Abstract
We report the properties of the new BseMII restriction and modification enzymes from Bacillus stearothermophilus Isl 15-111, which recognize the 5′-CTCAG sequence, and the nucleotide sequence of the genes encoding them. The restriction endonuclease R.BseMII makes a staggered cut at the tenth base pair downstream of the recognition sequence on the upper strand, producing a two base 3′-protruding end. Magnesium ions and S-adenosyl-l-methionine (AdoMet) are required for cleavage. S-adenosylhomocysteine and sinefungin can replace AdoMet in the cleavage reaction. The BseMII methyltransferase modifies unique adenine residues in both strands of the target sequence 5′-CTCAG-3′/5′-CTGAG-3′. Monomeric R.BseMII in addition to endonucleolytic activity also possesses methyltransferase activity that modifies the A base only within the 5′-CTCAG strand of the target duplex. The deduced amino acid sequence of the restriction endonuclease contains conserved motifs of DNA N6-adenine methylases involved in S-adenosyl-l-methionine binding and catalysis. According to its structure and enzymatic properties, R.BseMII may be regarded as a representative of the type IV restriction endonucleases.
INTRODUCTION
Restriction–modification (R–M) systems are traditionally divided into three classes, designated type I, II and III, on the basis of their enzyme subunit composition, cofactor requirements, substrate specificity and reaction products (1). The most prolific type II R–M systems comprise two separate enzymes, a homodimeric Mg2+-dependent endonuclease and a monomeric DNA methyltransferase, which only requires S-adenosyl-l-methionine (AdoMet) as a cofactor. Most known type II enzymes recognize palindromic nucleotide sequences 4–8 bp in length. Modification and cleavage occurs within the recognition site. Some restriction endonucleases, originally defined as type II, mostly those recognizing asymmetric continuous or interrupted sequences, which cleave within or at a short distance from those sequences on one or both sides, turned out to be substantially different from type II enzymes. It is now suggested that they may constitute new types or kinds (type IIS, type IIT, type IV, BcgI-like, HaeIV) of restriction enzymes (2–7).
Of the type IV R–M systems only the Eco57I R–M enzymes have been characterized so far, both with respect to biochemical properties and primary structure peculiarities (5,8). The single large polypeptide of the Eco57I endonuclease is a bifunctional enzyme that possesses both DNA cleavage and methylation activities (5). Only one strand of the asymmetric recognition sequence is methylated by the enzyme. The cleavage, which occurs at a fixed distance from the recognition sequence, is absolutely dependent on Mg2+ and is stimulated by AdoMet. In addition to the bifunctional restriction enzyme, the Eco57I R–M system also includes a separate Eco57I methyltransferase, which modifies both strands of the target sequence. Consistent with the endonucleolytic and methylation functions, the amino acid sequence of R.Eco57I contains the catalytic DNA cleavage/Mg2+-binding motif of restriction endonucleases (9) and conserved sequence motifs of DNA N6-adenine methyltransferases (8). On the basis of the structural organization and enzymatic properties of R.Eco57I, it has been suggested that its evolution involved fusion of Mod and Res subunits of type III enzymes and that type IV restriction endonucleases represent an intermediate form in the evolutionary pathway between the type III and type IIS enzymes (5,8).
Several other Eco57I-like endonucleases have been isolated from different bacteria: GsuI from Gluconobacter suboxydans (10); BspLU11III from Bacillus species LU11 (11); Bce83I from Bacillus cereus (12); BsgI from Bacillus sphaericus [H.Kong, cited by Roberts and Macelis as unpublished observations (13)]; and MmeI from Methylophilus methylotrophus (14). This suggests that type IV R–M systems may have a wider distribution than is currently recognized. The unique requirement for AdoMet may have limited their identification until now. Both the identification of new Eco57I-like enzymes as well as their detailed characterization are necessary for a better understanding of their evolution and to address the suggestion (8) that they represent a R–M system of a distinct class.
In this study, we purified a novel restriction endonuclease BseMII (R.BseMII) and its cognate methyltransferase (M.BseMII) with the new specificity 5′-CTCAG, and further cloned the genes of this R–M system. The properties of the BseMII restriction endonuclease and methyltransferase indicate similarity to Eco57I-like type IV R–M enzymes.
MATERIALS AND METHODS
Bacterial strains, plasmids and reagents
Bacillus stearothermophilus Isl 15-111, used for the isolation of the BseMII R–M enzymes and as the source of DNA for cloning of the BseMII R–M system, was provided by MBI Fermentas. Escherichia coli strain ER2267 (New England Biolabs) was used as a host in the cloning procedures. Plasmids pBR322 (15), pUC57 (MBI Fermentas), pACYC184 (16) and pMCL200 (17) were used as cloning vectors. Escherichia coli ER2267 was grown in LB medium (18) at 37°C; B.stearothermophilus Isl 11-115 was grown in Trypton soya broth (OXOID) at the same temperature. Ampicillin, kanamycin and chloramphenicol were added to Luria broth or LB-agar at concentrations of 60, 50 and 30 µg/ml, respectively, where needed. Escherichia coli ER2267 transformations were carried out either by electroporation or by the CaCl2-heat shock method (18). All enzymes, kits, DNAs of λ, pBR322, φX174, pUC18 and M13mp18, oligonucleotides, dNTPs, N4mdCyd, C5mdCyd and N6mdAdo were from MBI Fermentas. S-adenosyl-l-[3H-methyl]-methionine, [α-33P]dATP, [γ-33P]dATP and nucleic acid transfer membrane Hybond N+ were purchased from Amersham. Non-radioactive AdoMet was obtained from Serva. Molecular weight markers for sodium dodecyl sulfate (SDS) electrophoresis and gel filtration were from Boehringer Mannheim and Amersham Pharmacia Biotech, respectively.
Endonuclease and methylase assays
Restriction endonuclease activity was assayed in 50 µl reaction buffer (33 mM Tris-acetate pH 7.9, 10 mM Mg-acetate, 66 mM K-acetate, 0.01 mM AdoMet, 0.1 mg/ml BSA) and 1 µg of λ DNA. Either 3 µl of chromatographic fractions or various amounts (1, 3 or 5 µl) of cell-free extracts prepared as described (19) were added to the reaction mixture and incubated for 10–30 min at 55°C. The reaction products were resolved by electrophoresis on 0.8–1.4% agarose gels. One unit of the endonuclease is defined as the amount required to hydrolyze 1 µg of λ DNA in 1 h at 55°C. To determine the M.BseMII-specific modification generated in vivo, plasmid DNA isolated from transformants was challenged with an excess of R.BseMII followed by agarose gel electrophoresis. The in vitro modification activities of M.BseMII and R.BseMII were tested by the DNA protection assay where 1 µg of λ DNA was used as substrate in 50 µl of reaction mixture (25 mM Tris–HCl pH 8.0, 20 mM NaCl, 1 mM DTT, 0.1 mM AdoMet, 0.1 mg/ml BSA). The reaction was initiated by the addition of 2–3 µl aliquots of sample solution and incubated for 15–60 min at 55°C. MgCl2 solution was then added to the reaction mixture to a final concentration of 10 mM. The probes were challenged with an excess of R.BseMII for 1 h and the reaction products were then resolved by agarose gel electrophoresis. One unit of the modification activity is defined as the amount of either M.BseMII or R.BseMII that, in 1 h at 55°C, renders 1 µg of λ DNA resistant to cleavage by R.BseMII.
