Abstract
The molecule of the title compound, C12H16N2O4, is centrosymmetric and the amide group is twisted relative to the benzene ring by 14.40 (13)°. The molecules are hydrogen bonded into a three-dimensional framework, with the hydroxy O atoms acting as acceptors in N—H⋯O hydrogen bonds and as donors in O—H⋯O=C interactions.
Related literature
For the synthesis of the title compound, see: Sułkowski et al. (2000 ▶); Shukla & Harad (2006 ▶). For bond-length data, see: Allen (2002 ▶). For hydrogen bonding, see: Desiraju & Steiner (1999 ▶).
Experimental
Crystal data
C12H16N2O4
M r = 252.27
Monoclinic,

a = 4.9062 (4) Å
b = 13.6467 (10) Å
c = 8.8840 (7) Å
β = 97.262 (6)°
V = 590.04 (8) Å3
Z = 2
Mo Kα radiation
μ = 0.11 mm−1
T = 200 (1) K
0.26 × 0.22 × 0.18 mm
Data collection
Oxford Diffraction KM-4-CCD Sapphire3 diffractometer
Absorption correction: none
5655 measured reflections
2000 independent reflections
1599 reflections with I > 2σ(I)
R int = 0.013
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.097
S = 1.02
2000 reflections
114 parameters
All H-atom parameters refined
Δρmax = 0.34 e Å−3
Δρmin = −0.21 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2006 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808017467/gk2148sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808017467/gk2148Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H3⋯O2i | 0.879 (16) | 2.080 (16) | 2.9333 (10) | 163.3 (13) |
| O2—H8⋯O1ii | 0.863 (18) | 1.872 (18) | 2.7204 (9) | 167.1 (15) |
| C2—H2⋯O2i | 0.972 (15) | 2.412 (14) | 3.3458 (11) | 161.0 (11) |
| C5—H4⋯O1ii | 0.988 (12) | 2.523 (12) | 3.2738 (12) | 132.6 (9) |
| C5—H5⋯O1iii | 0.992 (13) | 2.612 (13) | 3.5671 (12) | 161.7 (11) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
supplementary crystallographic information
Comment
Polyethylene terephthalate (PET) is a very popular thermoplastic polyester. The chemical recycling of its wastes has been the subject of keen interest as a valuable material for different chemical processes. Aminolysis of PET yields N, N' - bis(2-hydroxyethyl)benzene-1,4-dicarboxamide, which can be a potential candidate for further reactions leading to obtain other useful products. To get information about the hydrogen bonding in this interesting material we determined its crystal structure. In crystal the title molecule is located around inversion center (Fig. 1.).
The value of the C2—C3—C4 angle of 123.58 (7)° is in agreement with a geometry of the Ph—C(=O)—NH—CH2 subunit. A search of the Cambridge Structural Database [version 5.28; Allen, 2002] shows that in similar compounds this angle is consistently greater than 120° with the mean value of 122.46 (8)°. The widening of this angle can be related to a steric hindrance between H3 of the amide group and H atom attached to C2, as the consequence of a small twist of the amide group relative to the benzene ring. The torsion angles around the C—C bond between the amide group and the benzene ring are: C1—C3—C4—O1 14.40 (13)° and C2—C3—C4—N1 14.74 (13)°.
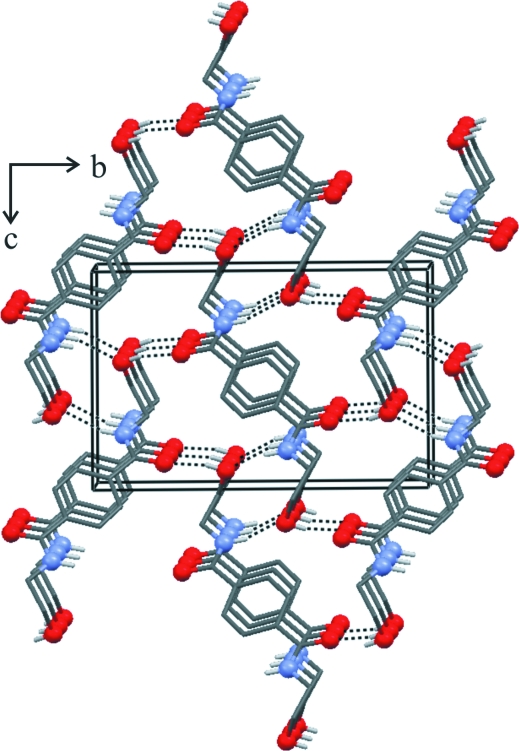
The molecules of the title compound are connected via N—H···O, O—H···O and C—H···O hydrogen bonds (Fig. 2; Table 1) into a three-dimensional framework. All N and O atoms participate in hydrogen bonding. The IR spectrum of the title compound shows bands corresponding to the N—H and O—H stretching vibrations in the 3370 - 2480 cm-1 region. The center of gravity of the νN—H and νO—H bands is located at ca 2960 cm-1. The relative shifts of about 440 cm-1 and 640 cm-1 for N—H and O—H bands allow to classify the N—H···O and O—H···O interactions in this crystal as strong hydrogen bonds (Desiraju & Steiner, 1999).
