Abstract
The title compound, C28H22N4, is the unexpected by-product of the reaction of 2-hydroxyacetophenone and 1,2-diaminobenzene under iodine catalysis, during which a carbon–carbon σ-bond between two quinoxaline units was formed. Although a fully oxidized title compound should sterically be possible, only one quinoxaline ring is fully oxidized while the second ring remains in the reduced form. As expected, the tetrahydroquinoxaline unit is not planar; it adopts a sofa conformation, whereby the atom joining the two heterocyclic systems lies out of the plane of the other atoms. The quinoxaline ring system makes a dihedral angle of 53.61 (4)° with its phenyl ring substituent. The crystal packing is determined by pairs of N—H⋯N, N—H⋯π and weak C—H⋯N hydrogen bonds, forming a chain parallel to the a axis.
Related literature
For related literature, see: Banik et al. (1999 ▶); Chen et al. (2005 ▶); Gazit et al. (1996 ▶); Hwang et al. (2005 ▶); Jones et al. (2006 ▶); Kim et al. (2004 ▶); Kulkarni et al. (2006 ▶); McGovern et al. (2005 ▶); More et al. (2005 ▶); Raw et al. (2004 ▶); Robinson & Taylor (2005 ▶); Shirota & Kageyama (2007 ▶).
Experimental
Crystal data
C28H22N4
M r = 414.50
Monoclinic,

a = 11.1601 (6) Å
b = 11.3987 (6) Å
c = 16.4638 (8) Å
β = 93.170 (2)°
V = 2091.17 (19) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 133 (2) K
0.35 × 0.35 × 0.32 mm
Data collection
Bruker SMART 1000 CCD diffractometer
Absorption correction: none
24311 measured reflections
6364 independent reflections
3306 reflections with I > 2σ(I)
R int = 0.109
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.152
S = 0.92
6364 reflections
297 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.42 e Å−3
Δρmin = −0.29 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 1998 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP (Siemens, 1994 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808015481/su2058sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808015481/su2058Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H02⋯N2i | 0.84 (2) | 2.49 (2) | 3.211 (2) | 144 (2) |
| C15—H15⋯N3ii | 1.00 | 2.69 | 3.499 (2) | 138 |
| N3—H01⋯Cent(C23–C28)ii | 0.85 (2) | 2.63 | 3.42 | 157 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
We thank the German Federal Ministry of Education and Research (BMBF) for financial support.
supplementary crystallographic information
Comment
Quinoxalines are a versatile class of heterocyclic compounds. This moiety is found in pharmaceutically and biologically active molecules e.g. as potential antibiotics (Kim et al., 2004), DNA cleavage agents (More et al., 2005) and for inhibition of tumor activity (Gazit et al., 1996). The electron-withdrawing property of quinoxalines leads to their use in electroluminescent devices as electron transporters (Shirota & Kageyama, 2007). Often these transporters are designed as a dipolar unit consisting of an acceptor (quinoxaline) and a donor (e.g. triarylamines) (Chen et al., 2005; Kulkarni et al., 2006). Lately quinoxalines have been used as ligands for metal complexes (Jones et al., 2006) that show efficient electroluminescence (Hwang et al., 2005) in organic light-emitting diodes (OLEDs).
Several routes for the synthesis of quinoxalines are described in the literature. For condensations, starting materials are 1,2-diketones and 1,2-diamines which are reacted in boiling ethanol (Gazit et al., 1996) or at room temperature in acetonitrile with iodine as catalyst (More et al., 2005). For 2-substituted quinoxalines, α-hydroxy ketones and 1,2-diamines are used together with different catalysts such as Pd(OAc)2/Et3N (Robinson & Taylor, 2005) or MnO2 (Raw et al., 2004). These catalysts are necessary to oxidize the alcohol from the a-hydroxy ketones. In the present study a synthesis for 2-phenylquinoxaline was planned under similiar conditions to those used by More et al. (2005) from 2-hydroxyacetophenone, 1,2-diaminobenzene and iodine without the use of an oxidization catalyst. As expected the yield of the reaction was low, but beside the anticipated product (I) the title compound (II) was formed. To the best of our knowledge neither the structure itself nor this type of formation have been described in the literature. Raw et al. (2004) describe the formation of an azobenzene derivative as the by-product. The most striking feature is the formation of a carbon-carbon σ-bond [C1—C15 1.536 (2) Å] between two quinoxaline moieties. Assuming that compound (I) is formed in the first place, either the attack on the C═ N bond or the reaction of (I) as a nucleophile with the starting materials could lead to the formation of a dimer. Subsequent reduction with 2 equivalents of hydrogen would form the title compound (II). Barik et al. (1999) demonstrated that dimeric structures starting from imines can be formed via a samarium-induced iodine-catalyzed reduction. The authors postulate a one electron transfer mechanism across the C═ N bond resulting in two carbon radicals merging in a pinacol type reaction. Even though these conditions cannot be found in our case, it is this reference that is most relevant to the formation of a dimer. The red color of compound (II) is remarkable and the origin is unclear, because the UV/VIS-spectrum shows no significant absorption above a maximum of 320 nm (ε = 9300 in CH3CN). In comparison McGovern et al. (2005) have shown that an intramolecular charge-transfer causes the red color of 9,14-dihydrodipyridophenazine, which possesses a moiety like the 1',2',3',4'-tetrahydroquinoxaline in the present study. The title compound potentially exists as two different diastereomers, but one of them is formed exclusively, as shown by spectroscopic evidence. We surmise that the other diastereomer is suppressed for steric reasons.