Cell growth and initial fractionation
Bacillus stearothermophilus Isl 11-115 cells were grown to late logarithmic phase with aeration, harvested by centrifugation and stored at –20°C. All further steps were carried out at 4°C. The frozen cells (250 g) were thawed in 500 ml of buffer A (10 mM K-phosphate pH 7.4, 1 mM EDTA, 7 mM 2-mercaptoethanol), containing 0.1 M NaCl, and the suspension was sonicated. Following cell rupture, insoluble material was removed by centrifugation at 30 000 g for 1 h. The supernatant was applied to a heparin–Sepharose column (5 × 27 cm) pre-equilibrated with buffer A, 0.1 M NaCl. The column was washed with the same buffer and eluted with a 3.5 l linear gradient of 0.1–0.8 M NaCl. Fractions were assayed for specific DNA cleavage and methylation activities. BseMII restriction endonuclease eluted at between 0.56 and 0.60 M NaCl. The peak fractions were pooled, dialyzed against buffer A, 0.1 M NaCl, and used for further purification of R.BseMII. The pooled fractions in parallel with R.BseMII contained another site-specific endonuclease BseMI (an isoschizomer of BsrDI), which eluted from the column together with R.BseMII. Two peaks of BseMII-specific methylase activity eluted from the heparin–Sepharose column. The first coincided with the endonuclease peak, while the second eluted at a lower salt concentration, between 0.45 and 0.48 M NaCl. Fractions of the second methylase peak were pooled, dialyzed against buffer A, 0.1 M NaCl, and used for purification of the M.BseMII modification enzyme.
Purification of the BseMII restriction endonuclease
The dialyzed endonuclease pool recovered from the heparin–Sepharose column was applied to an AH–Sepharose column (2.5 × 30 cm) equilibrated with the dialysis buffer. The column was washed with the same buffer and eluted with a 1.2 l linear gradient of 0.1–0.8 M NaCl in buffer A. R.BseMII eluted between 0.33 and 0.43 M NaCl, while activity of the BseMI endonuclease was found in the flow-through fraction. Peak fractions containing R.BseMII were pooled and dialyzed against buffer A containing 0.2 M NaCl. The enzyme pool from the AH–Sepharose column was applied to a Red–agarose column (2.5 × 20 cm) equilibrated with buffer A plus 0.2 M NaCl. The column was washed with the same buffer and subsequently developed with a 1 l linear gradient of 0.2–0.8 M NaCl in buffer A. The peak fractions containing the endonuclease (0.38–0.44 M NaCl) were pooled, dialyzed against storage buffer (10 mM Tris–HCl pH 7.4, 100 mM KCl, 1 mM EDTA, 1 mM DTT and 50% glycerol) and stored at –20°C.
Purification of the BseMII methyltransferase
The methylase preparation obtained after chromatography on heparin–Sepharose and subsequent dialysis was applied to an AH–Sepharose column (2.6 × 29 cm), equilibrated with buffer A containing 0.1 M NaCl. The enzyme was found in the flow-through fraction, which was applied to a Red–Sepharose (2.6 × 17 cm) column equilibrated with buffer A plus 0.1 M NaCl. The column was developed with a 650 ml linear gradient of 0.1–0.8 M NaCl. The methylase activity eluted at 0.58–0.65 M NaCl. Peak fractions were pooled, dialyzed against the same storage buffer used for BseMII endonuclease and stored at –20°C.
Molecular weight studies
Gel electrophoresis. Protein electrophoresis was carried out in 8% polyacrylamide gels in the presence of 1% SDS according to the method of Laemmli (20). Protein bands were visualized by staining with 0.1% Coomassie brilliant blue R-250. Aldolase (39.2 kDa), glutamate dehydrogenase (55.6 kDa), fructose-6-phosphate kinase (85.2 kDa), β-galactosidase (116.4 kDa) and α2-macroglobulin (170 kDa) were used as standard molecular weight markers.
Gel filtration. Size-exclusion chromatography on a Superdex 200 HR 10/30 column in 10 mM K2PO4 buffer pH 7.4, 1 mM EDTA, 7 mM 2-mercaptoethanol, 10% glycerol, was used to determine the molecular weight of the purified BseMII enzymes. Two concentrations of NaCl (0.2 and 1.0 M) in the above buffer were used in separate experiments. Ovalbumin (43 kDa), bovine serum albumin (67 kDa) and aldolase (158 kDa) were used as standard molecular weight markers.
Determination of the recognition sequence and cleavage site
The recognition sequence of R.BseMII was inferred by restriction mapping of recognition sites on DNAs of phages λ, φX174 and M13mp18 and plasmids pBR322 and pUC18. Then the fragments predicted by cleavage of the inferred recognition sites were compared with the observed restriction fragments from BseMII cleavage of the DNAs.
DNA of plasmid pBR322 was used as a template to characterize the cleavage position of R.BseMII. A 20mer oligodeoxyribonucleotide complementary to pBR322 at positions 2390–2370 (ccw primer) was used in reverse sequencing through the BseMII site located at position 2283. Four dideoxy sequencing reactions (A, T, G and C) using [α-33P]dATP were carried out (21). The same primer and template were used in a fifth non-terminating reaction, which also included T7 DNA polymerase, dNTP and [α-33P]dATP. The extension reaction was heat inactivated and then the radiolabeled DNA was digested by BseMII and the reaction mix was divided in half. One sample was treated with T4 DNA polymerase. Both samples were diluted with sequencing dye and loaded on a standard sequencing gel together with the dideoxy sequencing reactions.
Determination of the methylation specificity
The modification specificity of the BseMII restriction endonuclease and that of the separate BseMII methylase were determined using two oligodeoxyribonucleotides (26mer and 22mer) that form a duplex DNA fragment containing the target sequence (shown in bold) for the BseMII enzymes:
5′- CCTTGGCTCAGCCTATTTGGTT -3′ (1)
3′- GGCGGAACCGAGTCGGATAAACCAAT-5′ (2)
In some experiments the oligoduplex, containing sequence CTAAG instead of CTCAG, was used as a control. The modification reactions were performed in 100 µl of the R.BseMII reaction buffer containing 100 pmol of the synthetic substrate and 3 µCi of [3H-methyl]-AdoMet (67 Ci/mmol specific activity). Twenty methylase units of the R.BseMII or M.BseMII was added to the reaction mixture and incubated for 3 h at 55°C. Aliquots of the reaction mixture were enzymatically hydrolyzed to deoxynucleosides (22) and the nature of the modified products was determined by high-performance liquid chromatography (HPLC) analysis (23) using methylated deoxynucleosides N4mdCyd, C5mdCyd and N6mdAdo as standards. The fractions corresponding to those of the standards were collected and counted for 3H-radioactivity in scintillation fluid.
To determine the capacity of the BseMII enzymes to modify different strands of the substrate, the individual strands of the methylated oligodeoxynucleotide duplex were separated by polyacrylamide gel electrophoresis (PAGE) under denaturing conditions (18). Ethidium bromide-stained DNA strands were identified, excised, extracted from the gel into 150 µl of H2O and counted for their 3H-radioactivity.
Isolation of DNA and recombinant DNA techniques
Bacillus stearothermophilus Isl 15-111 genomic DNA was extracted and purified essentially as described previously (24). Plasmids were prepared by the alkaline lysis procedure (25) and purified additionally according to the technique of Marko et al. (26). Recombinant plasmid construction, restriction and deletion mapping, agarose gel electrophoresis and isolation of individual DNA restriction fragments from agarose gels were carried out using standard techniques (18). A library of nested deletions used for the sequencing of R–M genes was obtained using the ExoIII/S1 Deletion Kit (MBI Fermentas). The nucleotide sequence was determined by the Sanger dideoxynucleotide chain termination method (27) using the Cycle ReaderTM DNA Sequencing Kit (MBI Fermentas) and standard end-labeled M13/pUC sequencing primers.
