Abstract
In the title compound, C21H22N2O6S, the phenyl ring forms a dihedral angle of 83.17 (7)° with the indole ring system. The methyl group of the ester unit is disordered over two positions with site occupancies of 0.635 (6) and 0.365 (6). In the crystal structure, weak intramolecular C—H⋯O interactions and intermolecular C—H⋯O, C—H⋯N and C—H⋯π interactions are observed.
Related literature
For biological activity, see: Merck (1973 ▶, 1974 ▶); Hendi & Basangoudar (1981 ▶); Kolocouris et al. (1994 ▶); Uchida et al. (1989 ▶); Shaaban et al. (1977 ▶). For the structures of closely related compounds, see: Chakkaravarthi et al. (2007 ▶, 2008 ▶).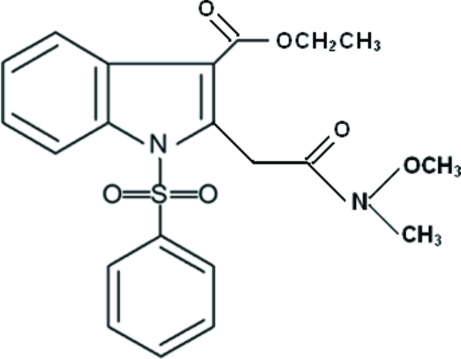
Experimental
Crystal data
C21H22N2O6S
M r = 430.47
Monoclinic,

a = 8.5827 (3) Å
b = 11.0783 (5) Å
c = 21.7433 (8) Å
β = 97.091 (2)°
V = 2051.58 (14) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 295 (2) K
0.30 × 0.20 × 0.20 mm
Data collection
Bruker Kappa APEX2 diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.914, T max = 0.961
29808 measured reflections
7666 independent reflections
3947 reflections with I > 2σ(I)
R int = 0.029
Refinement
R[F 2 > 2σ(F 2)] = 0.064
wR(F 2) = 0.238
S = 1.04
7666 reflections
285 parameters
20 restraints
H-atom parameters constrained
Δρmax = 0.82 e Å−3
Δρmin = −0.62 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808014979/is2294sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808014979/is2294Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O2 | 0.93 | 2.54 | 2.896 (3) | 103 |
| C6—H6⋯O3 | 0.93 | 2.37 | 3.218 (3) | 152 |
| C10—H10⋯O6 | 0.93 | 2.46 | 2.969 (4) | 114 |
| C13—H13⋯O2 | 0.93 | 2.51 | 3.039 (4) | 117 |
| C15—H15A⋯O5 | 0.97 | 2.39 | 2.844 (3) | 108 |
| C15—H15B⋯O1 | 0.97 | 2.17 | 2.847 (3) | 126 |
| C17—H17B⋯O5i | 0.96 | 2.47 | 3.420 (3) | 173 |
| C20—H20C⋯N1ii | 0.97 | 2.45 | 3.351 (4) | 155 |
| C20—H20C⋯O2ii | 0.97 | 2.53 | 3.343 (4) | 142 |
| C21A—H21F⋯Cgiii | 0.96 | 2.80 | 3.196 (13) | 105 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  . Cg is the centroid of the C1–C6 phenyl ring.
. Cg is the centroid of the C1–C6 phenyl ring.
Acknowledgments
The authors acknowledge the Sophisticated Analytical Instrument Facility, Indian Institute of Technology, Madras, for the data collection.
supplementary crystallographic information
Comment
In continuation of our studies of indole derivatives, which are found to possess antihypertensive (Merck, 1973), muscle-relaxant (Hendi & Basangoudar, 1981), antiviral (Kolocouris et al., 1994) antiulcer (Uchida et al., 1989) and analgesic (Shaaban et al., 1977) activities, we determined the crystal structure of the title compound, (I). The geometric parameters of the molecule of (I) (Fig. 1) agree well with those reported for similar structures (Chakkaravarthi et al., 2007, 2008).