The molecular structure of compound (II) is illustrated in Fig. 1. Bond lengths and angles in the two phenyl rings and in the quinoxaline unit are normal. As expected the tetrahydroquinoxaline unit is not planar; it adopts a sofa conformation, whereby the atom joining the two heterocyclic systems lies out of the plane of the other atoms. Atoms C15 and C16 show sp3 hybridization (angles ranged from 107.9° to 113.9°). The relative configurations at atoms C15 and C16 are S,R. The bond length C15—C16 [1.549 (2) Å] indicates a C—C single bond, whereas bond C1—C2 [1.444 (2) Å] shows the aromatic character of an oxidized ring. The phenyl rings subtend interplanar angles of 53.61 (4)° with the quinoxaline ring system [C1, N1, C8, C3, N2 and C2], and 79.01 (4)° with the tetrahydroquinoxaline ring [C16, N4, C17, C22 and N3] (atom C15 lies 0.655 (2) Å out of this plane).
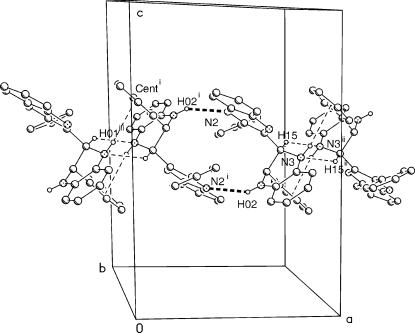
In the crystal structure of (II) the packing of the molecules is determined by weak N4—H02···N2 and C15—H15···N3 hydrogen bonds (Fig 2 and Table 1). Pairs of alternating C—H···N and N—H···N hydrogen bonds are formed across inversion centres. Additionally, there is an N—H···π contact from N3—H01 to the centroid of the phenyl ring [C23–C28]. The overall effect is to form a chain parallel to the a axis.
Experimental
A 100 ml round-bottomed flask was charged with 2-hydroxyacetophenone (3.00 g, 22.034 mmol), 1,2-diaminobenzene (2.86 g, 26.441 mmol), iodine (559 mg, 2.203 mmol), and acetonitrile (30 ml). The reaction was stirred for 23h at room temperature, monitored by thin-layer chromatography and finally concentrated to dryness under reduced pressure. The dark crude product obtained was then subjected to flash column chromatography using silica gel (eluent: 6:1 n-hexane–EtOAc). 630 mg (14%) of 2-phenylquinoxaline, the expected product, and 390 mg (9%) of the unexpected title compound, (II), were obtained. Red crystals of (II) grew overnight from the eluted fractions of the flash column chromatography.
Refinement
Amide H atoms were freely refined [N—H = 0.84 (1), 0.85 (1) Å]. The other H-atoms were included in calculated positions and refined using a riding model: C—H = 0.95 - 1.0 Å with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of compound (II), showing the atom numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
The crystal packing of compound (II), viewed approximately parallel to the b axis, showing the formation of a chain of molecules. Dashed lines indicate C—H···N, N—H···π (thin) and N—H···N (thick) hydrogen bonds. Symmetry operator (iii): -1 + x,y,z.
Fig. 3.
The formation of the title compound.