Cloning of the BseMII methyltransferase gene
The methyltransferase selection method (28) was used to clone the gene for the BseMII methylase. The B.stearothermophilus Isl 15-111 gene library was constructed by partially digesting genomic DNA with Bsp143I and ligating the fragments into BamHI cleaved and dephosphorylated pBR322. The ligation mixture was used to electrotransform E.coli ER2267. Plasmid DNA was isolated from the pooled ampicillin-resistant colonies, digested with an excess of R.BseMII and used to transform E.coli ER2267. Plasmids of the resulting transformants were subjected to the next round of selection and then randomly picked transformants were screened for resistance to R.BseMII digestion.
Cloning and expression of the BseMII restriction endonuclease gene
The restriction map of the DNA surrounding the BseMII methyltransferase gene in the B.stearothermophilus Isl 15-111 genome was constructed by Southern blotting. Transfer of the DNA, digested with various restriction enzymes, from agarose gels to Hybond N+ nylon membrane was performed as described previously (18). A HexaLabelTM DNA Labeling Kit (MBI Fermentas) was used for the preparation of the radioactive probe containing the BglII–SmiI fragment of the cloned 3′-terminal part of the putative R.BseMII gene (see Fig. 6). To clone the Bst1107I fragment located downstream from the BseMII methyltransferase gene, B.stearothermophilus Isl 15-111 DNA was digested with Bst1107I, the reaction products were resolved on an agarose gel and DNA fragments of ~5 kb in length were isolated. They were ligated to Eco32I cleaved and dephosphorylated pACYC184. The ligation mixture was used to transform competent E.coli ER2267 cells possessing the functionally active gene for the BseMII methylase (plasmid pBR-BseMIIM17). Plasmids of the resulting 350 transformants were tested by restriction mapping for the presence of the desired Bst1107I fragment. Two plasmids carrying the expected fragment in the same orientation were isolated. One of them, denoted pAC-BseMIIR5, was used for subsequent experiments. In order to enhance the transcription of the open reading frame (ORF) of the putative BseMII endonuclease found within the cloned and sequenced Bst1107I fragment (Results), the BcuI–PvuII fragment, 3.5 kb in size (see Fig. 6), was subcloned from pAC-BseMIIR5 into the BcuI and Eco32I cleaved vector pMCL200 downstream of the Plac promoter. The resulting plasmid was used to transform E.coli ER2267[pBR-BseMIIM17]. The selected transformants were grown in the presence of 0.5 mM of the Plac inducer isopropyl-β-d-thiogalactopyranoside and assayed for BseMII restriction endonuclease activity.
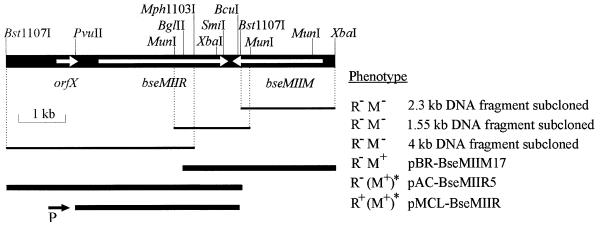
Figure 6.
Restriction endonuclease and subcloning mapping of genes encoding the BseMII R–M system. The fragment of the B.stearothermophilus Isl 15-111 DNA cloned and sequenced in this work is shown. Thin lines indicate DNA fragments that were subcloned into the pUC57 and sequenced. Thick lines represent DNA fragments present in recombinant plasmids pBR-BseMIIM17, pAC-BseMIIR5 and pMCL-BseMIIR. The determination of R–M phenotypes is described in Materials and Methods. M+, methylase activity; M–, no activity; R+, endonuclease activity; R–, no activity. Designations for genetic markers are: orfX, ORF of unknown function; bseMIIR, gene encoding the BseMII restriction endonuclease; bseMIIM, gene encoding the BseMII methyltransferase. Direction of transcription from the vector lac promoter P in plasmid pMCL-BseMIIR is indicated by the black arrow. The asterisks and parentheses denote plasmid provided in trans (pBR-BseMIIM17) and its phenotype (M+).
Comparison of deduced amino acid sequences
The deduced amino acid sequences were compared with sequences deposited with the EMBL and SWISS-PROT sequence databases using the FASTA (29) program. The similarity of the BseMII restriction endonuclease and methylase sequences was tested using the HR-SEARCH program (30).
RESULTS
Purification of BseMII restriction endonuclease
Bacillus stearothermophilus strain Isl 15-111 produces the type II restriction endonuclease BseMI, an isoschisomer of BsrDI (J.Vitkute, Institute of Biotechnology, personal communication), and another site-specific endodeoxyribonuclease BseMII, which unlike other type II enzymes was found to be activated by AdoMet. Fractionation of the B.stearothermophilus Isl 15-111 crude extract on a heparin–Sepharose column did not result in separation of R.BseMI and R.BseMII activities: both eluted together between 0.56 and 0.66 M NaCl. The enzymes were separated on an AH–Sepharose column: R.BseMI was found in the flow-through fraction, while R.BseMII activity appeared between 0.33 and 0.43 M NaCl. The additional purification of the R.BseMII enzyme on a Red–agarose column (activity eluted between 0.38 and 0.44 M NaCl) resulted in a homogeneous preparation of R.BseMII (Fig. 1). During the purification of the restriction endonuclease, chromatographic fractions were also assessed for BseMII-specific DNA modification using the DNA protection assay. BseMII endonuclease and methylation activities copurified during all isolation steps (data not shown). In addition, another BseMII-specific DNA methylation activity was detected, which eluted between 0.45 and 0.48 M NaCl on heparin–Sepharose, was found in the flow-through fraction on AH–Sepharose and eluted between 0.58 and 0.65 M NaCl on Red–agarose. The same chromatographic sorbents were used for purification of the BseMII restriction endonuclease and the separate methylase M.BseMII. The methylase activity associated with the endonuclease and that of M.BseMII were very different in their elution profiles making it unlikely that M.BseMII would contaminate the final preparation of R.BseMII.
Figure 1.
