Abstract
Cytenamide forms a 1:1 solvate with trifluoroacetic acid (systematic name: 5H-dibenzo[a,d]cycloheptatriene-5-carboxamide trifluoroacetic acid solvate), C16H13NO·C2HF3O2. The compound crystallizes with one molecule of cytenamide and one of trifluoroacetic acid in the asymmetric unit; these are linked by O—H⋯O and N—H⋯O hydrogen bonds to form an R 2 2(8) motif. The trifluoromethyl group of the solvent molecule displays rotational disorder over two sites, with site-occupancy factors of 0.964 (4) and 0.036 (4).
Related literature
For details on the experimental methods used to obtain this form, see: Davis et al. (1964 ▶); Florence et al. (2003 ▶); Florence, Johnston, Fernandes et al. (2006 ▶). For literature on carbamazepine and other related structures, see: Cyr et al. (1987 ▶); Fleischman et al. (2003 ▶); Florence, Johnston, Price et al. (2006 ▶); Florence, Leech et al. (2006 ▶); Bandoli et al. (1992 ▶); Harrison et al. (2006 ▶); Leech et al. (2007 ▶); Florence, Bedford et al. (2008 ▶); Florence, Shankland et al. (2008 ▶); Fernandes et al. (2007 ▶). For hydrogen-bond motifs, see: Etter (1990 ▶); Bernstein et al. (1995 ▶).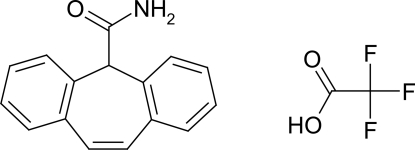
Experimental
Crystal data
C16H13NO·C2HF3O2
M r = 349.31
Monoclinic,

a = 12.1673 (11) Å
b = 6.3235 (6) Å
c = 21.4525 (15) Å
β = 101.932 (8)°
V = 1614.9 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.12 mm−1
T = 160 K
0.16 × 0.13 × 0.08 mm
Data collection
Oxford Diffraction Gemini S diffractometer
Absorption correction: multi-scan (ABSPACK/CrysAlis RED; Oxford Diffraction, 2006 ▶) T min = 0.83, T max = 0.99
10796 measured reflections
2995 independent reflections
1423 reflections with I > 2σ(I)
R int = 0.094
Refinement
R[F 2 > 2σ(F 2)] = 0.080
wR(F 2) = 0.178
S = 1.04
2984 reflections
236 parameters
24 restraints
H-atom parameters not refined
Δρmax = 0.73 e Å−3
Δρmin = −0.60 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2006 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: CRYSTALS (Betteridge et al., 2003 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2008 ▶) and PLATON (Spek, 2003 ▶).
Supplementary Material
Crystal structure: contains datablocks I. DOI: 10.1107/S1600536808016577/cf2202sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808016577/cf2202Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H5⋯O2i | 0.89 | 1.58 | 2.462 (4) | 173 |
| N1—H11⋯O1ii | 0.86 | 2.23 | 2.976 (4) | 144 |
| N1—H12⋯O1iii | 0.87 | 2.16 | 2.982 (5) | 159 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
The authors thank the Basic Technology Programme of the UK Research Councils for funding this work under the project Control and Prediction of the Organic Solid State (www.cposs.org.uk).
supplementary crystallographic information
Comment
Cytenamide (CYT) is an analogue of carbamazepine (CBZ), a dibenzazepine drug used to control seizures (Cyr et al., 1987). Cytenamide-trifluoroacetic acid solvate (CYT-TFAA) was produced during an automated parallel crystallization study (Florence, Johnston, Fernandes et al., 2006) of CYT as part of a wider investigation that couples automated parallel crystallization with crystal structure prediction methodology to investigate the basic science underlying the solid-state diversity of CBZ (Florence, Johnston, Price et al., 2006; Florence, Leech et al., 2006) and its closely related analogues: CYT (Florence, Bedford et al., 2008), 10,11-dihydrocarbamazepine (Bandoli et al., 1992; Harrison et al., 2006; Leech et al., 2007) and cyheptamide (Florence, Shankland et al., 2008). The sample was identified as a new form using multi-sample foil transmission X-ray powder diffraction analysis (Florence et al., 2003). Subsequent manual recrystallization from a saturated TFAA solution by slow evaporation at 278 K yielded a sample suitable for single-crystal X-ray diffraction (Fig. 1).
