Abstract
The title compound, C21H18O7, is an important intermediate in the synthesis of 3-(4-hydroxyphenyl)-4-methyl-6-methoxy-7-hydroxycoumarin, which is a nonsteroidal analogue of 2-methoxyestradiol (2-ME). The substituent benzene ring is not in the same plane as the coumarin ring system, with a dihedral angle of 66.88 (10)°. There are some weak intermolecular C—H⋯O interactions. One carbonyl O atom is disordered over two sites, with occupancies of 0.6 and 0.4.
Related literature
For related literature, see: Gibanananda et al. (2006 ▶); Sutherland et al. (2007 ▶).
Experimental
Crystal data
C21H18O7
M r = 382.35
Triclinic,
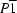
a = 8.142 (3) Å
b = 11.167 (4) Å
c = 11.756 (4) Å
α = 65.130 (4)°
β = 75.392 (4)°
γ = 79.055 (4)°
V = 934.1 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.10 mm−1
T = 293 (2) K
0.15 × 0.12 × 0.04 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.985, T max = 0.996
3893 measured reflections
3245 independent reflections
2279 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.074
wR(F 2) = 0.254
S = 1.09
3245 reflections
266 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.28 e Å−3
Δρmin = −0.50 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SMART; data reduction: SAINT (Bruker, 2000 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808012890/cf2199sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808012890/cf2199Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C20—H20B⋯O2i | 0.96 | 2.47 | 3.362 (4) | 154 |
| C20—H20C⋯O4Bi | 0.96 | 2.55 | 3.297 (9) | 134 |
| C11—H11B⋯O7ii | 0.96 | 2.74 | 3.349 (4) | 122 |
| C13—H13⋯O2iii | 0.93 | 2.74 | 3.331 (4) | 122 |
| C19—H19A⋯O7iii | 0.96 | 2.50 | 3.392 (5) | 154 |
| C17—H17⋯O2iv | 0.93 | 2.66 | 3.246 (3) | 122 |
Symmetry codes: (i) 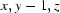 ; (ii)
; (ii) 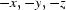 ; (iii)
; (iii) 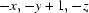 ; (iv)
; (iv) 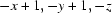 .
.
Acknowledgments
This work was financed by the National Natural Science Foundation of China (grant No. 30500631, awarded to Qian Zhang) and the Postgraduate Innovative Research Foundation of Fudan University (grant awarded to Hao Jiang). The authors acknowledge Professor Minqin Chen, Center of Analysis and Measurement, Fudan University, for his kind help with the data analysis and his professional advice.
supplementary crystallographic information
Comment
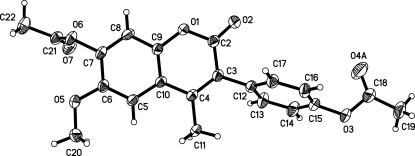
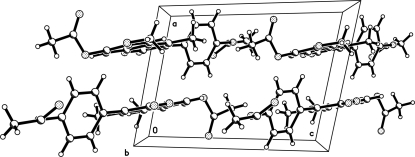
2-ME, an endogenous metabolite of estrogen, was proved to be a potent antitumor and antiangiogenic compound (Gibanananda et al., 2006). Currently 2-ME is in phase I–III clinical trials for treating a variety of solid cancers, especially breast cancer, prostate cancer and multiple myeloma (Sutherland et al., 2007). Based on the structure and the bioactivity of 2-ME, 3-(p-hydroxyphenyl)-4-methyl-6-methoxyl-7-hydroxycoumarin, an non-steroidal analog of 2-ME, was designed, synthesized and evaluated on Human Umbilical Vein Endothelial Cells (HUVEC). The compound showed higher activity and much lower toxicity (EC50 = 5.69 µM; TI = 45.01) than 2-ME (EC50 = 8.59 µM; TI = 8.25) in the biological assay. Here we report the crystal structure of 3-(p-acetoxyphenyl)-4-methyl-6-methoxyl-7-acetoxycoumarin, which is an important intermediate in the synthesis of 3-(p-hydroxyphenyl)-4-methyl-6-methoxyl-7-hydroxycoumarin. The molecular structure of (I) is illustrated in Fig.1. The coumarin ring system (C1—C10) is essentially planar, with a mean deviation of 0.0153 Å from the least-squares plane defined by the ten constituent atoms. The coumarin ring system and the 3-aryl ring make a dihedral angle of 66.88 (10)°. The fact that the of C3—C12 bond [length 1.480 (4) Å] is a single bond also confirms that the coumarin ring system and the 3-substituent are not conjugated. The molecular packing (Fig. 2) is stabilized by weak intermolecular C—H···O hydrogen bonds.