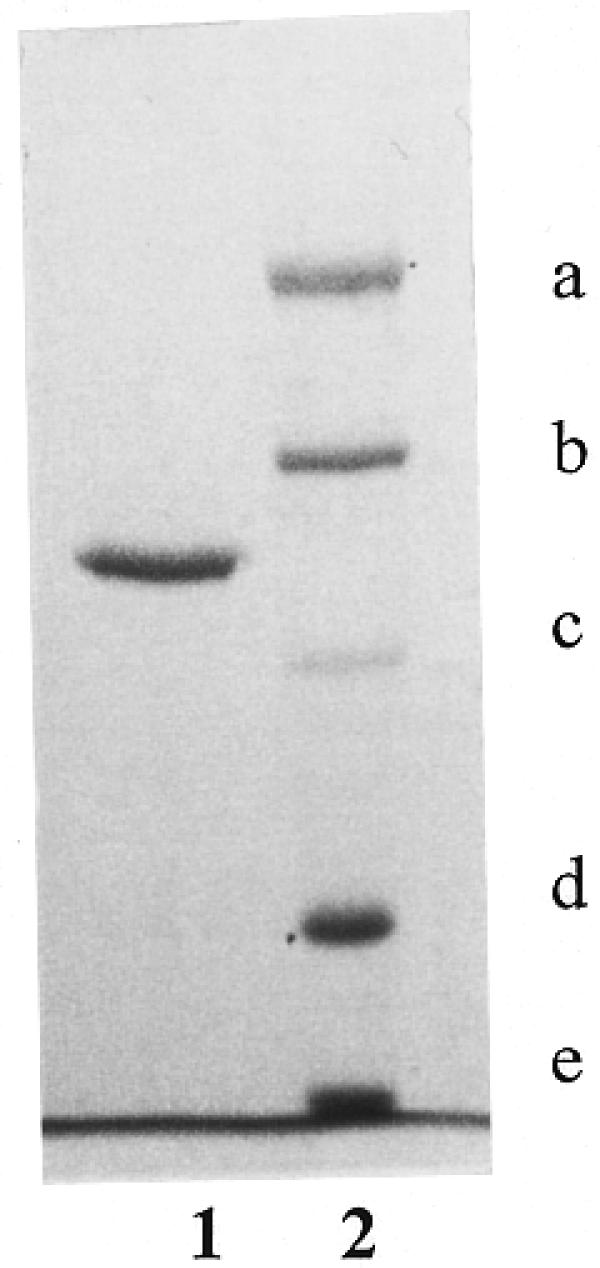
SDS–PAGE analysis of the purified BseMII endonuclease (lane 1) with size standards (lane 2) as follows: (a) α2-macroglobulin (170 kDa), (b) β-galactosidase (116.4 kDa), (c) fructose-6-phosphate kinase (85.2 kDa), (d) glutamate dehydrogenase (55.6 kDa) and (e) aldolase (39.2 kDa).
Data presented in Figure 2 illustrate that a homogeneous preparation of R.BseMII possesses not only endonucleolytic activity (lane 2) but also methylation activity, which renders DNA resistant to R.BseMII cleavage (lane 4). A control experiment confirms (lane 5) that this observation is not due to the experimental artifact, since λ DNA added to the methylated λ DNA yields the same R.BseMII cleavage pattern as in lane 2. Preincubation of λ DNA with the enzyme in the reaction mixture without cofactors AdoMet and Mg2+ did not reveal any DNA protection (lane 3). This observation indicates that the enzyme is free of endogenous AdoMet, which could copurify with R.BseMII.
Figure 2.
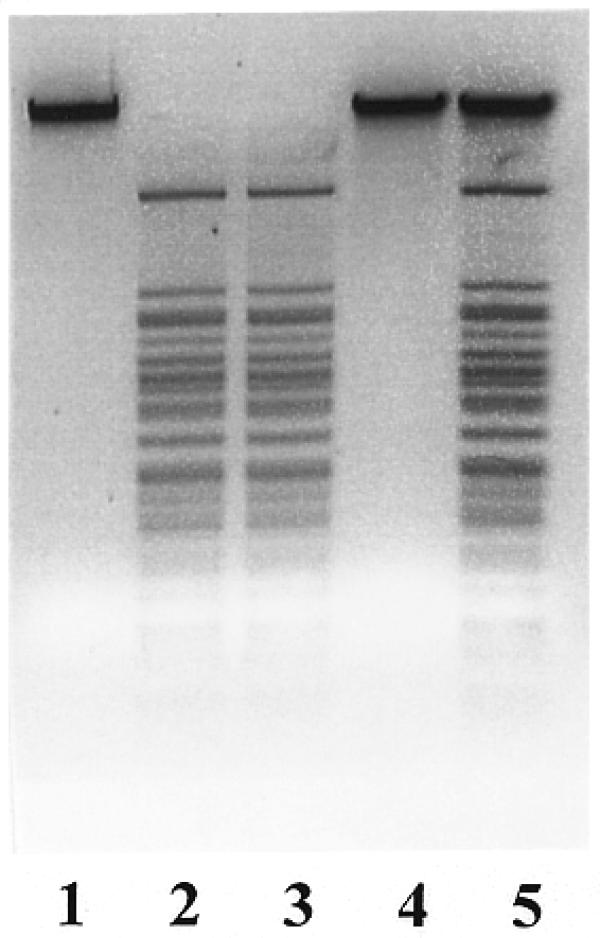
Bifunctionality of R.BseMII. Lane 1, λ DNA; lane 2, λ DNA incubated with 5 U of R.BseMII in a reaction mixture containing 10 mM Mg2+ and 10 µM AdoMet; lane 3, λ DNA incubated with 5 U of R.BseMII in reaction mixture without Mg2+ and AdoMet and, following addition of Mg2+ and AdoMet, cleaved with R.BseMII; lane 4, λ DNA incubated with 5 U of R.BseMII in a reaction mixture with 10 µM AdoMet without Mg2+ and, following addition of Mg2+, cleaved with R.BseMII; lane 5, λ DNA incubated with 5 U of R.BseMII in the reaction mixture with 10 µM AdoMet without Mg2+ and, following addition of Mg2+ and 1 µg λ DNA, cleaved with R.BseMII.
The recognition sequence and cleavage site of R.BseMII
To determine the recognition sequence, R.BseMII sites were mapped on pBR322 DNA (approximate positions 1570, 1740, 2280, 3320 and 3860) by double digestion with HindIII, BamHI, Eco88I, Kpn2I, NdeI, CaiI, Eam1105I and PstI (data not shown). In addition, after digestion of φX174 DNA with R.BseMII in combination with Eco47I and Eco147I (data not shown) two R.BseMII sites at approximate positions 4250 and 4730 were localized. A computer-aided search of homologous nucleotide sequences at the mapped R.BseMII sites and areas surrounding them revealed only one common sequence, 5′-CTCAG, for all the positions mapped, which differed from those of all known restriction endonucleases. The number and sizes of fragments generated by R.BseMII digestion of pBR322 (seven fragments), φX174 (10 fragments), pUC18 (five fragments) and M13mp18 (23 fragments) DNAs were consistent with this recognition site assignment. Additional evidence supporting this conclusion was obtained after double digestion of λ DNA with BseMII and DdeI. The R.BseMII recognition sequence is also recognized by DdeI, whose recognition sequence is 5′-CTNAG. The double digestion of any DNA substrate using these two enzymes should therefore yield the DdeI fragmentation pattern. Data presented in Figure 3 show that this was the case.
Figure 3.
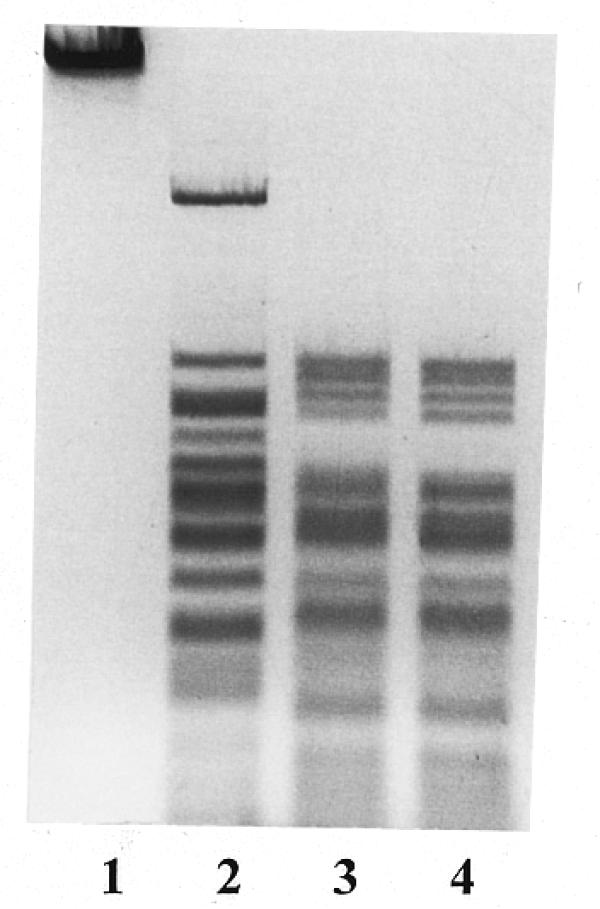
Double digestion of λ DNA with R.BseMII and R.DdeI. Lane 1, λ DNA; lane 2, λ DNA + R.BseMII; lane 3, λ DNA + R.BseMII + R.DdeI; lane 4, λ DNA + R.DdeI.