The compound crystallizes in space group P21/n with one molecule of CBZ and one molecule of TFAA in the asymmetric unit. As in the structure of CBZ-TFAA solvate (Fernandes et al., 2007) the solvent molecule displays rotational disorder and the fluorine atoms were refined over two sites yielding site occupancy factors 0.964 (4), 0.036 (4) and 0.53 (1), 0.47 (1) for CYT-TFAA and CBZ-TFAA respectively. The molecules also adopt a hydrogen-bonded arrangement similar to that observed in CBZ-TFAA solvate whereby the amide group of each CYT molecule is connected to the carboxylic acid group of a TFAA molecule by N–H···O and O–H···O contacts (Table 1) to form an R22(8) hydrogen-bonded motif (Etter, 1990; Bernstein et al., 1995). CYT also forms a second N—H···O contact with an adjacent solvent molecule to form a chain extending along the [010] direction.
Experimental
A sample of cytenamide was synthesized according to a modification of the published method (Davis et al., 1964). A single-crystal sample of cytenamide-TFAA was grown from a saturated TFAA solution by isothermal solvent evaporation at 278 K.
Refinement
Owing to the weak scattering, data were integrated applying a theta cut off of 25°. All non-hydrogen atoms were modelled with anisotropic displacement parameters with the exception of the minor component of the disordered site in the TFAA molecule, for which one common isotropic displacement parameter was calculated and fixed during refinement. Bond-length restraints were applied to C—F bond lengths involving atoms F1, F4, F5 and F6. 3-Fold symmetry was imposed on the disordered minor site of the the TFAA molecule by the use of restraints. H-atoms were found in a difference Fourier map and were initially refined with soft restraints on the bond lengths and angles to regularize their geometry and Uiso(H) (in the range 1.2–1.5 times Ueq of the parent atom), after which the positions were fixed. Eleven reflections were suppressed as outliers in an analysis of the data.
Figures
Fig. 1.
The molecular structure of CYT-TFAA, showing 50% probablility displacement ellipsoids. The lower occupancy fluorine atoms have beem omitted for clarity.
Fig. 2.
Hydrogen-bonded contacts in CYT-TFAA, showing the adjacent R22(8) CYT-TFAA units further linked by an N—H···O interaction. Minor ocupancy components have been omitted for clarity. CYT and TFAA molecules are coloured according to symmetry equivalence (green and blue respectively) and hydrogen bonds are shown as dashed lines.
Crystal data
| C16H13NO·C2HF3O2 | F000 = 720 |
| Mr = 349.31 | Dx = 1.437 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1137 reflections |
| a = 12.1673 (11) Å | θ = 3–25º |
| b = 6.3235 (6) Å | µ = 0.12 mm−1 |
| c = 21.4525 (15) Å | T = 160 K |
| β = 101.932 (8)º | Block, colourless |