Experimental
A mixture of 1-(2,4-dihydroxyl-5-methoxyphenyl)ethanone (300 mg, 1.65 mmol), 4-hydroxyphenylacetic acid (501 mg, 3.29 mmol), Et3N (6 ml) and Ac2O (10 ml) was refluxed for 10 h. After cooling, the mixture was poured into 2 N HCl (20 ml) and extracted with acetyl acetate. The organic layer was dried over Na2SO4, filtered, and concentrated under reduced pressure to give a yellow oil, which was purified via chromatography on silica gel column with petroleum ether/acetone (10:3) as eluent. The title compound was recrystallized from acetyl acetate to give colorless crystals for the single-crystal X-ray diffraction analysis.
Refinement
All H atoms were positioned geometrically and refined using a riding model with C—H = 0.93 Å for aromatic H atoms and 0.96 Å for methyl H atoms, and refined in riding mode with Uiso(H) = 1.2 Ueq(C) for aromatic H atoms and Uiso(H) = 1.5 Ueq(C) for methyl H atoms.
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 30% probability displacement ellipsoids for non-H atoms. The minor disorder component is not shown.
Fig. 2.
Packing diagram, viewed down the b axis.
Crystal data
| C21H18O7 | Z = 2 |
| Mr = 382.35 | F000 = 400 |
| Triclinic, P1 | Dx = 1.359 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 8.142 (3) Å | Cell parameters from 954 reflections |
| b = 11.167 (4) Å | θ = 2.6–26.3º |
| c = 11.756 (4) Å | µ = 0.10 mm−1 |
| α = 65.130 (4)º | T = 293 (2) K |
| β = 75.392 (4)º | Sheet, colorless |
| γ = 79.055 (4)º | 0.15 × 0.12 × 0.04 mm |
| V = 934.1 (5) Å3 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 3245 independent reflections |
| Radiation source: fine-focus sealed tube | 2279 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.031 |
| T = 293(2) K | θmax = 25.1º |
| φ and ω scans | θmin = 2.0º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −9→9 |
| Tmin = 0.985, Tmax = 0.996 | k = −13→10 |
| 3893 measured reflections | l = −13→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.074 | H-atom parameters constrained |
| wR(F2) = 0.254 | w = 1/[σ2(Fo2) + (0.1741P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max < 0.001 |
| 3245 reflections | Δρmax = 0.28 e Å−3 |
| 266 parameters | Δρmin = −0.50 e Å−3 |