To determine the cleavage position of R.BseMII, the T7 DNA polymerase extension products through the recognition sequence of the enzyme on pBR322 DNA were digested by R.BseMII and compared with sequencing ladders. The fragment generated by R.BseMII digestion co-migrated with the C-band of the sequence ladder obtained using the same ccw primer through the R.BseMII recognition site (Fig. 4, lane 1). From this result it can be inferred that R.BseMII cleaves DNA 10 bases away from the 3′-end of the R.BseMII recognition sequence 5′-CTCAG. Lane 2 shows the result obtained when the R.BseMII restriction product was further treated with T4 DNA polymerase. This procedure resulted in a single band that co-migrates with the eighth base (T band) from the recognition sequence, indicating that DNA cleavage by R.BseMII generates a two base 3′-protruding end that is removed by the exonucleolytic activity of T4 DNA polymerase. Similar experiments were carried out using the cw primer, which yielded the same results (data not shown). These results would therefore indicate that DNA cleavage specificity of R.BseMII is 5′-CTCAG(N)10/8↓.
Figure 4.
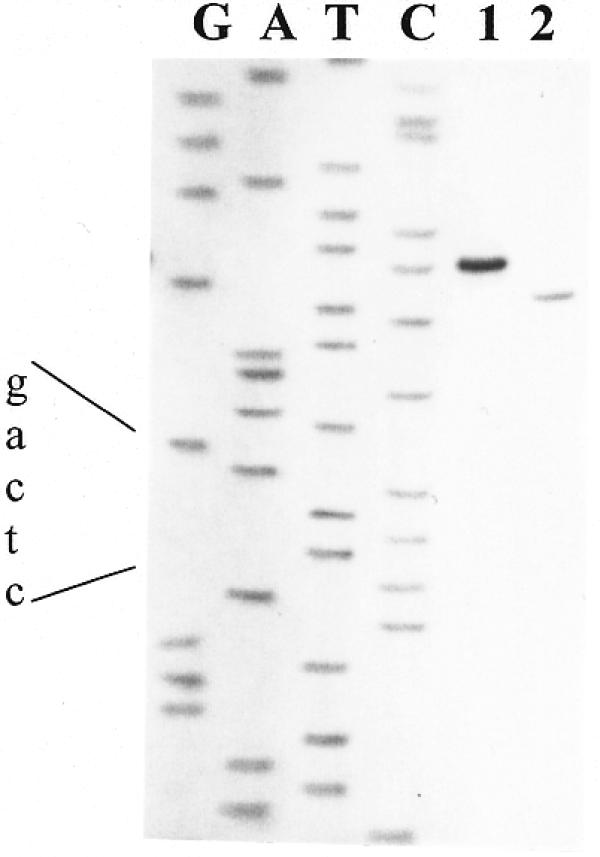
Determination of BseMII cleavage site. Lanes G, A, T and C, the sequence ladders through the R.BseMII site; lane 1, the product of the primed synthesis reaction cleaved with R.BseMII; lane 2, T4 DNA polymerase action on the R.BseMII digest.
Methylation specificity
In order to characterize the methylation specificity of R.BseMII and that of the separate M.BseMII, the nature of the modified base and the modification of different strands of the target duplex were investigated. The nature of the modified products that resulted after treatment of the synthetic substrate containing the sequence 5′-CTCAG (Materials and Methods) with either M.BseMII or R.BseMII in the presence of [3H-methyl]-AdoMet was determined by HPLC analysis using N4mdCyd, C5mdCyd and N6mdAdo as standards. Only one radioactivity peak corresponding to N6mdAdo standard was found (data not shown). This indicates that the enzymes modify the unique adenine residue present within the recognition sequence to yield N6-methyladenine.
The two strands of the same premethylated synthetic oligoduplex were separated by PAGE under denaturing conditions. It was found that M.BseMII modifies both strands of the duplex, while R.BseMII modifies only the strand containing the sequence 5′-CTCAG (data not shown). To exclude the possibility that M.BseMII is less specific than R.BseMII (e.g. recognizes interrupted symmetric DNA pentanucleotide 5′-CTNAG or the sequence 5′-YTCAR), the oligoduplex containing the 5′-CTAAG sequence instead of 5′-CTCAG, as well as the λ DNA precleaved with DdeI, were used as substrates for M.BseMII. No enzymatic methylation of these substrates was found. These observations indicate that R.BseMII and M.BseMII recognize the identical target sequence 5′-CTCAG.
Subunit structure and molecular weight
The apparent molecular mass of R.BseMII as determined by SDS–PAGE was found to be 98 kDa (Fig. 1). Gel filtration in buffers of different NaCl concentrations (0.2 and 1.0 M) containing 10% glycerol showed that the native endonuclease has a molecular mass of ~94 kDa (data not shown).
The yield of BseMII methyltransferase, isolated as described in Materials and Methods, was low. SDS–PAGE of the final preparation revealed two protein bands with apparent molecular masses of 48 and 78 kDa (data not shown). Gel filtration was carried out to identify which of the two proteins represents M.BseMII and to determine the molecular weight of the native enzyme. The peak activity of M.BseMII, eluted from the Superdex 200 HR column, corresponded to the protein of apparent molecular mass 67 kDa (data not shown).
Enzymatic properties
Using a purified preparation, the influence of different factors (cofactors, metal ions, pH, ionic strength) on the activity of the BseMII restriction endonuclease was investigated in more detail. Purified R.BseMII maintains an absolute requirement for Mg2+ and AdoMet for cleavage activity (Fig. 5, lanes 4 and 5). No specific DNA fragmentation was observed in the reaction mixture without AdoMet even after the incubation of 6 U of enzyme with 1 µg of λ DNA for 16 h. A concentration of 1 µM AdoMet was sufficient to achieve maximal R.BseMII activity. Increasing the cofactor concentration up to 50 µM did not influence the cleavage rate. S-adenosylhomocysteine and sinefungin were able to replace AdoMet as a cleavage cofactor for R.BseMII (optimal concentrations: 200 µM S-adenosylhomocysteine and 1 µM sinefungin). R.BseMII did not cut its ligated fragments if the restriction was performed in the presence of AdoMet, but did cleave the ligated DNA if sinefungin was used instead (data not shown). ATP did not affect the endonucleolytic activity of R.BseMII (Fig. 5, lane 3) at concentrations of up to 2 mM. The specific cleavage activity of R.BseMII was also independent of pH in the range from 7.5 to 8.5 and salt concentration in the range from 0 to 100 mM. The optimal temperature for cleavage was found to be 55°C. Incubation at 37°C resulted in only 30% activity.