| V = 1614.9 (2) Å3 | 0.16 × 0.13 × 0.08 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Gemini S diffractometer | 2995 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1423 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.094 |
| Detector resolution: 15.9745 pixels mm-1 | θmax = 25.5º |
| T = 160 K | θmin = 3.1º |
| φ and ω scans | h = −14→14 |
| Absorption correction: multi-scan(ABSPACK/CrysAlis RED; Oxford Diffraction, 2006) | k = 0→7 |
| Tmin = 0.83, Tmax = 0.99 | l = 0→25 |
| 10796 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.080 | H-atom parameters not refined |
| wR(F2) = 0.178 | Method = Modified Sheldrick w = 1/[σ2(F2) + ( 0.06P)2 + 0.42P] ,where P = (max(Fo2,0) + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.0004 |
| 2984 reflections | Δρmax = 0.73 e Å−3 |
| 236 parameters | Δρmin = −0.60 e Å−3 |
| 24 restraints | Extinction correction: None |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| C1 | 0.6284 (3) | 0.6720 (7) | 0.3296 (2) | 0.0461 | |
| C2 | 0.5488 (4) | 0.4854 (7) | 0.31163 (18) | 0.0431 | |
| C3 | 0.4806 (3) | 0.4971 (7) | 0.24379 (18) | 0.0380 | |
| C4 | 0.4965 (4) | 0.3481 (7) | 0.1996 (2) | 0.0507 | |
| C5 | 0.4406 (4) | 0.3618 (8) | 0.1366 (2) | 0.0578 | |
| C6 | 0.3726 (4) | 0.5332 (9) | 0.1180 (2) | 0.0576 | |
| C7 | 0.3553 (4) | 0.6826 (8) | 0.16104 (19) | 0.0519 | |
| C8 | 0.4077 (3) | 0.6646 (7) | 0.22567 (17) | 0.0393 | |
| C9 | 0.3794 (4) | 0.8214 (7) | 0.2691 (2) | 0.0475 | |
| C10 | 0.3773 (3) | 0.7968 (7) | 0.33147 (19) | 0.0456 | |
| C11 | 0.4028 (3) | 0.6095 (7) | 0.37105 (18) | 0.0369 | |
| C12 | 0.3488 (4) | 0.5854 (7) | 0.42250 (19) | 0.0470 | |
| C13 | 0.3646 (4) | 0.4094 (8) | 0.45999 (18) | 0.0515 | |
| C14 | 0.4337 (4) | 0.2524 (8) | 0.4474 (2) | 0.0570 | |
| C15 | 0.4912 (4) | 0.2755 (7) | 0.3988 (2) | 0.0497 | |
| C16 | 0.4774 (3) | 0.4536 (7) | 0.36104 (17) | 0.0374 | |
| C17 | 0.8909 (4) | 0.3112 (8) | 0.4387 (2) | 0.0572 | |
| C18 | 0.8197 (3) | 0.1386 (7) | 0.4008 (2) | 0.0461 | |
| N1 | 0.6679 (3) | 0.7697 (6) | 0.28446 (15) | 0.0533 | |
| O1 | 0.7936 (2) | 0.1490 (5) | 0.34338 (13) | 0.0564 | |
| O2 | 0.6593 (2) | 0.7215 (5) | 0.38684 (12) | 0.0591 | |
| O3 | 0.7904 (3) | −0.0019 (5) | 0.43735 (12) | 0.0618 | |
| F1 | 0.9217 (4) | 0.4524 (6) | 0.40116 (15) | 0.1040 | 0.964 (4) |
| F2 | 0.9828 (2) | 0.2331 (5) | 0.47540 (13) | 0.0714 | 0.964 (4) |
| F3 | 0.8361 (3) | 0.4082 (5) | 0.47792 (16) | 0.0845 | 0.964 (4) |
| F4 | 0.846 (3) | 0.502 (2) | 0.429 (3) | 0.0800* | 0.036 (4) |
| F5 | 0.992 (2) | 0.330 (8) | 0.425 (3) | 0.0800* | 0.036 (4) |
| F6 | 0.910 (5) | 0.282 (6) | 0.5010 (4) | 0.0800* | 0.036 (4) |
| H11 | 0.6461 | 0.7332 | 0.2450 | 0.0619* | |
| H12 | 0.7150 | 0.8730 | 0.2945 | 0.0619* | |
| H21 | 0.5969 | 0.3601 | 0.3139 | 0.0495* | |