| 1 restraint | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.3185 (3) | 0.17959 (18) | 0.16190 (17) | 0.0502 (6) | |
| O2 | 0.3094 (3) | 0.3939 (2) | 0.1010 (2) | 0.0586 (6) | |
| O3 | 0.2256 (3) | 0.8533 (2) | −0.4110 (2) | 0.0687 (7) | |
| O5 | 0.2670 (3) | −0.27608 (18) | 0.10771 (19) | 0.0563 (6) | |
| O6 | 0.3460 (2) | −0.28758 (18) | 0.31855 (17) | 0.0507 (6) | |
| O7 | 0.0661 (3) | −0.3060 (2) | 0.3850 (2) | 0.0736 (7) | |
| C2 | 0.2997 (4) | 0.3052 (3) | 0.0710 (3) | 0.0461 (7) | |
| C3 | 0.2680 (3) | 0.3215 (3) | −0.0519 (3) | 0.0422 (7) | |
| C4 | 0.2529 (3) | 0.2128 (3) | −0.0733 (2) | 0.0405 (6) | |
| C5 | 0.2552 (3) | −0.0362 (3) | 0.0142 (2) | 0.0425 (7) | |
| H5 | 0.2329 | −0.0317 | −0.0613 | 0.051* | |
| C6 | 0.2753 (3) | −0.1569 (3) | 0.1113 (3) | 0.0442 (7) | |
| C7 | 0.3133 (3) | −0.1642 (3) | 0.2232 (3) | 0.0440 (7) | |
| C8 | 0.3266 (4) | −0.0521 (3) | 0.2393 (3) | 0.0481 (7) | |
| H8 | 0.3507 | −0.0578 | 0.3148 | 0.058* | |
| C9 | 0.3036 (3) | 0.0699 (3) | 0.1407 (2) | 0.0413 (6) | |
| C10 | 0.2678 (3) | 0.0826 (3) | 0.0268 (2) | 0.0399 (6) | |
| C11 | 0.2208 (4) | 0.2260 (3) | −0.1983 (3) | 0.0527 (8) | |
| H11A | 0.2364 | 0.3147 | −0.2601 | 0.079* | |
| H11B | 0.1061 | 0.2074 | −0.1871 | 0.079* | |
| H11C | 0.2992 | 0.1644 | −0.2276 | 0.079* | |
| C12 | 0.2578 (3) | 0.4597 (3) | −0.1475 (3) | 0.0444 (7) | |
| C13 | 0.1053 (4) | 0.5256 (3) | −0.1852 (3) | 0.0592 (8) | |
| H13 | 0.0075 | 0.4810 | −0.1518 | 0.071* | |
| C14 | 0.0962 (4) | 0.6549 (3) | −0.2704 (3) | 0.0641 (9) | |
| H14 | −0.0067 | 0.6966 | −0.2953 | 0.077* | |
| C15 | 0.2384 (4) | 0.7232 (3) | −0.3192 (3) | 0.0524 (8) | |
| C16 | 0.3906 (4) | 0.6615 (3) | −0.2841 (3) | 0.0540 (8) | |
| H16 | 0.4875 | 0.7072 | −0.3181 | 0.065* | |
| C17 | 0.3995 (4) | 0.5314 (3) | −0.1982 (3) | 0.0497 (7) | |
| H17 | 0.5028 | 0.4907 | −0.1737 | 0.060* | |
| C18 | 0.2305 (6) | 0.9555 (4) | −0.3829 (4) | 0.0864 (13) | |
| O4A | 0.1775 (16) | 0.9331 (7) | −0.2672 (6) | 0.159 (4) | 0.55 |
| O4B | 0.3254 (11) | 0.9405 (7) | −0.3076 (8) | 0.094 (2) | 0.45 |
| C19 | 0.2078 (6) | 1.0852 (3) | −0.4860 (4) | 0.0876 (12) | |
| H19A | 0.1643 | 1.1509 | −0.4506 | 0.131* | |
| H19B | 0.1287 | 1.0822 | −0.5329 | 0.131* | |
| H19C | 0.3155 | 1.1075 | −0.5424 | 0.131* | |