Figure 5.
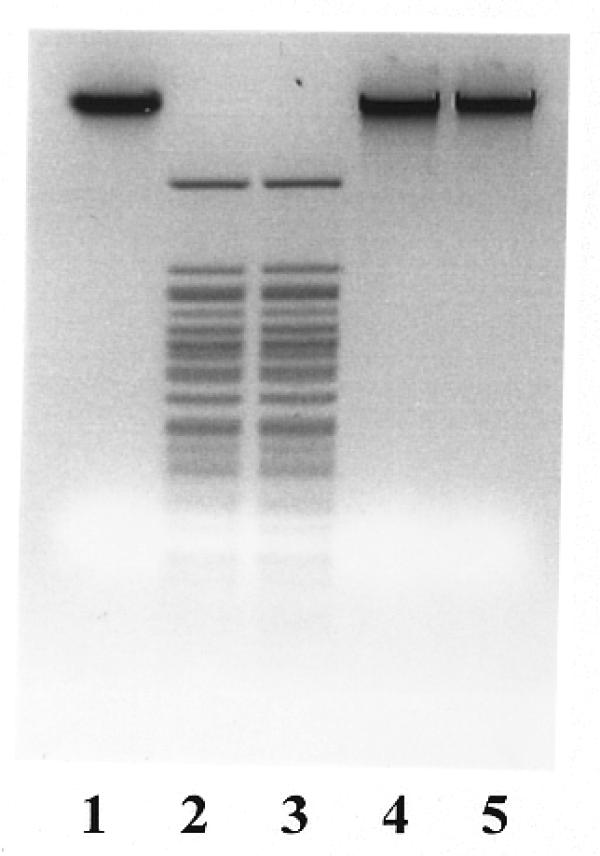
Assay of the cofactors that are required in R.BseMII cleavage reaction. Lane 1, λ DNA; lane 2, λ DNA digested with 5 U of R.BseMII in the presence of 1 mM ATP, 20 µM AdoMet and 10 mM Mg2+; lane 3, same components as in lane 2 without ATP; lane 4, same components as in lane 3 without AdoMet; lane 5, same components as in lane 4 without Mg2+.
Cloning of the genes encoding BseMII R–M enzymes
Selection of the gene for M.BseMII was based on the resistance of self-modifying recombinant plasmids to digestion by BseMII (28). Twenty-nine randomly picked transformants, obtained after the selection procedure described in Materials and Methods, were further screened for the presence of BseMII-specific modification. Plasmid DNAs of all 29 clones were found to be resistant to BseMII digestion, but none of these clones expressed the BseMII restriction endonuclease. The nucleotide sequence on both strands of the 1.55 kb MunI and 2.3 kb Bst1107I–XbaI fragments (Fig. 6), found by restriction mapping to be common for some M+ plasmids, was determined. DNA sequence analysis demonstrated the presence of two convergent ORFs, one complete and the other truncated. Phenotypes of plasmids carrying the various subcloned fragments showed that the complete ORF, bseMIIM, encoded M.BseMII (Fig. 6), while the truncated one could have been the 3′-terminal part of the restriction endonuclease gene. This speculation was based on the general observation that endonuclease genes are usually closely linked to the genes of the cognate methylases. A 5 kb Bst1107I fragment (Fig. 6) with the potential to encompass the full-length ORF, presumably encoding the restriction endonuclease and an upstream region, was identified by Southern restriction mapping of the B.stearothermophilus Isl 15-111 genomic R–M locus (data not shown). Cloning of the Bst1107I fragment was carried out using the recipient ER2267[pBR-BseMIIM17] possessing the functionally active gene for M.BseMII. Two transformants carrying the desired plasmid pAC-BseMIIR5 were found among 350 recombinant clones screened by restriction mapping. Unfortunately, none of these clones showed any R.BseMII activity. This indicated that the gene for the BseMII restriction endonuclease was either not located within the cloned fragment or was not expressed in E.coli. To analyze the DNA region upstream of the truncated ORF, the 4 kb Bst1107I–Mph1103I fragment (Fig. 6) was subcloned into the multicopy vector pUC57 and sequenced. DNA sequence analysis indicated that the ORF, which presumably encodes the BseMII restriction endonuclease, is flanked by targets for BcuI and PvuII (Fig. 6). Cloning of this BcuI–PvuII fragment into the vector pMCL200 downstream from the Plac promoter then resulted in the appearance of BseMII restriction endonuclease activity in crude extracts of cells carrying the recombinant plasmid pMCL-BseMIIR (data not shown). These findings indicate that expression of the BseMII endonuclease gene from its own promoter in E.coli is so weak that it could not be detected by the methods used.
Sequence analysis of the BseMII R–M system
The B.stearothermophilus Isl 15-111 genomic DNA region, flanked by targets for Bst1107I and XbaI restriction endonucleases, and encoding the complete BseMII R–M system was sequenced on both strands. Three ORFs were identified in the determined nucleotide sequence of 7276 bp (Fig. 6). The first ORF (orfX) starts with a TTG codon at position 1067, ends at position 1537 and is capable of coding for a protein of 156 amino acids (18.3 kDa). No amino acid sequences similar to that of the putative translation product of orfX were found, however, in the EMBL and SWISS-PROT databases, so the function of orfX remains unknown. The next ORF, bseMIIR, extended from position 1914 to the termination codon TAA at position 4721 and encodes the BseMII restriction endonuclease (see previous section). The first potential start codon ATG at position 1914 is not preceded by a sequence that might serve as a ribosome binding site (RBS; 31), while the second in-frame ATG (position 1953–1955) is preceded by the RBS-like sequence 5′-GGAGGT at positions 1943–1948. If the second ATG is used, bseMIIR encodes a BseMII restriction endonuclease of 922 amino acids with a calculated molecular mass of 105.6 kDa. This number is just slightly higher than that of the purified restriction endonuclease (∼94–98 kDa) as estimated by SDS–PAGE (Fig. 1) or by gel-filtration. The third ORF is located on the complementary strand. It starts with the translation initiation codon ATG at position 6724, ends at the termination codon TAA (position 4780–4778) and encodes the BseMII methyltransferase (see previous section) of 648 amino acids with a calculated molecular mass of 75.4 kDa (gene bseMIIM), which corresponds well with the molecular weight, as determined by SDS–PAGE, of one of the two proteins present in the final preparation of the enzyme, and is slightly larger than the 67 kDa determined for the native protein. The 5′-TAAGGAGGT sequence located –14 to –6 prior to the ATG of bseMIIM has the potential to bind E.coli ribosomes. The convergently transcribed genes of the BseMII R–M system are separated by an intergenic region of 56 nt.