The plane of phenyl ring forms 83.17 (7)° with the indole ring system. The plane of N1—S1—C1 forms the dihedral angles of 39.36 (9)° and 69.28 (9)°, respectively, with the phenyl ring and the indole ring. The carboxylate group and N-methoxy-N-methylcarbamide group are approximately orthogonal to each other [dihedral angle 88.45 (1)°] and makes the dihedral angles of 18.61 (14)° and 84.81 (7)°, respectively, with the indole ring. The sum of bond angles around N1 (359.69°) indicates that N1 is sp2-hybridized. The torsion angles O1—S1—N1—C7 and O2—S1—N1—C14 [-9.9 (2)° and 48.3 (2)°, respectively] indicate the syn conformation of the sulfonyl moiety.
The methyl C atom of the ester group is disordered over two positions with occupancies of 0.635 (6) and 0.365 (6). The molecular structure is stabilized by weak intramolecular C—H···O interactions and the crystal packing (Fig. 2) exhibits weak intermolecular C—H···O, C—H···N interactions and a C—H···π interaction involving the ring C1—C6 (centroid Cg) (Table 1).
Experimental
CO gas was passed through the stirred solution of Ethyl-1-phenylsulfonyl-2-bromomethylindole-3-carboxylate (5.0 g, 11.84 mmol) in CH3CN (60 ml). To this, PdCl2 (200 mg, 1.12 mmol) and an in situ prepared HN(OMe)Me solution [HCl.HN(OMe)Me (230 mg, 23.71 mmol) in CH3CN (30 ml)], K2CO3 (3.27 g, 23.69 mmol) and H2O (0.5 ml) were added, stirred for 1 h and filtered] were added. Usual work up followed by evaporation of solvent to give sticky product and the crude product was purified by column chromatography (silica gel) using hexane and ethyl acetate mixture (7:3). Crystals suitable for X-ray analysis were grown by slow evaporation of ethyl acetate solution.
Refinement
H atoms were positioned geometrically (C—H = 0.93 – 0.97 Å) and refined using riding model, with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C). The site occupancy factors for disordered C atom was refined as C21 = 0.635 (6) and C21A = 0.365 (6) during anisotropic refinement. The bond distances C20—C21 and C20—C21A were restrained to be 1.5 (3) Å and anisotropic displacement parameters of atoms C20, C21 and C21A were refined with a similar displacement restraint (SIMU) in the final cycles of refinement.
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 50% probability displacement ellipsoids for non-H atoms.
Fig. 2.
The packing of (I), viewed down the c axis. Intermolecular Hydrogen bonds are shown as dashed lines. H atoms not involved in hydrogen bonding have been omitted.