| C20 | 0.2417 (4) | −0.2736 (3) | −0.0087 (3) | 0.0571 (8) | |
| H20A | 0.1307 | −0.2306 | −0.0234 | 0.086* | |
| H20B | 0.2502 | −0.3627 | −0.0032 | 0.086* | |
| H20C | 0.3270 | −0.2258 | −0.0781 | 0.086* | |
| C21 | 0.2086 (4) | −0.3556 (3) | 0.3919 (3) | 0.0515 (8) | |
| C22 | 0.2624 (5) | −0.4899 (3) | 0.4777 (3) | 0.0720 (10) | |
| H22A | 0.2921 | −0.4860 | 0.5500 | 0.108* | |
| H22B | 0.3596 | −0.5265 | 0.4327 | 0.108* | |
| H22C | 0.1707 | −0.5449 | 0.5065 | 0.108* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0708 (14) | 0.0396 (11) | 0.0404 (11) | −0.0112 (9) | −0.0212 (9) | −0.0075 (9) |
| O2 | 0.0779 (15) | 0.0445 (12) | 0.0597 (13) | −0.0139 (10) | −0.0268 (11) | −0.0153 (10) |
| O3 | 0.113 (2) | 0.0374 (12) | 0.0452 (12) | −0.0105 (12) | −0.0248 (12) | 0.0010 (9) |
| O5 | 0.0827 (15) | 0.0348 (11) | 0.0463 (12) | −0.0121 (10) | −0.0177 (10) | −0.0054 (9) |
| O6 | 0.0515 (12) | 0.0402 (11) | 0.0424 (11) | −0.0096 (9) | −0.0125 (9) | 0.0050 (9) |
| O7 | 0.0529 (14) | 0.0680 (16) | 0.0697 (16) | −0.0088 (12) | −0.0070 (11) | 0.0002 (12) |
| C2 | 0.0489 (16) | 0.0437 (16) | 0.0429 (16) | −0.0153 (12) | −0.0139 (12) | −0.0068 (13) |
| C3 | 0.0420 (15) | 0.0379 (15) | 0.0410 (15) | −0.0089 (11) | −0.0109 (12) | −0.0064 (12) |
| C4 | 0.0398 (14) | 0.0402 (15) | 0.0345 (14) | −0.0074 (11) | −0.0094 (11) | −0.0052 (11) |
| C5 | 0.0492 (16) | 0.0401 (15) | 0.0345 (14) | −0.0087 (12) | −0.0120 (12) | −0.0072 (12) |
| C6 | 0.0464 (15) | 0.0364 (15) | 0.0426 (15) | −0.0107 (12) | −0.0077 (12) | −0.0062 (12) |
| C7 | 0.0437 (15) | 0.0378 (14) | 0.0361 (14) | −0.0081 (11) | −0.0069 (11) | 0.0005 (11) |
| C8 | 0.0555 (17) | 0.0502 (17) | 0.0336 (14) | −0.0113 (14) | −0.0154 (12) | −0.0053 (12) |
| C9 | 0.0469 (15) | 0.0372 (14) | 0.0371 (14) | −0.0089 (11) | −0.0092 (11) | −0.0093 (11) |
| C10 | 0.0402 (14) | 0.0402 (15) | 0.0336 (14) | −0.0096 (11) | −0.0089 (11) | −0.0057 (11) |
| C11 | 0.073 (2) | 0.0404 (15) | 0.0380 (15) | −0.0088 (14) | −0.0198 (14) | −0.0027 (12) |
| C12 | 0.0499 (16) | 0.0397 (15) | 0.0401 (15) | −0.0092 (12) | −0.0122 (12) | −0.0084 (12) |
| C13 | 0.0510 (17) | 0.0442 (17) | 0.065 (2) | −0.0106 (14) | −0.0162 (15) | 0.0009 (14) |