Experimental
The title compound was obtained according to the method described by Sułkowski et al. (2000) and Shukla & Harad (2006). Single crystal suitable for X-ray analysis was obtained from water solution. Analysis calculated: C 57.13, H 6.39, N 11.10%; found C 57.12, H 6.26, N 10.93%. IR spectra were recorded with the Perkin-Elmer Spectrum.
Refinement
All H atoms were located in a difference Fourier map and freely refined with isotropic displacement parameters.
Figures
Fig. 1.
Molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level. The H-atom radius is arbitrary. Symmetry code: (a) -x, -y + 1, -z + 1
Fig. 2.
Packing diagram for the title compound. Hydrogen bonds are shown with dashed lines. Hydrogen atoms not involved in hydrogen bonding are omitted for clarity.
Crystal data
| C12H16N2O4 | F000 = 268 |
| Mr = 252.27 | Dx = 1.420 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3594 reflections |
| a = 4.9062 (4) Å | θ = 3.0–32.8º |
| b = 13.6467 (10) Å | µ = 0.11 mm−1 |
| c = 8.8840 (7) Å | T = 200 (1) K |
| β = 97.262 (6)º | Needle, colourless |
| V = 590.04 (8) Å3 | 0.26 × 0.22 × 0.18 mm |
| Z = 2 |
Data collection
| Oxford Diffraction KM-4-CCD Sapphire3 diffractometer | 2000 independent reflections |
| Radiation source: fine-focus sealed tube | 1599 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.014 |
| Detector resolution: 16.0328 pixels mm-1 | θmax = 32.9º |
| T = 200(1) K | θmin = 3.0º |
| ω scans | h = −7→5 |
| Absorption correction: none | k = −19→19 |
| 5655 measured reflections | l = −13→12 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | All H-atom parameters refined |
| wR(F2) = 0.097 | w = 1/[σ2(Fo2) + (0.0542P)2 + 0.0884P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 2000 reflections | Δρmax = 0.34 e Å−3 |
| 114 parameters | Δρmin = −0.21 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.27008 (15) | 0.27588 (5) | 0.35222 (8) | 0.03059 (17) | |
| H8 | 0.422 (3) | 0.3500 (13) | −0.1314 (17) | 0.051 (4)* | |
| O2 | 0.50462 (15) | 0.40412 (5) | −0.10308 (8) | 0.02923 (17) | |
| N1 | 0.45385 (15) | 0.39818 (5) | 0.22776 (8) | 0.02234 (16) | |
| H3 | 0.470 (3) | 0.4611 (12) | 0.2100 (16) | 0.043 (4)* | |
| C1 | −0.0348 (2) | 0.59732 (6) | 0.45869 (11) | 0.02646 (19) | |
| H1 | −0.063 (3) | 0.6650 (11) | 0.4282 (15) | 0.037 (3)* | |
| C2 | 0.1105 (2) | 0.53512 (7) | 0.37400 (10) | 0.02685 (19) | |
| H2 | 0.183 (3) | 0.5608 (11) | 0.2851 (16) | 0.044 (4)* | |
| C3 | 0.14642 (16) | 0.43721 (6) | 0.41503 (9) | 0.01995 (16) | |
| C4 | 0.29698 (17) | 0.36475 (6) | 0.32869 (9) | 0.02059 (16) | |
| C5 | 0.61451 (18) | 0.33030 (6) | 0.14753 (10) | 0.02380 (17) | |
| H4 | 0.493 (2) | 0.2750 (9) | 0.1119 (13) | 0.028 (3)* | |
| H5 | 0.772 (3) | 0.3050 (10) | 0.2180 (15) | 0.037 (3)* | |
| C6 | 0.71991 (19) | 0.37877 (7) | 0.01299 (11) | 0.02745 (19) | |
| H6 | 0.851 (3) | 0.3331 (10) | −0.0254 (15) | 0.039 (3)* | |
| H7 | 0.816 (2) | 0.4390 (10) | 0.0435 (14) | 0.029 (3)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0417 (4) | 0.0180 (3) | 0.0354 (4) | 0.0064 (3) | 0.0175 (3) | 0.0056 (2) |
| O2 | 0.0438 (4) | 0.0192 (3) | 0.0259 (3) | −0.0063 (3) | 0.0089 (3) | 0.0005 (2) |
| N1 | 0.0275 (4) | 0.0175 (3) | 0.0238 (3) | 0.0003 (2) | 0.0099 (3) | −0.0013 (2) |
| C1 | 0.0365 (5) | 0.0179 (4) | 0.0276 (4) | 0.0047 (3) | 0.0145 (3) | 0.0056 (3) |