Crystal data
| C21H22N2O6S | F000 = 904 |
| Mr = 430.47 | Dx = 1.394 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 8381 reflections |
| a = 8.5827 (3) Å | θ = 2.4–29.8º |
| b = 11.0783 (5) Å | µ = 0.20 mm−1 |
| c = 21.7433 (8) Å | T = 295 (2) K |
| β = 97.091 (2)º | Block, colourless |
| V = 2051.58 (14) Å3 | 0.30 × 0.20 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker KappaAPEX2 diffractometer | 7666 independent reflections |
| Radiation source: fine-focus sealed tube | 3947 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.029 |
| T = 295(2) K | θmax = 32.9º |
| ω and φ scans | θmin = 1.9º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −13→12 |
| Tmin = 0.914, Tmax = 0.961 | k = −16→16 |
| 29808 measured reflections | l = −33→32 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.064 | H-atom parameters constrained |
| wR(F2) = 0.238 | w = 1/[σ2(Fo2) + (0.1138P)2 + 0.5762P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 7666 reflections | Δρmax = 0.82 e Å−3 |
| 285 parameters | Δρmin = −0.62 e Å−3 |
| 20 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | 0.67495 (6) | 0.03757 (6) | 0.79732 (2) | 0.05333 (19) | |
| O1 | 0.5718 (2) | 0.1330 (2) | 0.77694 (8) | 0.0696 (5) | |
| O2 | 0.6406 (2) | −0.08033 (19) | 0.77500 (8) | 0.0702 (5) | |
| O3 | 0.7525 (2) | 0.34032 (17) | 0.89517 (10) | 0.0685 (5) | |
| O4 | 0.36103 (19) | 0.42442 (18) | 0.88546 (8) | 0.0636 (5) | |
| O5 | 0.5791 (3) | 0.2410 (2) | 1.02766 (10) | 0.0887 (7) | |
| N1 | 0.6850 (2) | 0.03196 (17) | 0.87461 (8) | 0.0500 (4) | |
| N2 | 0.5241 (2) | 0.4359 (2) | 0.89484 (10) | 0.0581 (5) | |
| C1 | 0.8659 (2) | 0.0746 (2) | 0.78344 (10) | 0.0515 (5) | |
| C2 | 0.9197 (3) | 0.0215 (3) | 0.73297 (13) | 0.0733 (8) | |
| H2 | 0.8590 | −0.0352 | 0.7093 | 0.088* | |
| C3 | 1.0668 (4) | 0.0543 (4) | 0.71811 (17) | 0.0957 (11) | |
| H3 | 1.1061 | 0.0188 | 0.6845 | 0.115* | |
| C4 | 1.1538 (3) | 0.1389 (3) | 0.75290 (16) | 0.0841 (9) | |
| H4 | 1.2520 | 0.1606 | 0.7426 | 0.101* | |
| C5 | 1.0981 (3) | 0.1920 (3) | 0.80265 (15) | 0.0694 (7) | |
| H5 | 1.1588 | 0.2494 | 0.8258 | 0.083* | |
| C6 | 0.9522 (3) | 0.1609 (2) | 0.81871 (12) | 0.0602 (6) | |
| H6 | 0.9133 | 0.1971 | 0.8523 | 0.072* | |
| C7 | 0.6276 (2) | 0.1142 (2) | 0.91567 (10) | 0.0484 (5) | |
| C8 | 0.6784 (3) | 0.0778 (2) | 0.97433 (10) | 0.0508 (5) | |
| C9 | 0.7702 (3) | −0.0301 (2) | 0.97205 (10) | 0.0500 (5) | |