Crystal data
| C28H22N4 | F000 = 872 |
| Mr = 414.50 | Dx = 1.317 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3799 reflections |
| a = 11.1601 (6) Å | θ = 2–30º |
| b = 11.3987 (6) Å | µ = 0.08 mm−1 |
| c = 16.4638 (8) Å | T = 133 (2) K |
| β = 93.170 (2)º | Prism, red |
| V = 2091.17 (19) Å3 | 0.35 × 0.35 × 0.32 mm |
| Z = 4 |
Data collection
| Bruker SMART 1000 CCD diffractometer | 6364 independent reflections |
| Radiation source: fine-focus sealed tube | 3306 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.109 |
| Detector resolution: 8.192 pixels mm-1 | θmax = 30.5º |
| T = 133(2) K | θmin = 1.8º |
| ω and φ scans | h = −15→15 |
| Absorption correction: none | k = −16→16 |
| 24311 measured reflections | l = −23→23 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.057 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.152 | w = 1/[σ2(Fo2) + (0.0734P)2] |
| S = 0.92 | (Δ/σ)max = 0.001 |
| 6364 reflections | Δρmax = 0.42 e Å−3 |
| 297 parameters | Δρmin = −0.29 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.Least-squares planes (x,y,z in crystal coordinates) and deviations from them (* indicates atom used to define plane)7.0105 (0.0062) x + 1.5050 (0.0091) y - 13.1768 (0.0076) z = 0.4647 (0.0095)* -0.0196 (0.0009) N3 * 0.0131 (0.0012) C22 * 0.0135 (0.0011) C17 * -0.0327 (0.0012) N4 * 0.0257 (0.0009) C16 - 0.6550 (0.0023) C15Rms deviation of fitted atoms = 0.02227.1369 (0.0068) x - 0.2689 (0.0093) y + 12.0496 (0.0089) z = 10.3944 (0.0068)Angle to previous plane (with approximate e.s.d.) = 79.01 (0.04)* 0.0027 (0.0013) C23 * -0.0049 (0.0014) C24 * 0.0020 (0.0015) C25 * 0.0031 (0.0014) C26 * -0.0053 (0.0013) C27 * 0.0023 (0.0013) C28Rms deviation of fitted atoms = 0.00365.4792 (0.0064) x + 2.1540 (0.0079) y + 13.5331 (0.0066) z = 12.6941 (0.0020)Angle to previous plane (with approximate e.s.d.) = 15.63 (0.09)* -0.0411 (0.0011) C1 * 0.0366 (0.0011) C2 * 0.0019 (0.0011) N2 * -0.0370 (0.0012) C3 * 0.0330 (0.0012) C8 * 0.0067 (0.0011) N1Rms deviation of fitted atoms = 0.0304- 4.5755 (0.0081) x + 2.1574 (0.0088) y + 15.0404 (0.0054) z = 7.1104 (0.0101)Angle to previous plane (with approximate e.s.d.) = 53.61 (0.04)* -0.0098 (0.0013) C9 * 0.0103 (0.0013) C10 * -0.0010 (0.0014) C11 * -0.0088 (0.0014) C12 * 0.0092 (0.0014) C13 * 0.0001 (0.0013) C14Rms deviation of fitted atoms = 0.0078 |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.73023 (13) | 0.32241 (13) | 0.59153 (9) | 0.0174 (3) | |
| N2 | 0.55476 (13) | 0.48400 (13) | 0.63650 (9) | 0.0168 (3) | |
| N3 | 0.90846 (13) | 0.36907 (13) | 0.49171 (10) | 0.0173 (3) | |
| H01 | 0.9504 (18) | 0.3484 (17) | 0.5339 (12) | 0.019 (5)* | |
| N4 | 0.70665 (15) | 0.39968 (13) | 0.38883 (10) | 0.0187 (4) | |