| C14 | 0.0596 (19) | 0.0531 (19) | 0.063 (2) | −0.0022 (15) | −0.0246 (16) | −0.0007 (15) |
| C15 | 0.072 (2) | 0.0405 (16) | 0.0363 (15) | −0.0119 (14) | −0.0124 (14) | −0.0031 (12) |
| C16 | 0.0615 (19) | 0.0416 (16) | 0.0510 (17) | −0.0156 (14) | −0.0083 (14) | −0.0075 (13) |
| C17 | 0.0490 (16) | 0.0442 (16) | 0.0510 (17) | −0.0111 (13) | −0.0130 (13) | −0.0092 (13) |
| C18 | 0.153 (4) | 0.045 (2) | 0.057 (2) | −0.003 (2) | −0.040 (3) | −0.0074 (16) |
| O4A | 0.341 (14) | 0.060 (4) | 0.057 (4) | 0.018 (7) | −0.045 (6) | −0.017 (3) |
| O4B | 0.162 (7) | 0.044 (3) | 0.087 (5) | −0.015 (4) | −0.069 (5) | −0.009 (3) |
| C19 | 0.128 (4) | 0.0412 (19) | 0.070 (2) | −0.002 (2) | −0.020 (2) | −0.0011 (16) |
| C20 | 0.071 (2) | 0.0475 (17) | 0.0543 (18) | −0.0161 (14) | −0.0085 (15) | −0.0191 (14) |
| C21 | 0.0548 (19) | 0.0492 (17) | 0.0397 (16) | −0.0136 (14) | −0.0073 (13) | −0.0047 (13) |
| C22 | 0.077 (2) | 0.0501 (19) | 0.061 (2) | −0.0128 (17) | −0.0088 (17) | 0.0060 (16) |
Geometric parameters (Å, °)
| O1—C2 | 1.369 (3) | C11—H11C | 0.960 |
| O1—C9 | 1.380 (3) | C12—C17 | 1.389 (4) |
| O2—C2 | 1.205 (3) | C12—C13 | 1.392 (4) |
| O3—C18 | 1.324 (4) | C13—C14 | 1.369 (4) |
| O3—C15 | 1.402 (3) | C13—H13 | 0.930 |
| O5—C6 | 1.364 (3) | C14—C15 | 1.372 (5) |
| O5—C20 | 1.423 (4) | C14—H14 | 0.930 |
| O6—C21 | 1.363 (3) | C15—C16 | 1.369 (5) |
| O6—C7 | 1.392 (3) | C16—C17 | 1.378 (4) |
| O7—C21 | 1.194 (4) | C16—H16 | 0.930 |
| C2—C3 | 1.461 (4) | C17—H17 | 0.930 |
| C3—C4 | 1.371 (4) | C18—O4A | 1.248 (7) |
| C3—C12 | 1.480 (4) | C18—O4B | 1.257 (7) |
| C4—C10 | 1.442 (3) | C18—C19 | 1.461 (5) |
| C4—C11 | 1.498 (4) | C19—H19A | 0.960 |
| C5—C6 | 1.364 (4) | C19—H19B | 0.960 |
| C5—C10 | 1.421 (4) | C19—H19C | 0.960 |
| C5—H5 | 0.930 | C20—H20A | 0.960 |
| C6—C7 | 1.392 (4) | C20—H20B | 0.960 |
| C7—C8 | 1.369 (4) | C20—H20C | 0.960 |
| C8—C9 | 1.386 (4) | C21—C22 | 1.470 (4) |
| C8—H8 | 0.930 | C22—H22A | 0.960 |
| C9—C10 | 1.386 (4) | C22—H22B | 0.960 |
| C11—H11A | 0.960 | C22—H22C | 0.960 |
| C11—H11B | 0.960 | ||
| C2—O1—C9 | 121.4 (2) | C14—C13—H13 | 119.3 |
| C18—O3—C15 | 120.8 (2) | C12—C13—H13 | 119.3 |
| C6—O5—C20 | 116.9 (2) | C13—C14—C15 | 120.2 (3) |
| C21—O6—C7 | 116.7 (2) | C13—C14—H14 | 119.9 |
| O2—C2—O1 | 115.9 (2) | C15—C14—H14 | 119.9 |