| C10 | 0.8521 (3) | −0.1040 (3) | 1.01676 (12) | 0.0647 (7) | |
| H10 | 0.8514 | −0.0882 | 1.0587 | 0.078* | |
| C11 | 0.9338 (4) | −0.2009 (3) | 0.99756 (16) | 0.0754 (8) | |
| H11 | 0.9901 | −0.2504 | 1.0269 | 0.090* | |
| C12 | 0.9333 (4) | −0.2257 (3) | 0.93565 (16) | 0.0764 (8) | |
| H12 | 0.9885 | −0.2926 | 0.9242 | 0.092* | |
| C13 | 0.8548 (3) | −0.1556 (2) | 0.89043 (14) | 0.0668 (7) | |
| H13 | 0.8556 | −0.1728 | 0.8486 | 0.080* | |
| C14 | 0.7736 (3) | −0.0572 (2) | 0.90994 (11) | 0.0515 (5) | |
| C15 | 0.5211 (3) | 0.2171 (2) | 0.89660 (11) | 0.0553 (5) | |
| H15A | 0.4437 | 0.2243 | 0.9253 | 0.066* | |
| H15B | 0.4659 | 0.2011 | 0.8558 | 0.066* | |
| C16 | 0.6103 (3) | 0.3346 (2) | 0.89525 (10) | 0.0524 (5) | |
| C17 | 0.5823 (4) | 0.5537 (3) | 0.88116 (14) | 0.0696 (7) | |
| H17A | 0.6936 | 0.5570 | 0.8936 | 0.104* | |
| H17B | 0.5312 | 0.6139 | 0.9034 | 0.104* | |
| H17C | 0.5610 | 0.5689 | 0.8375 | 0.104* | |
| C18 | 0.2965 (4) | 0.4541 (3) | 0.94111 (15) | 0.0785 (8) | |
| H18A | 0.3349 | 0.3983 | 0.9732 | 0.118* | |
| H18B | 0.1841 | 0.4495 | 0.9338 | 0.118* | |
| H18C | 0.3273 | 0.5346 | 0.9537 | 0.118* | |
| C19 | 0.6408 (3) | 0.1433 (3) | 1.02879 (12) | 0.0629 (6) | |
| O6 | 0.6834 (2) | 0.0827 (2) | 1.08088 (8) | 0.0821 (6) | |
| C20 | 0.6451 (4) | 0.1324 (5) | 1.13690 (14) | 0.1112 (13) | |
| H20A | 0.6224 | 0.0677 | 1.1645 | 0.133* | 0.635 (6) |
| H20B | 0.5515 | 0.1816 | 1.1284 | 0.133* | 0.635 (6) |
| H20C | 0.5409 | 0.1042 | 1.1426 | 0.133* | 0.365 (6) |
| H20D | 0.6384 | 0.2193 | 1.1316 | 0.133* | 0.365 (6) |
| C21 | 0.7741 (6) | 0.2069 (7) | 1.1677 (3) | 0.1115 (15) | 0.635 (6) |
| H21A | 0.8666 | 0.1582 | 1.1767 | 0.167* | 0.635 (6) |
| H21B | 0.7445 | 0.2389 | 1.2056 | 0.167* | 0.635 (6) |
| H21C | 0.7952 | 0.2722 | 1.1409 | 0.167* | 0.635 (6) |
| C21A | 0.7493 (10) | 0.1083 (14) | 1.1950 (3) | 0.1106 (15) | 0.365 (6) |
| H21D | 0.7271 | 0.0296 | 1.2102 | 0.166* | 0.365 (6) |
| H21E | 0.7315 | 0.1680 | 1.2254 | 0.166* | 0.365 (6) |
| H21F | 0.8568 | 0.1118 | 1.1872 | 0.166* | 0.365 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0435 (3) | 0.0740 (4) | 0.0426 (3) | −0.0057 (2) | 0.0057 (2) | −0.0026 (2) |
| O1 | 0.0498 (9) | 0.1027 (14) | 0.0557 (9) | 0.0104 (9) | 0.0048 (7) | 0.0120 (9) |
| O2 | 0.0692 (11) | 0.0873 (13) | 0.0553 (10) | −0.0237 (10) | 0.0127 (8) | −0.0188 (9) |
| O3 | 0.0493 (9) | 0.0661 (11) | 0.0919 (13) | 0.0024 (8) | 0.0161 (8) | −0.0005 (10) |