| H02 | 0.639 (2) | 0.408 (2) | 0.3641 (15) | 0.043 (7)* | |
| C1 | 0.73376 (15) | 0.43303 (15) | 0.56896 (10) | 0.0147 (4) | |
| C2 | 0.64804 (15) | 0.51743 (15) | 0.59598 (10) | 0.0158 (4) | |
| C3 | 0.54617 (16) | 0.36728 (15) | 0.65568 (10) | 0.0157 (4) | |
| C4 | 0.44601 (16) | 0.32623 (17) | 0.69629 (10) | 0.0191 (4) | |
| H4 | 0.3847 | 0.3792 | 0.7103 | 0.023* | |
| C5 | 0.43791 (18) | 0.20925 (17) | 0.71539 (11) | 0.0228 (4) | |
| H5 | 0.3694 | 0.1811 | 0.7411 | 0.027* | |
| C6 | 0.53032 (18) | 0.13035 (17) | 0.69726 (11) | 0.0239 (4) | |
| H6 | 0.5238 | 0.0498 | 0.7113 | 0.029* | |
| C7 | 0.62894 (18) | 0.16901 (17) | 0.65979 (11) | 0.0225 (4) | |
| H7 | 0.6919 | 0.1160 | 0.6493 | 0.027* | |
| C8 | 0.63732 (17) | 0.28797 (16) | 0.63657 (10) | 0.0173 (4) | |
| C9 | 0.66016 (16) | 0.64540 (15) | 0.58036 (11) | 0.0174 (4) | |
| C10 | 0.56469 (17) | 0.70799 (16) | 0.54368 (11) | 0.0205 (4) | |
| H10 | 0.4918 | 0.6686 | 0.5286 | 0.025* | |
| C11 | 0.57501 (19) | 0.82717 (17) | 0.52897 (12) | 0.0270 (5) | |
| H11 | 0.5099 | 0.8690 | 0.5030 | 0.032* | |
| C12 | 0.6806 (2) | 0.88530 (18) | 0.55223 (13) | 0.0305 (5) | |
| H12 | 0.6881 | 0.9668 | 0.5416 | 0.037* | |
| C13 | 0.77457 (19) | 0.82530 (17) | 0.59063 (13) | 0.0280 (5) | |
| H13 | 0.8459 | 0.8659 | 0.6077 | 0.034* | |
| C14 | 0.76546 (17) | 0.70532 (17) | 0.60446 (12) | 0.0229 (4) | |
| H14 | 0.8309 | 0.6640 | 0.6303 | 0.027* | |
| C15 | 0.82767 (15) | 0.46513 (15) | 0.50791 (11) | 0.0161 (4) | |
| H15 | 0.8761 | 0.5330 | 0.5299 | 0.019* | |
| C16 | 0.76314 (16) | 0.50187 (15) | 0.42612 (11) | 0.0170 (4) | |
| H16 | 0.6983 | 0.5586 | 0.4385 | 0.020* | |
| C17 | 0.75484 (15) | 0.28781 (15) | 0.39818 (11) | 0.0154 (4) | |
| C18 | 0.70330 (17) | 0.19060 (16) | 0.35817 (11) | 0.0204 (4) | |
| H18 | 0.6338 | 0.2006 | 0.3229 | 0.025* | |
| C19 | 0.75227 (18) | 0.07975 (17) | 0.36928 (12) | 0.0254 (5) | |
| H19 | 0.7159 | 0.0143 | 0.3420 | 0.030* | |
| C20 | 0.85415 (18) | 0.06402 (17) | 0.42005 (12) | 0.0262 (5) | |
| H20 | 0.8884 | −0.0118 | 0.4272 | 0.031* | |
| C21 | 0.90571 (17) | 0.15974 (17) | 0.46026 (11) | 0.0226 (4) | |
| H21 | 0.9756 | 0.1487 | 0.4950 | 0.027* | |
| C22 | 0.85742 (15) | 0.27129 (15) | 0.45090 (10) | 0.0157 (4) | |
| C23 | 0.84954 (16) | 0.56456 (16) | 0.37228 (10) | 0.0166 (4) | |
| C24 | 0.92716 (18) | 0.50358 (18) | 0.32431 (12) | 0.0261 (5) | |
| H24 | 0.9247 | 0.4203 | 0.3231 | 0.031* | |
| C25 | 1.00828 (19) | 0.56274 (18) | 0.27815 (12) | 0.0300 (5) | |
| H25 | 1.0612 | 0.5195 | 0.2462 | 0.036* | |
| C26 | 1.01289 (18) | 0.68356 (18) | 0.27821 (12) | 0.0279 (5) | |
| H26 | 1.0687 | 0.7236 | 0.2466 | 0.033* | |
| C27 | 0.93552 (18) | 0.74584 (18) | 0.32474 (12) | 0.0263 (5) | |