| O2—C2—C3 | 125.5 (3) | C16—C15—C14 | 120.0 (3) |
| O1—C2—C3 | 118.6 (2) | C16—C15—O3 | 121.5 (3) |
| C4—C3—C2 | 120.2 (2) | C14—C15—O3 | 118.4 (3) |
| C4—C3—C12 | 124.4 (2) | C15—C16—C17 | 119.7 (3) |
| C2—C3—C12 | 115.4 (2) | C15—C16—H16 | 120.1 |
| C3—C4—C10 | 119.3 (2) | C17—C16—H16 | 120.1 |
| C3—C4—C11 | 121.6 (2) | C16—C17—C12 | 121.6 (3) |
| C10—C4—C11 | 119.1 (2) | C16—C17—H17 | 119.2 |
| C6—C5—C10 | 121.0 (2) | C12—C17—H17 | 119.2 |
| C6—C5—H5 | 119.5 | O4A—C18—O3 | 113.0 (5) |
| C10—C5—H5 | 119.5 | O4B—C18—O3 | 117.6 (5) |
| O5—C6—C5 | 125.2 (2) | O4A—C18—C19 | 123.9 (5) |
| O5—C6—C7 | 115.1 (2) | O4B—C18—C19 | 119.1 (5) |
| C5—C6—C7 | 119.6 (2) | O3—C18—C19 | 114.7 (3) |
| C8—C7—O6 | 119.0 (2) | C18—C19—H19A | 109.5 |
| C8—C7—C6 | 121.2 (2) | C18—C19—H19B | 109.5 |
| O6—C7—C6 | 119.7 (2) | H19A—C19—H19B | 109.5 |
| C7—C8—C9 | 118.6 (2) | C18—C19—H19C | 109.5 |
| C7—C8—H8 | 120.7 | H19A—C19—H19C | 109.5 |
| C9—C8—H8 | 120.7 | H19B—C19—H19C | 109.5 |
| O1—C9—C10 | 121.3 (2) | O5—C20—H20A | 109.5 |
| O1—C9—C8 | 116.2 (2) | O5—C20—H20B | 109.5 |
| C10—C9—C8 | 122.5 (2) | H20A—C20—H20B | 109.5 |
| C9—C10—C5 | 117.0 (2) | O5—C20—H20C | 109.5 |
| C9—C10—C4 | 119.1 (2) | H20A—C20—H20C | 109.5 |
| C5—C10—C4 | 123.8 (2) | H20B—C20—H20C | 109.5 |
| C4—C11—H11A | 109.5 | O7—C21—O6 | 121.7 (3) |
| C4—C11—H11B | 109.5 | O7—C21—C22 | 127.3 (3) |
| H11A—C11—H11B | 109.5 | O6—C21—C22 | 111.0 (3) |
| C4—C11—H11C | 109.5 | C21—C22—H22A | 109.5 |
| H11A—C11—H11C | 109.5 | C21—C22—H22B | 109.5 |
| H11B—C11—H11C | 109.5 | H22A—C22—H22B | 109.5 |
| C17—C12—C13 | 117.1 (3) | C21—C22—H22C | 109.5 |
| C17—C12—C3 | 120.9 (2) | H22A—C22—H22C | 109.5 |
| C13—C12—C3 | 121.9 (2) | H22B—C22—H22C | 109.5 |
| C14—C13—C12 | 121.4 (3) | ||
| C9—O1—C2—O2 | 177.5 (2) | C8—C9—C10—C4 | −177.3 (2) |
| C9—O1—C2—C3 | −1.8 (4) | C6—C5—C10—C9 | 0.9 (4) |
| O2—C2—C3—C4 | −177.4 (3) | C6—C5—C10—C4 | 178.1 (2) |
| O1—C2—C3—C4 | 1.8 (4) | C3—C4—C10—C9 | −2.5 (4) |
| O2—C2—C3—C12 | 3.8 (4) | C11—C4—C10—C9 | 177.7 (2) |
| O1—C2—C3—C12 | −177.0 (2) | C3—C4—C10—C5 | −179.6 (2) |
| C2—C3—C4—C10 | 0.3 (4) | C11—C4—C10—C5 | 0.6 (4) |
| C12—C3—C4—C10 | 179.0 (2) | C4—C3—C12—C17 | −113.2 (3) |
| C2—C3—C4—C11 | −179.9 (2) | C2—C3—C12—C17 | 65.6 (4) |