| O4 | 0.0513 (9) | 0.0826 (12) | 0.0564 (9) | 0.0145 (8) | 0.0052 (7) | −0.0038 (8) |
| O5 | 0.1006 (16) | 0.0973 (17) | 0.0711 (13) | 0.0049 (13) | 0.0215 (11) | −0.0281 (12) |
| N1 | 0.0524 (10) | 0.0554 (11) | 0.0425 (9) | 0.0009 (8) | 0.0065 (7) | −0.0044 (8) |
| N2 | 0.0512 (10) | 0.0622 (12) | 0.0624 (12) | 0.0070 (9) | 0.0126 (9) | 0.0021 (9) |
| C1 | 0.0411 (10) | 0.0663 (14) | 0.0475 (11) | −0.0022 (9) | 0.0073 (8) | 0.0010 (10) |
| C2 | 0.0582 (14) | 0.100 (2) | 0.0646 (15) | −0.0152 (14) | 0.0181 (12) | −0.0212 (15) |
| C3 | 0.0676 (18) | 0.140 (3) | 0.086 (2) | −0.015 (2) | 0.0361 (16) | −0.026 (2) |
| C4 | 0.0518 (14) | 0.110 (3) | 0.093 (2) | −0.0117 (15) | 0.0209 (14) | 0.0050 (19) |
| C5 | 0.0463 (12) | 0.0724 (17) | 0.0877 (19) | −0.0074 (11) | 0.0009 (12) | −0.0014 (14) |
| C6 | 0.0494 (12) | 0.0692 (15) | 0.0617 (14) | −0.0006 (10) | 0.0059 (10) | −0.0083 (11) |
| C7 | 0.0442 (10) | 0.0533 (12) | 0.0487 (11) | −0.0063 (9) | 0.0099 (8) | −0.0046 (9) |
| C8 | 0.0495 (11) | 0.0593 (13) | 0.0441 (10) | −0.0119 (9) | 0.0079 (8) | −0.0049 (9) |
| C9 | 0.0477 (10) | 0.0534 (12) | 0.0486 (11) | −0.0140 (9) | 0.0045 (8) | 0.0002 (9) |
| C10 | 0.0635 (14) | 0.0711 (16) | 0.0574 (13) | −0.0178 (12) | −0.0003 (11) | 0.0120 (12) |
| C11 | 0.0720 (17) | 0.0644 (17) | 0.086 (2) | −0.0079 (13) | −0.0068 (14) | 0.0207 (15) |
| C12 | 0.0746 (18) | 0.0573 (15) | 0.096 (2) | 0.0052 (13) | 0.0070 (15) | 0.0031 (15) |
| C13 | 0.0689 (15) | 0.0629 (15) | 0.0688 (16) | 0.0048 (12) | 0.0093 (12) | −0.0071 (12) |
| C14 | 0.0495 (11) | 0.0527 (12) | 0.0522 (12) | −0.0051 (9) | 0.0052 (9) | 0.0000 (9) |
| C15 | 0.0435 (10) | 0.0647 (14) | 0.0580 (13) | 0.0024 (10) | 0.0067 (9) | −0.0059 (11) |
| C16 | 0.0504 (11) | 0.0612 (13) | 0.0462 (11) | 0.0039 (10) | 0.0080 (8) | −0.0028 (10) |
| C17 | 0.0807 (18) | 0.0645 (16) | 0.0678 (16) | 0.0088 (13) | 0.0265 (13) | 0.0075 (12) |
| C18 | 0.0718 (17) | 0.091 (2) | 0.0787 (19) | 0.0057 (15) | 0.0315 (14) | −0.0056 (16) |
| C19 | 0.0539 (13) | 0.0821 (18) | 0.0542 (13) | −0.0131 (12) | 0.0123 (10) | −0.0155 (12) |
| O6 | 0.0712 (12) | 0.1331 (19) | 0.0432 (9) | −0.0085 (12) | 0.0115 (8) | −0.0107 (10) |
| C20 | 0.0693 (16) | 0.209 (4) | 0.0573 (15) | 0.010 (2) | 0.0169 (12) | −0.032 (2) |
| C21 | 0.0722 (19) | 0.205 (4) | 0.0585 (18) | 0.007 (2) | 0.0138 (15) | −0.036 (2) |
| C21A | 0.071 (2) | 0.206 (4) | 0.057 (2) | 0.008 (2) | 0.0164 (17) | −0.034 (2) |
Geometric parameters (Å, °)
