C | 109.5 |
| C2—C3—H3 | 119.9 | N1—C11—H11A | 109.5 |
| C3—C4—C5 | 121.04 (14) | N1—C11—H11B | 109.5 |
| C3—C4—H4 | 119.5 | H11A—C11—H11B | 109.5 |
| C5—C4—H4 | 119.5 | N1—C11—H11C | 109.5 |
| C4—C5—C6 | 119.79 (14) | H11A—C11—H11C | 109.5 |
| C4—C5—H5 | 120.1 | H11B—C11—H11C | 109.5 |
| C6—C5—H5 | 120.1 | C8—N1—C11 | 114.26 (11) |
| C5—C6—C1 | 118.30 (13) | C8—N1—S1 | 112.75 (10) |
| C5—C6—C7 | 121.47 (13) | C11—N1—S1 | 116.79 (10) |
| C1—C6—C7 | 120.23 (13) | C7—O3—H3A | 109.5 |
| O3—C7—C8 | 122.30 (13) | O2—S1—O1 | 119.39 (7) |
| O3—C7—C6 | 114.96 (13) | O2—S1—N1 | 108.47 (7) |
| C8—C7—C6 | 122.73 (13) | O1—S1—N1 | 107.24 (6) |
| C7—C8—N1 | 120.44 (12) | O2—S1—C1 | 109.27 (7) |
| C7—C8—C9 | 120.64 (14) | O1—S1—C1 | 108.93 (7) |
| N1—C8—C9 | 118.91 (13) | N1—S1—C1 | 102.15 (6) |
| O4—C9—C8 | 119.92 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3A···O4 | 0.84 | 1.78 | 2.525 (2) | 146 |
| C4—H4···O2i | 0.95 | 2.36 | 3.193 (2) | 146 |
Symmetry codes: (i) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2072).
References
- Bihovsky, R., Tao, M., Mallamo, J. P. & Wells, G. J. (2004). Bioorg. Med. Chem. Lett.14, 1035–1038. [DOI] [PubMed]
- Fabiola, G. F., Pattabhi, V., Manjunatha, S. G., Rao, G. V. & Nagarajan, K. (1998). Acta Cryst. C54, 2001–2003.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Golič, L. & Leban, I. (1987). Acta Cryst. C43, 280–282.
- Nonius (1998). COLLECT Nonius BV, Delft,The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr. & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Pearce, L. J. & Watkin, D. J. (1993). CAMERON Chemical Crystallography Laboratory, University of Oxford, England.
- Sheldrick, G. M. (2007). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Turck, D., Busch, U., Heinzel, G., Narjes, H. & Nehmiz, G. (1996). Clin. Pharm.36, 79–84. [DOI] [PubMed]
- Zia-ur-Rehman, M. Z., Choudary, J. A. & Ahmad, S. (2005). Bull. Korean Chem. Soc.26, 1771–1175.
- Zia-ur-Rehman, M. Z., Choudary, J. A., Ahmad, S. & Siddiqui, H. L. (2006). Chem. Pharm. Bull.54, 1175–1178. [DOI] [PubMed]
- Zia-ur-Rehman, M., Choudary, J. A., Elsegood, M. R. J., Siddiqui, H. L. & Weaver, G. W. (2007). Acta Cryst. E63, o4215–o4216.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808007721/pv2072sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808007721/pv2072Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report