| H41 | 0.5449 | 0.2331 | 0.2125 | 0.0596* | |
| H51 | 0.4499 | 0.2559 | 0.1077 | 0.0680* | |
| H61 | 0.3386 | 0.5487 | 0.0754 | 0.0651* | |
| H71 | 0.3068 | 0.7953 | 0.1473 | 0.0581* | |
| H91 | 0.3579 | 0.9563 | 0.2521 | 0.0539* | |
| H101 | 0.3556 | 0.9169 | 0.3516 | 0.0541* | |
| H121 | 0.3022 | 0.6959 | 0.4312 | 0.0539* | |
| H131 | 0.3278 | 0.3947 | 0.4943 | 0.0619* | |
| H141 | 0.4423 | 0.1260 | 0.4713 | 0.0647* | |
| H151 | 0.5414 | 0.1676 | 0.3914 | 0.0557* | |
| H5 | 0.7404 | −0.0935 | 0.4169 | 0.0888* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.033 (2) | 0.065 (3) | 0.040 (3) | −0.002 (2) | 0.005 (2) | 0.003 (2) |
| C2 | 0.036 (2) | 0.047 (3) | 0.044 (2) | 0.008 (2) | 0.002 (2) | −0.006 (2) |
| C3 | 0.028 (2) | 0.046 (3) | 0.041 (2) | −0.007 (2) | 0.0095 (19) | −0.007 (2) |
| C4 | 0.044 (3) | 0.056 (3) | 0.052 (3) | 0.000 (2) | 0.011 (2) | −0.009 (2) |
| C5 | 0.056 (3) | 0.072 (4) | 0.048 (3) | −0.012 (3) | 0.015 (2) | −0.021 (3) |
| C6 | 0.053 (3) | 0.081 (4) | 0.036 (3) | −0.011 (3) | 0.002 (2) | −0.003 (3) |
| C7 | 0.044 (3) | 0.070 (3) | 0.041 (3) | −0.004 (2) | 0.004 (2) | 0.002 (3) |
| C8 | 0.030 (2) | 0.049 (3) | 0.038 (2) | −0.003 (2) | 0.0052 (19) | 0.000 (2) |
| C9 | 0.047 (3) | 0.041 (3) | 0.055 (3) | 0.002 (2) | 0.009 (2) | −0.001 (2) |
| C10 | 0.047 (3) | 0.045 (3) | 0.047 (3) | 0.003 (2) | 0.013 (2) | −0.006 (2) |
| C11 | 0.036 (2) | 0.037 (3) | 0.035 (2) | −0.006 (2) | 0.0022 (19) | −0.006 (2) |
| C12 | 0.047 (3) | 0.056 (3) | 0.038 (2) | −0.005 (2) | 0.008 (2) | −0.011 (2) |
| C13 | 0.050 (3) | 0.069 (4) | 0.035 (2) | −0.014 (3) | 0.007 (2) | 0.006 (3) |
| C14 | 0.058 (3) | 0.057 (3) | 0.050 (3) | −0.010 (3) | −0.004 (2) | 0.011 (3) |
| C15 | 0.043 (3) | 0.055 (3) | 0.048 (3) | −0.003 (2) | 0.001 (2) | 0.002 (2) |
| C16 | 0.040 (3) | 0.036 (3) | 0.032 (2) | −0.002 (2) | −0.0011 (19) | −0.003 (2) |
| C17 | 0.054 (3) | 0.064 (4) | 0.055 (3) | −0.003 (3) | 0.014 (3) | −0.002 (3) |
| C18 | 0.034 (3) | 0.060 (3) | 0.044 (3) | 0.002 (2) | 0.007 (2) | 0.008 (3) |
| N1 | 0.043 (2) | 0.078 (3) | 0.038 (2) | −0.017 (2) | 0.0045 (17) | −0.009 (2) |
| O1 | 0.0488 (19) | 0.082 (2) | 0.0375 (17) | −0.0082 (17) | 0.0064 (14) | 0.0091 (17) |
| O2 | 0.054 (2) | 0.090 (3) | 0.0300 (17) | −0.0283 (18) | 0.0024 (14) | −0.0002 (16) |
| O3 | 0.060 (2) | 0.085 (2) | 0.0362 (17) | −0.0297 (19) | −0.0009 (15) | 0.0067 (17) |
| F1 | 0.132 (3) | 0.101 (3) | 0.074 (2) | −0.059 (3) | 0.008 (2) | 0.020 (2) |
| F2 | 0.0448 (18) | 0.091 (2) | 0.0715 (19) | −0.0066 (16) | −0.0045 (14) | −0.0160 (17) |
| F3 | 0.075 (2) | 0.086 (2) | 0.095 (2) | 0.0090 (19) | 0.0230 (19) | −0.027 (2) |
Geometric parameters (Å, °)
| C1—C2 | 1.525 (6) | C11—C16 | 1.386 (5) |
| C1—N1 | 1.321 (5) | C12—C13 | 1.363 (6) |
| C1—O2 | 1.247 (4) | C12—H121 | 0.942 |