| S1—O2 | 1.412 (2) | C10—H10 | 0.9300 |
| S1—O1 | 1.415 (2) | C11—C12 | 1.373 (5) |
| S1—N1 | 1.6732 (19) | C11—H11 | 0.9300 |
| S1—C1 | 1.751 (2) | C12—C13 | 1.364 (4) |
| O3—C16 | 1.222 (3) | C12—H12 | 0.9300 |
| O4—N2 | 1.395 (3) | C13—C14 | 1.388 (4) |
| O4—C18 | 1.430 (3) | C13—H13 | 0.9300 |
| O5—C19 | 1.204 (4) | C15—C16 | 1.513 (3) |
| N1—C7 | 1.407 (3) | C15—H15A | 0.9700 |
| N1—C14 | 1.414 (3) | C15—H15B | 0.9700 |
| N2—C16 | 1.343 (3) | C17—H17A | 0.9600 |
| N2—C17 | 1.442 (4) | C17—H17B | 0.9600 |
| C1—C2 | 1.374 (4) | C17—H17C | 0.9600 |
| C1—C6 | 1.383 (3) | C18—H18A | 0.9600 |
| C2—C3 | 1.390 (4) | C18—H18B | 0.9600 |
| C2—H2 | 0.9300 | C18—H18C | 0.9600 |
| C3—C4 | 1.366 (5) | C19—O6 | 1.328 (4) |
| C3—H3 | 0.9300 | O6—C20 | 1.412 (3) |
| C4—C5 | 1.368 (5) | C20—C21 | 1.474 (3) |
| C4—H4 | 0.9300 | C20—C21A | 1.479 (3) |
| C5—C6 | 1.385 (4) | C20—H20A | 0.9700 |
| C5—H5 | 0.9300 | C20—H20B | 0.9700 |
| C6—H6 | 0.9300 | C20—H20C | 0.9700 |
| C7—C8 | 1.358 (3) | C20—H20D | 0.9700 |
| C7—C15 | 1.487 (3) | C21—H21A | 0.9600 |
| C8—C9 | 1.435 (3) | C21—H21B | 0.9600 |
| C8—C19 | 1.458 (3) | C21—H21C | 0.9600 |
| C9—C14 | 1.387 (3) | C21A—H21D | 0.9600 |
| C9—C10 | 1.393 (3) | C21A—H21E | 0.9600 |
| C10—C11 | 1.375 (5) | C21A—H21F | 0.9600 |
| O2—S1—O1 | 119.21 (13) | C14—C13—H13 | 121.7 |
| O2—S1—N1 | 107.07 (11) | C9—C14—C13 | 122.6 (2) |
| O1—S1—N1 | 107.15 (10) | C9—C14—N1 | 107.6 (2) |
| O2—S1—C1 | 108.50 (12) | C13—C14—N1 | 129.7 (2) |
| O1—S1—C1 | 109.49 (12) | C7—C15—C16 | 111.70 (18) |
| N1—S1—C1 | 104.39 (10) | C7—C15—H15A | 109.3 |
| N2—O4—C18 | 110.0 (2) | C16—C15—H15A | 109.3 |
| C7—N1—C14 | 108.36 (18) | C7—C15—H15B | 109.3 |
| C7—N1—S1 | 129.40 (16) | C16—C15—H15B | 109.3 |
| C14—N1—S1 | 121.93 (15) | H15A—C15—H15B | 107.9 |
| C16—N2—O4 | 117.8 (2) | O3—C16—N2 | 120.4 (2) |
| C16—N2—C17 | 123.6 (2) | O3—C16—C15 | 123.5 (2) |
| O4—N2—C17 | 114.8 (2) | N2—C16—C15 | 116.1 (2) |
| C2—C1—C6 | 121.9 (2) | N2—C17—H17A | 109.5 |
| C2—C1—S1 | 116.83 (19) | N2—C17—H17B | 109.5 |
| C6—C1—S1 | 121.00 (18) | H17A—C17—H17B | 109.5 |
| C1—C2—C3 | 118.6 (3) | N2—C17—H17C | 109.5 |
| C1—C2—H2 | 120.7 | H17A—C17—H17C | 109.5 |
| C3—C2—H2 | 120.7 | H17B—C17—H17C | 109.5 |
| C4—C3—C2 | 120.0 (3) | O4—C18—H18A | 109.5 |
| C4—C3—H3 | 120.0 | O4—C18—H18B | 109.5 |
| C2—C3—H3 | 120.0 | H18A—C18—H18B | 109.5 |
| C3—C4—C5 | 120.8 (3) | O4—C18—H18C | 109.5 |
| C3—C4—H4 | 119.6 | H18A—C18—H18C | 109.5 |