| H27 | 0.9373 | 0.8292 | 0.3246 | 0.032* | |
| C28 | 0.85472 (17) | 0.68660 (16) | 0.37190 (11) | 0.0207 (4) | |
| H28 | 0.8026 | 0.7302 | 0.4042 | 0.025* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0193 (8) | 0.0180 (8) | 0.0151 (8) | −0.0017 (6) | 0.0013 (6) | −0.0004 (6) |
| N2 | 0.0183 (8) | 0.0170 (8) | 0.0151 (7) | −0.0004 (6) | 0.0007 (6) | 0.0009 (6) |
| N3 | 0.0133 (8) | 0.0209 (8) | 0.0176 (8) | 0.0021 (6) | −0.0016 (6) | 0.0006 (7) |
| N4 | 0.0156 (8) | 0.0164 (8) | 0.0234 (9) | 0.0010 (6) | −0.0053 (7) | 0.0001 (6) |
| C1 | 0.0146 (9) | 0.0171 (9) | 0.0122 (8) | 0.0002 (7) | −0.0019 (7) | −0.0019 (7) |
| C2 | 0.0153 (9) | 0.0177 (9) | 0.0141 (9) | −0.0007 (7) | −0.0024 (7) | 0.0001 (7) |
| C3 | 0.0164 (9) | 0.0160 (9) | 0.0144 (9) | −0.0005 (7) | −0.0017 (7) | 0.0007 (7) |
| C4 | 0.0176 (9) | 0.0245 (10) | 0.0151 (9) | −0.0013 (8) | 0.0009 (7) | 0.0018 (8) |
| C5 | 0.0251 (11) | 0.0251 (11) | 0.0182 (10) | −0.0085 (8) | 0.0008 (8) | 0.0049 (8) |
| C6 | 0.0350 (11) | 0.0190 (10) | 0.0177 (10) | −0.0048 (9) | 0.0002 (8) | 0.0033 (8) |
| C7 | 0.0275 (11) | 0.0199 (10) | 0.0200 (10) | 0.0014 (8) | 0.0013 (8) | 0.0005 (8) |
| C8 | 0.0211 (9) | 0.0192 (9) | 0.0115 (8) | −0.0020 (7) | −0.0001 (7) | 0.0008 (7) |
| C9 | 0.0198 (9) | 0.0179 (9) | 0.0150 (9) | −0.0004 (7) | 0.0047 (7) | −0.0003 (7) |
| C10 | 0.0222 (10) | 0.0198 (10) | 0.0196 (9) | −0.0014 (8) | 0.0016 (8) | −0.0004 (8) |
| C11 | 0.0371 (12) | 0.0182 (10) | 0.0260 (11) | 0.0032 (9) | 0.0039 (9) | 0.0009 (8) |
| C12 | 0.0445 (13) | 0.0159 (10) | 0.0322 (12) | −0.0033 (9) | 0.0117 (10) | −0.0005 (9) |
| C13 | 0.0296 (11) | 0.0207 (11) | 0.0345 (12) | −0.0098 (9) | 0.0089 (9) | −0.0085 (9) |
| C14 | 0.0192 (10) | 0.0218 (10) | 0.0277 (11) | −0.0009 (8) | 0.0015 (8) | −0.0040 (8) |
| C15 | 0.0147 (9) | 0.0168 (9) | 0.0170 (9) | 0.0002 (7) | 0.0016 (7) | −0.0012 (7) |
| C16 | 0.0163 (9) | 0.0169 (9) | 0.0178 (9) | −0.0006 (7) | 0.0010 (7) | −0.0025 (7) |
| C17 | 0.0157 (9) | 0.0155 (9) | 0.0152 (9) | −0.0007 (7) | 0.0041 (7) | 0.0009 (7) |
| C18 | 0.0230 (10) | 0.0226 (10) | 0.0158 (9) | −0.0028 (8) | 0.0019 (8) | −0.0007 (8) |
| C19 | 0.0339 (12) | 0.0174 (10) | 0.0253 (11) | −0.0042 (9) | 0.0057 (9) | −0.0035 (8) |
| C20 | 0.0335 (12) | 0.0201 (10) | 0.0258 (11) | 0.0075 (9) | 0.0081 (9) | 0.0021 (8) |
| C21 | 0.0206 (10) | 0.0252 (11) | 0.0223 (10) | 0.0079 (8) | 0.0036 (8) | 0.0021 (8) |
| C22 | 0.0141 (9) | 0.0183 (9) | 0.0151 (9) | 0.0000 (7) | 0.0040 (7) | 0.0004 (7) |
| C23 | 0.0178 (9) | 0.0180 (9) | 0.0139 (9) | −0.0001 (7) | −0.0004 (7) | 0.0019 (7) |
| C24 | 0.0328 (12) | 0.0214 (10) | 0.0249 (11) | −0.0028 (9) | 0.0091 (9) | −0.0006 (8) |
| C25 | 0.0378 (13) | 0.0285 (12) | 0.0250 (11) | 0.0005 (10) | 0.0137 (9) | 0.0007 (9) |