The deduced amino acid sequence of BseMII methyltransferase contains all nine conserved motifs of amino DNA methyltransferases (32). They are arranged in the order N-X, I-VIII-C (where N and C denote the two ends of M.BseMII). The longest variable sequence region (putative target recognition domain) occupies the C-terminal part of the enzyme. The structure of the M.BseMII conserved motif IV (NPPY) is typical for N6-adenine methyltransferases (30,33). The mutual location of the conserved motifs and of the putative target recognition domain suggests that M.BseMII could be assigned to the γ group of amino DNA methyltransferases. The same pattern of conserved motifs (except for the absence of the conserved motifs II and III) is found in the amino acid sequence of the BseMII restriction endonuclease, which is located in the middle part of the enzyme. The structure of the conserved motif IV (NLPF) in R.BseMII deviates from the canonical structure, NPP(Y/F). In addition, the endonuclease contains the amino acid sequence motif 61PD(X)16ELK, which matches well with the catalytic/Mg2+ binding motif of restriction endonucleases (9). The putative catalytic motif is located in the N-terminal part of the enzyme. The similarity between R.BseMII and M.BseMII is limited to the conserved motifs X, I, IV, V and VI, which comprise the motifs involved in AdoMet binding and catalysis. Comparison of the R.BseMII amino acid sequence with amino acid sequences of other restriction endonucleases or methylases (including M.BseMII) revealed no statistically significant similarities beyond conserved motifs common to DNA N6-adenine methyltransferases.
DISCUSSION
BseMII is a novel restriction endonuclease that recognizes the nucleotide sequence 5′-CTCAG and makes a staggered cut at the tenth base pair downstream of the recognition sequence on the upper strand, producing a two base 3′-protruding end. R.BseMII is accompanied by a separate BseMII methyltransferase. The properties of BseMII enzymes including substrate specificity (asymmetric recognition sequence), DNA cleavage mode (complete DNA cleavage at a fixed distance from the recognition sequence) and the presence of separate BseMII methyltransferase when taken alone would indicate that they belong to the type IIS R–M system. However R.BseMII, unlike monofunctional type IIS endonucleases, is bifunctional. Our data strongly suggest that the methylase activity of R.BseMII is an intrinsic property of the enzyme represented by a single large polypeptide. This conclusion is based on the following observations: (i) endonuclease and methylase activities copurify in all chromatographic steps of R.BseMII isolation; (ii) R.BseMII and M.BseMII are eluted from the same chromatographic sorbents at such different ionic strengths that their cross-contamination seems unlikely; (iii) the deduced amino acid sequence of the R.BseMII contains conserved sequence motifs involved in AdoMet binding and catalysis, which are common to all N6-adenine methylases (30,33); (iv) R.BseMII and M.BseMII DNA methylation activities differ in their strand specificity such that M.BseMII modifies A bases in both strands, while R.BseMII only acts upon the strand containing the 5′-CTCAG sequence. The properties of R.BseMII make it similar to other type IV restriction endonucleases, except for the different effect of AdoMet on the cleavage reaction. The endonucleolytic activity of other known type IV endonucleases is only stimulated by AdoMet (5,10–12,14), while R.BseMII reveals an absolute requirement for this cofactor. We cannot exclude though that when other type IV enzymes are purified from bacterial cells, some amount of AdoMet may remain bound as has been shown for type III enzymes (34). Therefore differences between R.BseMII and other known type IV restriction endonucleases, with respect to the AdoMet effect (absolute requirement versus stimulation) on endonucleolytic reaction, may reflect differences in copurification of the enzymes and AdoMet. This notion is supported by the observation that some purified preparations of R.BseMII manifest trace endonucleolytic activity in the absence of AdoMet. In addition, unlike the Eco57I restriction endonuclease, R.BseMII yields complete cleavage of DNA in the presence of AdoMet. The same is true for other Eco57I-like restriction enzymes GsuI, BspLU11III and MmeI that have been characterized in this respect (10,11,14). Further, R.BseMII stimulation by the AdoMet analogs sinefungin and S-adenosylhomocysteine indicates that binding of the cofactor rather than methyl group transfer is essential for cleavage. The effect of sinefungin has also been characterized in the case of R.MmeI (14) and R.Eco57I (R.Rimseliene, Institute of Biotechnology, personal communication), where it was shown to substitute for AdoMet as a cofactor. Of note are the practical implications of this observation. Ligated R.BseMII and R.Eco57I fragments are susceptible to the corresponding endonucleases only if digestion before ligation is carried out in the presence of sinefungin, but not AdoMet (data not shown). This observation indicates, however, that in the presence of AdoMet modification of recognition sequence occurs in parallel with or after DNA cleavage.
Type IV enzymes are more similar to the type III endonucleases (35–37) than to the other known AdoMet-stimulated restriction endonucleases i.e. type I and BcgI (6). R.BseMII, like other type IV endonucleases, shares with the type III enzymes such properties as asymmetric recognition sequence, cleavage position in the vicinity of the recognition sequence, methylation of a single strand of the substrate and stimulation of endonuclease activity by AdoMet. However, unlike type III enzymes, the type IV endonucleases including R.BseMII do not require ATP and combine cleavage and methylation activities in a single polypeptide, while type III restriction endonucleases are hetero-oligomers, which use the Mod subunit for both sequence recognition and methylation and the Res subunit for DNA cleavage.
The R.BseMII polypeptide can be subdivided into three structural/functional parts. The central part contains conserved motifs of DNA N-methyltransferases involved in AdoMet binding and catalysis. The C-terminal section corresponds to the putative target recognition domain of the γ group of DNA methylases (33), while the N-terminal part contains the putative catalytic/Mg2+ binding motif 61PD(X)16ELK. The same organization is a characteristic of R.Eco57I (8) and R.GsuI (R.Vaisvila, New England Biolabs, personal communication). In the case of R.Eco57I, direct evidence has been obtained that the 77PD(X)13EAK motif located in the N-terminal part of the enzyme is involved in DNA cleavage catalysis (R.Rimseliene, personal communication).
DNA methylation by R.BseMII itself is not sufficient to protect host DNA from the enzyme and consequently a separate methylase is also needed. Recognition and modification of the different DNA strands of the asymmetric target duplex by type IIS methylases requires two methylases, which exist as separate enzymes (38,39) or are represented by a fused protein containing two sets of conserved motifs and two target recognition domains (40). Strand-specific type IIS and type III methyltransferases, which modify only one DNA strand, contain only one set of conserved motifs (30,38–40). The same is true for R.BseMII. Surprisingly, M.BseMII, like M.Eco57I (8) and M.GsuI (R.Vaisvila, personal communication), also contains only one set of conserved motifs although it modifies both strands of the recognition sequence. M.Eco57I also modifies both DNA strands, while M.GsuI has not been investigated with regard to this as yet. Given that the BseMII recognition sequence has elements of symmetry, it can be argued that M.BseMII recognizes the sequence as symmetric (i.e. 5′-CTSAG) like type II methylases, while R.BseMII recognizes the sequence as asymmetric (5′-CTCAG). The above reasoning could not be applied to M.Eco57I and M.GsuI, whose hexanucleotide recognition sequences have no symmetry in their central dinucleotide. It cannot be excluded that type IV methylases use an as yet unknown mechanism for recognition and modification of the different strands of the target duplex (e.g. type IV methyltransferases may combine one set of conserved motifs and two target recognition domains, each for a different strand). The recognition sequences of the putative type IV methylases MmeI and BspLU11III do not contain elements of symmetry. Cloning and sequencing of their genes would help to establish whether they are type IIS-like, or are similar to type IV methylases.
In summarizing the above observations, it can be concluded that both components of the type IV R–M system, the endonuclease and the methylase, are different from other known R–M systems. Indeed the evolution of type IV restriction endonucleases is intriguing. R.BseMII does not share significant similarity with R.Eco57I and R.GsuI beyond the conserved motifs of N6-adenine methylases and the catalytic motifs. This observation indicates that they evolved independently. However structural/functional organization suggests that the evolution of the type IV endonucleases involved the same mechanism: fusion of a methylase and an endonuclease. It has been suggested that the progenitor of R.Eco57I was generated by the fusion of Mod and Res subunits of a type III endonuclease (8). Alternatively it cannot be excluded that the evolutionary scenario of type IV restriction endonucleases involved the fusion of a type IIS single strand-specific methylase and an endonucleolytic domain. Activation by AdoMet favors the Mod–Res fusion hypothesis, while the absence of an ATP effect favors the type IIS single strand-specific methylase–endonucleolytic domain fusion scenario.