| C2—C3 | 1.521 (5) | C13—C14 | 1.364 (6) |
| C2—C16 | 1.517 (6) | C13—H131 | 0.942 |
| C2—H21 | 0.980 | C14—C15 | 1.378 (6) |
| C3—C4 | 1.379 (6) | C14—H141 | 0.945 |
| C3—C8 | 1.384 (5) | C15—C16 | 1.376 (5) |
| C4—C5 | 1.383 (6) | C15—H151 | 0.951 |
| C4—H41 | 0.941 | C17—C18 | 1.520 (5) |
| C5—C6 | 1.372 (7) | C17—F1 | 1.307 (4) |
| C5—H51 | 0.936 | C17—F2 | 1.322 (5) |
| C6—C7 | 1.368 (6) | C17—F3 | 1.328 (5) |
| C6—H61 | 0.928 | C17—C18 | 1.520 (5) |
| C7—C8 | 1.406 (5) | C17—F4 | 1.323 (7) |
| C7—H71 | 0.934 | C17—F5 | 1.323 (7) |
| C8—C9 | 1.451 (5) | C17—F6 | 1.323 (7) |
| C9—C10 | 1.352 (5) | C18—O1 | 1.209 (4) |
| C9—H91 | 0.944 | C18—O3 | 1.283 (5) |
| C10—C11 | 1.453 (6) | N1—H11 | 0.865 |
| C10—H101 | 0.938 | N1—H12 | 0.867 |
| C11—C12 | 1.405 (5) | O3—H5 | 0.887 |
| C2—C1—N1 | 119.0 (4) | C12—C11—C16 | 118.2 (4) |
| C2—C1—O2 | 119.3 (4) | C11—C12—C13 | 121.4 (4) |
| N1—C1—O2 | 121.6 (4) | C11—C12—H121 | 118.3 |
| C1—C2—C3 | 113.4 (4) | C13—C12—H121 | 120.4 |
| C1—C2—C16 | 110.5 (3) | C12—C13—C14 | 119.6 (4) |
| C3—C2—C16 | 113.4 (3) | C12—C13—H131 | 120.7 |
| C1—C2—H21 | 105.8 | C14—C13—H131 | 119.7 |
| C3—C2—H21 | 106.9 | C13—C14—C15 | 120.2 (4) |
| C16—C2—H21 | 106.2 | C13—C14—H141 | 120.7 |
| C2—C3—C4 | 119.9 (4) | C15—C14—H141 | 119.0 |
| C2—C3—C8 | 119.8 (4) | C14—C15—C16 | 120.8 (4) |
| C4—C3—C8 | 120.2 (4) | C14—C15—H151 | 119.6 |
| C3—C4—C5 | 121.2 (4) | C16—C15—H151 | 119.6 |
| C3—C4—H41 | 119.6 | C2—C16—C11 | 120.1 (4) |
| C5—C4—H41 | 119.2 | C2—C16—C15 | 120.1 (4) |
| C4—C5—C6 | 118.6 (4) | C11—C16—C15 | 119.7 (4) |
| C4—C5—H51 | 120.0 | C18—C17—F1 | 111.5 (4) |
| C6—C5—H51 | 121.3 | C18—C17—F2 | 111.6 (4) |
| C5—C6—C7 | 121.1 (4) | F1—C17—F2 | 107.9 (4) |
| C5—C6—H61 | 119.4 | C18—C17—F3 | 111.4 (4) |
| C7—C6—H61 | 119.5 | F1—C17—F3 | 108.7 (4) |
| C6—C7—C8 | 120.6 (4) | F2—C17—F3 | 105.6 (4) |
| C6—C7—H71 | 119.3 | C18—C17—F4 | 113.56 (6) |
| C8—C7—H71 | 120.1 | C18—C17—F5 | 113.57 (6) |
| C7—C8—C3 | 118.2 (4) | F4—C17—F5 | 105.08 (7) |
| C7—C8—C9 | 117.4 (4) | C18—C17—F6 | 113.57 (6) |
| C3—C8—C9 | 124.4 (4) | F4—C17—F6 | 105.08 (7) |
| C8—C9—C10 | 127.8 (4) | F5—C17—F6 | 105.08 (7) |
| C8—C9—H91 | 116.8 | C17—C18—O1 | 120.4 (4) |
| C10—C9—H91 | 115.3 | C17—C18—O3 | 111.8 (4) |
| C9—C10—C11 | 128.8 (4) | O1—C18—O3 | 127.8 (4) |
| C9—C10—H101 | 115.3 | C1—N1—H11 | 120.6 |
| C11—C10—H101 | 115.9 | C1—N1—H12 | 119.5 |
| C10—C11—C12 | 118.0 (4) | H11—N1—H12 | 119.9 |
| C10—C11—C16 | 123.9 (4) | C18—O3—H5 | 113.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H5···O2i | 0.89 | 1.58 | 2.462 (4) | 173 |
| N1—H11···O1ii | 0.86 | 2.23 | 2.976 (4) | 144 |
| N1—H12···O1iii | 0.87 | 2.16 | 2.982 (5) | 159 |
Symmetry codes: (i) x, y−1, z; (ii) −x+3/2, y+1/2, −z+1/2; (iii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CF2202).