| C12—C3—C4—C11 | −1.2 (4) | C4—C3—C12—C13 | 70.4 (4) |
| C20—O5—C6—C5 | 2.5 (4) | C2—C3—C12—C13 | −110.8 (3) |
| C20—O5—C6—C7 | −175.2 (2) | C17—C12—C13—C14 | 1.2 (5) |
| C10—C5—C6—O5 | −179.4 (2) | C3—C12—C13—C14 | 177.8 (3) |
| C10—C5—C6—C7 | −1.8 (4) | C12—C13—C14—C15 | −1.0 (5) |
| C21—O6—C7—C8 | 106.7 (3) | C13—C14—C15—C16 | 0.9 (5) |
| C21—O6—C7—C6 | −76.1 (3) | C13—C14—C15—O3 | 176.9 (3) |
| O5—C6—C7—C8 | 179.6 (2) | C18—O3—C15—C16 | −74.4 (5) |
| C5—C6—C7—C8 | 1.7 (4) | C18—O3—C15—C14 | 109.6 (4) |
| O5—C6—C7—O6 | 2.5 (4) | C14—C15—C16—C17 | −1.0 (5) |
| C5—C6—C7—O6 | −175.4 (2) | O3—C15—C16—C17 | −176.9 (3) |
| O6—C7—C8—C9 | 176.3 (2) | C15—C16—C17—C12 | 1.1 (5) |
| C6—C7—C8—C9 | −0.8 (4) | C13—C12—C17—C16 | −1.2 (4) |
| C2—O1—C9—C10 | −0.4 (4) | C3—C12—C17—C16 | −177.8 (3) |
| C2—O1—C9—C8 | 179.4 (2) | C15—O3—C18—O4A | −27.7 (9) |
| C7—C8—C9—O1 | −180.0 (2) | C15—O3—C18—O4B | 35.1 (8) |
| C7—C8—C9—C10 | −0.1 (4) | C15—O3—C18—C19 | −177.3 (3) |
| O1—C9—C10—C5 | 179.9 (2) | C7—O6—C21—O7 | −8.4 (4) |
| C8—C9—C10—C5 | 0.0 (4) | C7—O6—C21—C22 | 173.1 (3) |
| O1—C9—C10—C4 | 2.6 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C20—H20B···O2i | 0.96 | 2.47 | 3.362 (4) | 154 |
| C20—H20C···O4Bi | 0.96 | 2.55 | 3.297 (9) | 134 |
| C11—H11B···O7ii | 0.96 | 2.74 | 3.349 (4) | 122 |
| C13—H13···O2iii | 0.93 | 2.74 | 3.331 (4) | 122 |
| C19—H19A···O7iii | 0.96 | 2.50 | 3.392 (5) | 154 |
| C17—H17···O2iv | 0.93 | 2.66 | 3.246 (3) | 122 |
Symmetry codes: (i) x, y−1, z; (ii) −x, −y, −z; (iii) −x, −y+1, −z; (iv) −x+1, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CF2199).
References
- Bruker (2000). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Gibanananda, R., Gopal, D., Veladhuizen, P. J. V., Banerjee, S., Saxena, N. K., Sengupta, K. & Banerjee, S. K. (2006). Biochemistry, 45, 3703–3713.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sutherland, T. E., Anderson, R. L., Hughes, A. R., Altmann, E., Schliga, M., Ziogas, J. & Stewart, A. G. (2007). Drug Discov. Today, 12, 577–584. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808012890/cf2199sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808012890/cf2199Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report