Acknowledgments
ACKNOWLEDGEMENTS
We thank MBI Fermentas for providing enzymes and kits. We are also grateful to Yoshio Nakano for kindly providing plasmid pMCL200, to Barbara Richmond-Smith for linguistic help and to Egle Cesnaviciene for critical reading of this manuscript. The work was supported by grants from the Lithuanian national research program ‘Molecular basis of Biotechnology’ and the MBI Fermentas.
DDBJ/EMBL/GenBank accession no. AF306668
References
- 1.Yuan R. (1981) Structure and mechanism of multifunctional restriction endonucleases. Annu. Rev. Biochem., 150, 285–315. [DOI] [PubMed] [Google Scholar]
- 2.Pingoud A. and Jeltch,A. (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 3.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes—a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 4.Stankevicius K., Lubys,A., Timinskas,A., Vaitkevicius,D. and Janulaitis,A. (1996) Cloning and analysis of the genes encoding the type IIS restriction–modification system HphI from Haemophilus parahaemolyticus. Nucleic Acids Res., 26, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janulaitis A., Petrusyte,M., Maneliene,Z., Klimasauskas,S and Butkus,V. (1992) Purification and properties of the Eco57I restriction endonuclease and methylase—prototypes of a new class (type IV). Nucleic Acids Res., 20, 6043–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong H., Morgan,R.D., Maunus,R.E., and Schildkraut,I. (1993) A unique restriction endonuclease, BcgI, from Bacillus coagulans. Nucleic Acids Res., 21, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piekarowicz A., Golaszewska,M., Sunday,A.O., Siwinska,M. and Stein,D.C. (1999) The HaeIV restriction modification system of Haemophilus aegyptius is encoded by a single polypeptide. J. Mol. Biol., 293, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 8.Janulaitis A.A., Vaisvila,R., Timinskas,A., Klimasauskas,S. and Butkus,V. (1992) Cloning and sequence analysis of the genes coding for Eco57I type IV restriction–modification enzymes. Nucleic Acids Res., 20, 6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson J.E. (1993) Restriction endonucleases and modification methylases. Curr. Opin. Struct. Biol., 3, 24–30. [Google Scholar]
- 10.Petrusyte M., Bitinaite,J., Menkevicius,S., Klimasauskas,S., Butkus,V. and Janulaitis,A. (1988) Restriction endonucleases of a new type. Gene, 74, 89–91. [DOI] [PubMed] [Google Scholar]
- 11.Chernov A.V., Matvienko,N.N., Zheleznaya,L.A. and Matvienko,N.I. (1994) A new site specific endonuclease-methylase from a thermophilic strain of Bacillus species LU11. Biokhimiya (in Russian), 59, 1714–1729. [PubMed] [Google Scholar]
- 12.Matvienko N.N., Kramarov,V.M., Ivanov,L.I. and Matvienko,N.I. (1992) Bce83I, a restriction endonuclease from Bacillus cereus 83 which recognizes novel non-palindromic sequence 5′-CTTGAG-3′ and is stimulated by S-adenosylmethionine. Nucleic Acids Res., 20, 1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts R.J. and Macelis,D. (2000) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucholski J., Zmijewski,J.W. and Podhajska,A.J. (1998) Two intertwined methylation activities of the MmeI restriction–modification class-IIS system from Methylophilus methylotrophus. Gene, 223, 293–302. [DOI] [PubMed] [Google Scholar]
- 15.Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Heyneker,H.L., Boyer,H.W., Crosa,J.H. and Falkow,S. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- 16.Chang A.C.Y. and Cohen,S.N. (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol ., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano Y., Yoshida,Y., Yamashita,Y. and Koga,T. (1995) Construction of a series of pACYC-derived plasmid vectors. Gene, 162, 157–158. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Whitehead P.R. and Brown,N.L. (1985) A simple and rapid method for screening bacteria for type II restriction endonucleases: enzymes in Aphanothece halophytica. Arch. Microbiol., 141, 70–74. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrke C.H., McCune,R., Gama-Sosa,M.A., Ehrlich,M. and Kuo,K. (1984) Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J. Chromatogr., 301, 199–219 . [DOI] [PubMed] [Google Scholar]
- 23.Klimasauskas S., Steponaviciene,D., Maneliene,Z., Petrusyte,M., Butkus,V. and Janulaitis,A. (1990) M.SmaI is an N4-methylcytosine specific DNA-methylase. Nucleic Acids Res., 18, 6607–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur J. (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol., 3, 208–218. [Google Scholar]
- 25.Birnboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marko M.A., Chipperfield,R. and Birnboim,H.C. (1982) A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal. Biochem., 121, 382–387. [DOI] [PubMed] [Google Scholar]
- 27.Chen E.Y. and Seeburg,P.H. (1985) Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA, 4, 165–170. [DOI] [PubMed] [Google Scholar]
- 28.Szomolanyi I., Kiss,A. and Venetianer,P. (1980) Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene, 10, 219–225. [DOI] [PubMed] [Google Scholar]
- 29.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timinskas A., Butkus,V. and Janulaitis,A. (1995) Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene, 157, 3–11. [DOI] [PubMed] [Google Scholar]
- 31.Shine J. and Dalgarno,L. (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA, 71, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 33.Wilson G.G. (1991) Organization of restriction-modification systems. Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piekarowicz A. and Brzezinski,R. (1980) Cleavage and methylation of DNA by the restriction endonuclease HinfIII isolated from Haemophilus influenzae Rf. J. Mol. Biol., 144, 415–429. [DOI] [PubMed] [Google Scholar]
- 35.Haberman A. (1974) The bacteriophage P1 restriction endonuclease. J. Mol. Biol., 89, 545–563. [DOI] [PubMed] [Google Scholar]
- 36.Kauc L. and Piekarowicz,A. (1978) Purification and properties of a new restriction endonuclease from Haemophilus influenzae Rf. Eur. J. Biochem., 92, 417–426. [DOI] [PubMed] [Google Scholar]
- 37.Reiser J. and Yuan,R. (1977) Purification and properties of the P15 specific restriction endonuclease from Escherichia coli. J. Biol. Chem., 252, 451–456. [PubMed] [Google Scholar]
- 38.Sugisaki H., Yamamoto,K. and Takanami,M. (1991) The HgaI restriction–modification system contains two cytosine methylase genes responsible for modification of different DNA strands. J. Biol. Chem., 266, 13952–13957. [PubMed] [Google Scholar]
- 39.Lubys A., Lubiene,J., Kulakauskas,S., Stankevicius,K., Timinskas,A. and Janulaitis,A. (1996) Cloning and analysis of the genes encoding the type IIS restriction–modification system HphI from Haemophilus parahaemolyticus. Nucleic Acids Res., 24, 2760–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Looney M.C., Moran,L.S., Jack,W.E., Feehery,G.R., Benner,J.S., Slatko,B.E. and Wilson,G.G. (1989) Nucleotide sequence of the FokI restriction–modification system: separate strand-specificity domains in the methyltransferase. Gene, 80, 193–208. [DOI] [PubMed] [Google Scholar]