| C2 | 0.0372 (5) | 0.0203 (4) | 0.0260 (4) | 0.0041 (3) | 0.0155 (3) | 0.0052 (3) |
| C3 | 0.0229 (4) | 0.0182 (3) | 0.0196 (3) | 0.0017 (3) | 0.0057 (3) | 0.0014 (3) |
| C4 | 0.0234 (3) | 0.0188 (4) | 0.0200 (3) | 0.0025 (3) | 0.0044 (3) | 0.0014 (3) |
| C5 | 0.0270 (4) | 0.0214 (4) | 0.0243 (4) | 0.0037 (3) | 0.0083 (3) | −0.0013 (3) |
| C6 | 0.0288 (4) | 0.0252 (4) | 0.0310 (4) | −0.0020 (3) | 0.0140 (3) | −0.0027 (3) |
Geometric parameters (Å, °)
| O1—C4 | 1.2404 (10) | C3—C1i | 1.3913 (12) |
| O2—H8 | 0.863 (18) | C3—C4 | 1.5017 (11) |
| N1—C4 | 1.3338 (11) | C5—H4 | 0.988 (12) |
| N1—C5 | 1.4594 (11) | C5—H5 | 0.992 (13) |
| N1—H3 | 0.879 (16) | C6—O2 | 1.4224 (12) |
| C1—H1 | 0.967 (14) | C6—C5 | 1.5133 (12) |
| C2—C1 | 1.3896 (12) | C6—H6 | 0.989 (14) |
| C2—C3 | 1.3902 (12) | C6—H7 | 0.970 (13) |
| C2—H2 | 0.972 (15) | ||
| C3—C2—C1 | 120.12 (8) | C2—C3—C4 | 123.58 (7) |
| C3—C2—H2 | 120.8 (9) | C1i—C3—C4 | 117.25 (7) |
| C1—C2—H2 | 119.0 (9) | N1—C5—C6 | 111.58 (7) |
| O2—C6—C5 | 112.48 (7) | N1—C5—H4 | 107.5 (7) |
| O2—C6—H7 | 106.7 (7) | C6—C5—H4 | 109.4 (7) |
| C5—C6—H7 | 110.7 (7) | N1—C5—H5 | 109.7 (8) |
| O2—C6—H6 | 111.1 (8) | C6—C5—H5 | 109.6 (8) |
| C5—C6—H6 | 107.4 (8) | H4—C5—H5 | 108.9 (10) |
| H7—C6—H6 | 108.4 (10) | O1—C4—N1 | 122.04 (8) |
| C6—O2—H8 | 106.3 (10) | O1—C4—C3 | 119.21 (7) |
| C4—N1—C5 | 120.29 (7) | N1—C4—C3 | 118.75 (7) |
| C4—N1—H3 | 122.0 (9) | C2—C1—C3i | 120.73 (8) |
| C5—N1—H3 | 117.7 (9) | C2—C1—H1 | 119.7 (8) |
| C2—C3—C1i | 119.16 (7) | C3i—C1—H1 | 119.6 (8) |
| C1—C2—C3—C1i | 0.09 (16) | C2—C3—C4—O1 | −164.62 (9) |
| C1—C2—C3—C4 | 179.10 (8) | C1i—C3—C4—O1 | 14.40 (13) |
| C4—N1—C5—C6 | 165.93 (8) | C2—C3—C4—N1 | 14.74 (13) |
| O2—C6—C5—N1 | −66.81 (10) | C1i—C3—C4—N1 | −166.23 (8) |
| C5—N1—C4—O1 | −3.87 (13) | C3—C2—C1—C3i | −0.09 (16) |
| C5—N1—C4—C3 | 176.78 (7) |
Symmetry codes: (i) −x, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H3···O2ii | 0.879 (16) | 2.080 (16) | 2.9333 (10) | 163.3 (13) |
| O2—H8···O1iii | 0.863 (18) | 1.872 (18) | 2.7204 (9) | 167.1 (15) |
| C2—H2···O2ii | 0.972 (15) | 2.412 (14) | 3.3458 (11) | 161.0 (11) |
| C5—H4···O1iii | 0.988 (12) | 2.523 (12) | 3.2738 (12) | 132.6 (9) |
| C5—H5···O1iv | 0.992 (13) | 2.612 (13) | 3.5671 (12) | 161.7 (11) |
Symmetry codes: (ii) −x+1, −y+1, −z; (iii) x, −y+1/2, z−1/2; (iv) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GK2148).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Desiraju, G. R. & Steiner, T. (1999). The Weak Hydrogen Bond in Structural Chemistry and Biology New York: Oxford University Press.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Oxford Diffraction (2006). CrysAlis CCD and CrysAlis RED Oxford Diffraction, Abingdon, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shukla, S. R. & Harad, A. M. (2006). Polym. Degrad. Stabil.91, 1850–1854.
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Sułkowski, W. W., Ossowski, J., Sułkowska, A. & Bajdur, W. (2000). Abstract. 1st International Conference ‘Modification, Degradation and Stabilization of Polymers’, p. P11, Palermo, Italy.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808017467/gk2148sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808017467/gk2148Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report