| C26 | 0.0279 (11) | 0.0299 (12) | 0.0264 (11) | −0.0059 (9) | 0.0059 (9) | 0.0075 (9) |
| C27 | 0.0315 (12) | 0.0212 (11) | 0.0259 (11) | −0.0033 (9) | −0.0010 (9) | 0.0054 (8) |
| C28 | 0.0225 (10) | 0.0197 (10) | 0.0200 (9) | 0.0026 (8) | 0.0006 (8) | 0.0007 (8) |
Geometric parameters (Å, °)
| N1—C1 | 1.316 (2) | C21—C22 | 1.386 (3) |
| N1—C8 | 1.365 (2) | C23—C24 | 1.390 (3) |
| N2—C2 | 1.323 (2) | C23—C28 | 1.392 (3) |
| N2—C3 | 1.372 (2) | C24—C25 | 1.389 (3) |
| N3—C22 | 1.406 (2) | C25—C26 | 1.378 (3) |
| N3—C15 | 1.453 (2) | C26—C27 | 1.382 (3) |
| N4—C17 | 1.389 (2) | C27—C28 | 1.396 (3) |
| N4—C16 | 1.445 (2) | N3—H01 | 0.85 (2) |
| C1—C2 | 1.444 (2) | N4—H02 | 0.84 (2) |
| C1—C15 | 1.536 (2) | C4—H4 | 0.9500 |
| C2—C9 | 1.489 (2) | C5—H5 | 0.9500 |
| C3—C8 | 1.409 (2) | C6—H6 | 0.9500 |
| C3—C4 | 1.414 (2) | C7—H7 | 0.9500 |
| C4—C5 | 1.374 (3) | C10—H10 | 0.9500 |
| C5—C6 | 1.413 (3) | C11—H11 | 0.9500 |
| C6—C7 | 1.364 (3) | C12—H12 | 0.9500 |
| C7—C8 | 1.413 (3) | C13—H13 | 0.9500 |
| C9—C10 | 1.392 (3) | C14—H14 | 0.9500 |
| C9—C14 | 1.398 (3) | C15—H15 | 1.0000 |
| C10—C11 | 1.386 (3) | C16—H16 | 1.0000 |
| C11—C12 | 1.387 (3) | C18—H18 | 0.9500 |
| C12—C13 | 1.376 (3) | C19—H19 | 0.9500 |
| C13—C14 | 1.391 (3) | C20—H20 | 0.9500 |
| C15—C16 | 1.549 (2) | C21—H21 | 0.9500 |
| C16—C23 | 1.523 (2) | C24—H24 | 0.9500 |
| C17—C18 | 1.397 (3) | C25—H25 | 0.9500 |
| C17—C22 | 1.411 (2) | C26—H26 | 0.9500 |
| C18—C19 | 1.385 (3) | C27—H27 | 0.9500 |
| C19—C20 | 1.385 (3) | C28—H28 | 0.9500 |
| C20—C21 | 1.385 (3) | ||
| C1—N1—C8 | 117.67 (15) | C25—C26—C27 | 119.30 (19) |
| C2—N2—C3 | 117.53 (15) | C26—C27—C28 | 120.16 (18) |
| C22—N3—C15 | 116.50 (15) | C23—C28—C27 | 120.93 (18) |
| C17—N4—C16 | 122.39 (15) | C22—N3—H01 | 111.2 (13) |
| N1—C1—C2 | 121.38 (16) | C15—N3—H01 | 112.2 (13) |
| N1—C1—C15 | 116.44 (15) | C17—N4—H02 | 119.4 (16) |
| C2—C1—C15 | 122.04 (15) | C16—N4—H02 | 118.0 (16) |
| N2—C2—C1 | 121.10 (16) | C5—C4—H4 | 120.2 |
| N2—C2—C9 | 116.80 (15) | C3—C4—H4 | 120.2 |
| C1—C2—C9 | 122.10 (16) | C4—C5—H5 | 119.6 |
| N2—C3—C8 | 120.67 (16) | C6—C5—H5 | 119.6 |
| N2—C3—C4 | 119.80 (16) | C7—C6—H6 | 119.8 |
| C8—C3—C4 | 119.52 (16) | C5—C6—H6 | 119.8 |
| C5—C4—C3 | 119.55 (17) | C6—C7—H7 | 120.0 |
| C4—C5—C6 | 120.76 (17) | C8—C7—H7 | 120.0 |
| C7—C6—C5 | 120.43 (18) | C11—C10—H10 | 119.7 |
| C6—C7—C8 | 119.95 (18) | C9—C10—H10 | 119.7 |
| N1—C8—C3 | 121.07 (16) | C10—C11—H11 | 120.1 |
| N1—C8—C7 | 119.10 (17) | C12—C11—H11 | 120.1 |
| C3—C8—C7 | 119.71 (17) | C13—C12—H12 | 119.9 |
| C10—C9—C14 | 118.95 (17) | C11—C12—H12 | 119.9 |
| C10—C9—C2 | 120.16 (16) | C12—C13—H13 | 119.9 |
| C14—C9—C2 | 120.86 (16) | C14—C13—H13 | 119.9 |