| C5—C4—H4 | 119.6 | H18B—C18—H18C | 109.5 |
| C4—C5—C6 | 120.5 (3) | O5—C19—O6 | 123.1 (3) |
| C4—C5—H5 | 119.8 | O5—C19—C8 | 124.9 (3) |
| C6—C5—H5 | 119.8 | O6—C19—C8 | 112.1 (3) |
| C1—C6—C5 | 118.1 (2) | C19—O6—C20 | 118.0 (3) |
| C1—C6—H6 | 120.9 | O6—C20—C21 | 111.7 (3) |
| C5—C6—H6 | 120.9 | O6—C20—C21A | 119.1 (5) |
| C8—C7—N1 | 107.8 (2) | C21—C20—C21A | 51.1 (6) |
| C8—C7—C15 | 127.3 (2) | O6—C20—H20A | 109.3 |
| N1—C7—C15 | 124.8 (2) | C21—C20—H20A | 109.3 |
| C7—C8—C9 | 109.2 (2) | O6—C20—H20B | 109.3 |
| C7—C8—C19 | 122.5 (2) | C21—C20—H20B | 109.3 |
| C9—C8—C19 | 128.3 (2) | H20A—C20—H20B | 108.0 |
| C14—C9—C10 | 118.8 (2) | O6—C20—H20C | 107.5 |
| C14—C9—C8 | 106.96 (19) | C21A—C20—H20C | 107.5 |
| C10—C9—C8 | 134.2 (2) | O6—C20—H20D | 107.5 |
| C11—C10—C9 | 118.7 (3) | C21A—C20—H20D | 107.5 |
| C11—C10—H10 | 120.7 | H20C—C20—H20D | 107.0 |
| C9—C10—H10 | 120.7 | C20—C21—H21A | 109.5 |
| C12—C11—C10 | 120.9 (3) | C20—C21—H21B | 109.5 |
| C12—C11—H11 | 119.6 | C20—C21—H21C | 109.5 |
| C10—C11—H11 | 119.6 | C20—C21A—H21D | 109.5 |
| C13—C12—C11 | 122.3 (3) | C20—C21A—H21E | 109.5 |
| C13—C12—H12 | 118.9 | H21D—C21A—H21E | 109.5 |
| C11—C12—H12 | 118.9 | C20—C21A—H21F | 109.5 |
| C12—C13—C14 | 116.7 (3) | H21D—C21A—H21F | 109.5 |
| C12—C13—H13 | 121.7 | H21E—C21A—H21F | 109.5 |
| O2—S1—N1—C7 | −138.9 (2) | C19—C8—C9—C10 | −1.3 (4) |
| O1—S1—N1—C7 | −9.9 (2) | C14—C9—C10—C11 | 0.3 (3) |
| C1—S1—N1—C7 | 106.2 (2) | C8—C9—C10—C11 | −177.9 (2) |
| O2—S1—N1—C14 | 48.3 (2) | C9—C10—C11—C12 | −0.9 (4) |
| O1—S1—N1—C14 | 177.31 (18) | C10—C11—C12—C13 | 0.9 (5) |
| C1—S1—N1—C14 | −66.6 (2) | C11—C12—C13—C14 | −0.3 (4) |
| C18—O4—N2—C16 | 111.0 (3) | C10—C9—C14—C13 | 0.3 (3) |
| C18—O4—N2—C17 | −90.2 (3) | C8—C9—C14—C13 | 178.9 (2) |
| O2—S1—C1—C2 | 29.4 (3) | C10—C9—C14—N1 | −178.86 (19) |
| O1—S1—C1—C2 | −102.2 (2) | C8—C9—C14—N1 | −0.3 (2) |
| N1—S1—C1—C2 | 143.3 (2) | C12—C13—C14—C9 | −0.3 (4) |
| O2—S1—C1—C6 | −155.9 (2) | C12—C13—C14—N1 | 178.7 (3) |
| O1—S1—C1—C6 | 72.4 (2) | C7—N1—C14—C9 | 0.2 (2) |
| N1—S1—C1—C6 | −42.0 (2) | S1—N1—C14—C9 | 174.29 (15) |
| C6—C1—C2—C3 | 1.4 (5) | C7—N1—C14—C13 | −178.9 (2) |
| S1—C1—C2—C3 | 176.0 (3) | S1—N1—C14—C13 | −4.8 (3) |
| C1—C2—C3—C4 | −0.8 (6) | C8—C7—C15—C16 | 85.8 (3) |
| C2—C3—C4—C5 | 0.2 (6) | N1—C7—C15—C16 | −98.8 (2) |
| C3—C4—C5—C6 | 0.0 (5) | O4—N2—C16—O3 | 170.5 (2) |
| C2—C1—C6—C5 | −1.2 (4) | C17—N2—C16—O3 | 13.6 (4) |
| S1—C1—C6—C5 | −175.6 (2) | O4—N2—C16—C15 | −10.3 (3) |