References
- Bandoli, G., Nicolini, M., Ongaro, A., Volpe, G. & Rubello, A. (1992). J. Chem. Crystallogr.22, 177–183.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Betteridge, P. W., Carruthers, J. R., Cooper, R. I., Prout, K. & Watkin, D. J. (2003). J. Appl. Cryst.36, 1487.
- Blessing, R. H. (1997). J. Appl. Cryst.30, 421–426.
- Cyr, T. D., Matsui, F., Sears, R. W., Curran, N. M. & Lovering, E. G. (1987). J. Assoc. Off. Anal. Chem.30, 421–426. [PubMed]
- Davis, M. A., Winthrop, S. O., Thomas, R. A., Herr, F., Charest, M.-P. & Gaudry, R. (1964). J. Med. Chem.7, 88–94. [DOI] [PubMed]
- Etter, M. C. (1990). Acc. Chem. Res.23, 120–126.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Fernandes, P., Bardin, J., Johnston, A., Florence, A. J., Leech, C. K., David, W. I. F. & Shankland, K. (2007). Acta Cryst. E63, o4269.
- Fleischman, S. G., Kuduva, S. S., McMahon, J. A., Moulton, B., Walsh, R. D. B., Rodriguez-Hornedo, N. & Zaworotko, M. J. (2003). Cryst. Growth Des.3, 909–919.
- Florence, A. J., Baumgartner, B., Weston, C., Shankland, N., Kennedy, A. R., Shankland, K. & David, W. I. F. (2003). J. Pharm. Sci.92, 1930–1938. [DOI] [PubMed]
- Florence, A. J., Bedford, C. T., Fabbiani, F. P. A., Shankland, K., Gelbrich, T., Hursthouse, M. B., Shankland, N., Johnston, A. & Fernandes, P. (2008). CrystEngComm, DOI: 10.1039/b719717a.
- Florence, A. J., Johnston, A., Fernandes, P., Shankland, N. & Shankland, K. (2006). J. Appl. Cryst.39, 922–924.
- Florence, A. J., Johnston, A., Price, S. L., Nowell, H., Kennedy, A. R. & Shankland, N. (2006). J. Pharm. Sci.95, 1918–1930. [DOI] [PubMed]
- Florence, A. J., Leech, C. K., Shankland, N., Shankland, K. & Johnston, A. (2006). CrystEngComm, 8, 746–747.
- Florence, A. J., Shankland, K., Gelbrich, T., Hursthouse, M. B., Shankland, N., Johnston, A., Fernandes, P. & Leech, C. K. (2008). CrystEngComm, 10, 26–28.
- Harrison, W. T. A., Yathirajan, H. S. & Anilkumar, H. G. (2006). Acta Cryst. C62, o240–o242. [DOI] [PubMed]
- Leech, C. K., Florence, A. J., Shankland, K., Shankland, N. & Johnston, A. (2007). Acta Cryst. E63, o675–o677. [DOI] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Oxford Diffraction (2006). CrysAlis CCD, CrysAlis RED and ABSPACK Oxford Diffraction Ltd, Abingdon, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Westrip, S. P. (2008). publCIF. In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I. DOI: 10.1107/S1600536808016577/cf2202sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808016577/cf2202Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