| C11—C10—C9 | 120.61 (18) | C13—C14—H14 | 119.9 |
| C10—C11—C12 | 119.83 (19) | C9—C14—H14 | 119.9 |
| C13—C12—C11 | 120.24 (19) | N3—C15—H15 | 108.8 |
| C12—C13—C14 | 120.20 (19) | C1—C15—H15 | 108.8 |
| C13—C14—C9 | 120.13 (18) | C16—C15—H15 | 108.8 |
| N3—C15—C1 | 113.18 (14) | N4—C16—H16 | 107.7 |
| N3—C15—C16 | 107.91 (14) | C23—C16—H16 | 107.7 |
| C1—C15—C16 | 109.39 (14) | C15—C16—H16 | 107.7 |
| N4—C16—C23 | 113.96 (15) | C19—C18—H18 | 119.6 |
| N4—C16—C15 | 108.80 (14) | C17—C18—H18 | 119.6 |
| C23—C16—C15 | 110.72 (14) | C18—C19—H19 | 119.9 |
| N4—C17—C18 | 121.95 (16) | C20—C19—H19 | 119.9 |
| N4—C17—C22 | 119.13 (16) | C21—C20—H20 | 120.3 |
| C18—C17—C22 | 118.91 (16) | C19—C20—H20 | 120.3 |
| C19—C18—C17 | 120.83 (18) | C20—C21—H21 | 119.3 |
| C18—C19—C20 | 120.17 (18) | C22—C21—H21 | 119.3 |
| C21—C20—C19 | 119.48 (18) | C25—C24—H24 | 119.5 |
| C20—C21—C22 | 121.39 (18) | C23—C24—H24 | 119.5 |
| C21—C22—N3 | 121.97 (17) | C26—C25—H25 | 119.7 |
| C21—C22—C17 | 119.21 (17) | C24—C25—H25 | 119.7 |
| N3—C22—C17 | 118.81 (16) | C25—C26—H26 | 120.3 |
| C24—C23—C28 | 118.01 (17) | C27—C26—H26 | 120.3 |
| C24—C23—C16 | 122.03 (16) | C26—C27—H27 | 119.9 |
| C28—C23—C16 | 119.94 (16) | C28—C27—H27 | 119.9 |
| C25—C24—C23 | 120.91 (18) | C23—C28—H28 | 119.5 |
| C26—C25—C24 | 120.69 (19) | C27—C28—H28 | 119.5 |
| C8—N1—C1—C2 | 4.7 (2) | N1—C1—C15—N3 | −6.8 (2) |
| C8—N1—C1—C15 | −171.04 (15) | C2—C1—C15—N3 | 177.41 (15) |
| C3—N2—C2—C1 | 3.5 (2) | N1—C1—C15—C16 | 113.51 (17) |
| C3—N2—C2—C9 | −176.16 (15) | C2—C1—C15—C16 | −62.2 (2) |
| N1—C1—C2—N2 | −8.0 (3) | C17—N4—C16—C23 | 91.8 (2) |
| C15—C1—C2—N2 | 167.59 (16) | C17—N4—C16—C15 | −32.3 (2) |
| N1—C1—C2—C9 | 171.65 (16) | N3—C15—C16—N4 | 54.42 (18) |
| C15—C1—C2—C9 | −12.8 (2) | C1—C15—C16—N4 | −69.11 (18) |
| C2—N2—C3—C8 | 3.4 (2) | N3—C15—C16—C23 | −71.57 (18) |
| C2—N2—C3—C4 | −177.68 (16) | C1—C15—C16—C23 | 164.91 (14) |
| N2—C3—C4—C5 | −179.96 (16) | C16—N4—C17—C18 | −176.29 (16) |
| C8—C3—C4—C5 | −1.1 (3) | C16—N4—C17—C22 | 5.0 (3) |
| C3—C4—C5—C6 | 2.1 (3) | N4—C17—C18—C19 | −179.23 (17) |
| C4—C5—C6—C7 | −0.5 (3) | C22—C17—C18—C19 | −0.6 (3) |
| C5—C6—C7—C8 | −2.0 (3) | C17—C18—C19—C20 | −0.5 (3) |
| C1—N1—C8—C3 | 2.2 (2) | C18—C19—C20—C21 | 0.7 (3) |
| C1—N1—C8—C7 | 178.17 (16) | C19—C20—C21—C22 | 0.1 (3) |
| N2—C3—C8—N1 | −6.6 (3) | C20—C21—C22—N3 | 179.64 (17) |
| C4—C3—C8—N1 | 174.52 (16) | C20—C21—C22—C17 | −1.1 (3) |
| N2—C3—C8—C7 | 177.48 (16) | C15—N3—C22—C21 | −152.96 (17) |
| C4—C3—C8—C7 | −1.4 (3) | C15—N3—C22—C17 | 27.8 (2) |
| C6—C7—C8—N1 | −173.07 (17) | N4—C17—C22—C21 | −179.94 (16) |
| C6—C7—C8—C3 | 2.9 (3) | C18—C17—C22—C21 | 1.3 (3) |
| N2—C2—C9—C10 | −53.0 (2) | N4—C17—C22—N3 | −0.7 (2) |