| C4—C5—C6—C1 | 0.5 (4) | C17—N2—C16—C15 | −167.1 (2) |
| C14—N1—C7—C8 | 0.0 (2) | C7—C15—C16—O3 | 16.2 (3) |
| S1—N1—C7—C8 | −173.54 (16) | C7—C15—C16—N2 | −163.0 (2) |
| C14—N1—C7—C15 | −176.2 (2) | C7—C8—C19—O5 | −9.2 (4) |
| S1—N1—C7—C15 | 10.3 (3) | C9—C8—C19—O5 | 170.6 (3) |
| N1—C7—C8—C9 | −0.2 (2) | C7—C8—C19—O6 | 171.1 (2) |
| C15—C7—C8—C9 | 175.9 (2) | C9—C8—C19—O6 | −9.1 (3) |
| N1—C7—C8—C19 | 179.7 (2) | O5—C19—O6—C20 | 4.1 (4) |
| C15—C7—C8—C19 | −4.3 (4) | C8—C19—O6—C20 | −176.2 (2) |
| C7—C8—C9—C14 | 0.3 (2) | C19—O6—C20—C21 | −93.0 (5) |
| C19—C8—C9—C14 | −179.6 (2) | C19—O6—C20—C21A | −149.5 (7) |
| C7—C8—C9—C10 | 178.6 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O2 | 0.93 | 2.54 | 2.896 (3) | 103 |
| C6—H6···O3 | 0.93 | 2.37 | 3.218 (3) | 152 |
| C10—H10···O6 | 0.93 | 2.46 | 2.969 (4) | 114 |
| C13—H13···O2 | 0.93 | 2.51 | 3.039 (4) | 117 |
| C15—H15A···O5 | 0.97 | 2.39 | 2.844 (3) | 108 |
| C15—H15B···O1 | 0.97 | 2.17 | 2.847 (3) | 126 |
| C17—H17B···O5i | 0.96 | 2.47 | 3.420 (3) | 173 |
| C20—H20C···N1ii | 0.97 | 2.45 | 3.351 (4) | 155 |
| C20—H20C···O2ii | 0.97 | 2.53 | 3.343 (4) | 142 |
| C21A—H21F···Cgiii | 0.96 | 2.80 | 3.196 (13) | 105 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x+1, −y, −z+2; (iii) −x, −y+2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2294).
References
- Bruker (2004). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Chakkaravarthi, G., Dhayalan, V., Mohanakrishnan, A. K. & Manivannan, V. (2007). Acta Cryst. E63, o4724.
- Chakkaravarthi, G., Dhayalan, V., Mohanakrishnan, A. K. & Manivannan, V. (2008). Acta Cryst. E64, o392. [DOI] [PMC free article] [PubMed]
- Hendi, S. & Basangoudar, L. D. (1981). Indian J. Chem.208, 285–288.
- Kolocouris, N., Foscolos, G. B., Kolocouris, A., Marakos, P., Pouli, N., Fytas, G., Ikeda, S. & De Clercq, E. (1994). J. Med. Chem.37, 2896–2902. [DOI] [PubMed]
- Merck (1973). French Patent 2 163 554.
- Merck (1974). Chem. Abstr 80, 27298.
- Shaaban, M. A., Ghoneim, K. M. & Khalifa, M. (1977). Pharmazie, 32, 90–92. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Uchida, M., Chihiro, M., Morita, S., Kanbe, T., Yamashita, H., Yamasaki, K., Yabuuchi, Y. & Nakagawa, K. (1989). Chem. Pharm. Bull. (Tokyo), 37, 2109–2116. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808014979/is2294sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808014979/is2294Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