| C1—C2—C9—C10 | 127.41 (18) | C18—C17—C22—N3 | −179.40 (16) |
| N2—C2—C9—C14 | 124.95 (19) | N4—C16—C23—C24 | −40.5 (2) |
| C1—C2—C9—C14 | −54.7 (2) | C15—C16—C23—C24 | 82.5 (2) |
| C14—C9—C10—C11 | 2.0 (3) | N4—C16—C23—C28 | 141.30 (17) |
| C2—C9—C10—C11 | 179.92 (17) | C15—C16—C23—C28 | −95.7 (2) |
| C9—C10—C11—C12 | −1.1 (3) | C28—C23—C24—C25 | 0.7 (3) |
| C10—C11—C12—C13 | −0.7 (3) | C16—C23—C24—C25 | −177.50 (18) |
| C11—C12—C13—C14 | 1.7 (3) | C23—C24—C25—C26 | −0.7 (3) |
| C12—C13—C14—C9 | −0.8 (3) | C24—C25—C26—C27 | −0.1 (3) |
| C10—C9—C14—C13 | −1.0 (3) | C25—C26—C27—C28 | 0.8 (3) |
| C2—C9—C14—C13 | −178.92 (17) | C24—C23—C28—C27 | 0.0 (3) |
| C22—N3—C15—C1 | 66.7 (2) | C16—C23—C28—C27 | 178.24 (16) |
| C22—N3—C15—C16 | −54.46 (19) | C26—C27—C28—C23 | −0.8 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H02···N2i | 0.84 (2) | 2.49 (2) | 3.211 (2) | 144 (2) |
| C15—H15···N3ii | 1.00 | 2.69 | 3.499 (2) | 138 |
| N3—H01···Cent(C23–C28)ii | 0.85 (2) | 2.63 | 3.42 | 157 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2058).
References
- Banik, B. K., Zegrocka, O., Banik, I., Hackfeld & L., Becker, F. F. (1999). Tetrahedron Lett.40, 6731–6734.
- Bruker (1998). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, C.-T., Lin, J.-S., Moturu, M. V. R. K., Lin, Y.-W., Yi, W., Tao, Y.-T. & Chien, C.-H. (2005). Chem. Commun. pp. 3980–3982. [DOI] [PubMed]
- Gazit, A., App, H., McMahon, G., Chen, J., Levitzki, A. & Bohmer, F. D. (1996). J. Med. Chem.39, 2170–2177. [DOI] [PubMed]
- Hwang, F.-M., Chen, H.-Y., Chen, P.-S., Liu, C.-S., Chi, Y., Shu, C.-F., Wu, F.-I., Chou, P.-T., Peng, S.-M. & Lee, G.-H. (2005). Inorg. Chem.44, 1344–1353. [DOI] [PubMed]
- Jones, P. G., Ammermann, S., Daniliuc, C., du Mont, W.-W., Kowalsky, W. & Johannes, H.-H. (2006). Acta Cryst. E62, m2202–m2204.
- Kim, Y. B., Kim, Y. H., Park, J. & Kim, S. K. (2004). Bioorg. Med. Chem.14, 541–544.
- Kulkarni, A. P., Kong, X. & Jenekhe, S. A. (2006). Adv. Funct. Mater.16, 1057–1066.
- McGovern, D. A., Selmi, A., O’Brien, J. E., Kelly, J. M. & Long, C. (2005). Chem. Commun. pp. 1402–1404. [DOI] [PubMed]
- More, S. V., Sastry, M. N. V., Wang, C.-C. & Yao, C.-F. (2005). Tetrahedron Lett.46, 6345–6348.
- Raw, S. A., Wilfred, C. D. & Taylor, J. K. (2004). Org. Biomol. Chem.2, 788–796. [DOI] [PubMed]
- Robinson, R. S. & Taylor, J. K. (2005). Synlett, pp. 1003–1005.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shirota, Y. & Kageyama, H. (2007). Chem. Rev.107, 953–1010. [DOI] [PubMed]
- Siemens (1994). XP Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808015481/su2058sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808015481/su2058Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report