Abstract
A total of 904 consecutive nosocomial isolates of Escherichia coli and Klebsiella pneumoniae collected from 28 Russian hospitals were screened for production of extended-spectrum β-lactamases (ESBLs). The ESBL phenotype was detected in 78 (15.8%) E. coli and 248 (60.8%) K. pneumoniae isolates. One hundred fifteen isolates carried the genes for CTX-M-type β-lactamases, which, as shown by PCR-restriction fragment length polymorphism analysis, were distributed into the two genetic groups of CTX-M-1 (93%)- and CTX-M-2 (7%)-related enzymes. Isolates producing the enzymes of the first group were found in 20 hospitals from geographically distant regions of the country and were characterized by considerable diversity of genetic types, as was demonstrated by enterobacterial repetitive consensus PCR typing. Within this group the CTX-M-3 and the CTX-M-15 β-lactamases were identified. In contrast, the enzymes of the CTX-M-2 group (namely, CTX-M-5) were detected only in eight clonally related E. coli isolates from a single hospital. Notably, the levels of resistance to ceftazidime were remarkably variable among the CTX-M producers. This study provides further evidence of the global dissemination of CTX-M type ESBLs and emphasizes the need for their epidemiological monitoring.
During the past decade extended-spectrum β-lactamases (ESBLs) of the CTX-M type emerged in many countries of the world. The first organisms producing β-lactamases of this type were identified both as single and epidemic clinical isolates in very distant geographic regions (Germany and France and Argentina) in the early 1990s (3, 5, 7). More recently, a rapid increase in the proportion of multiple CTX-M variants to the TEM- and SHV-derived ESBLs has been reported in many hospitals in Spain (9, 12, 15, 35), the United Kingdom (I. Alobwede, F. H. M'Zali, D. M. Livermore, J. Heritage, N. Todd, and P. M. Hawkey, Letter, J. Antimicrob. Chemother. 51:470-471, 2003), Canada (M. P. Muller, D. Boyd, A. Ashi-Sulaiman, C. Larocque, M. Mulvey, D. Reynolds, A. McGeer, and B. M. Willey, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-233, 2001; M. R. Mulvey, E. Bryce, D. Boyd, M. Ofner-Agostini, A. Simor, and S. Paton, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1877, 2002), China (14), and Korea (28). Furthermore, CTX-M β-lactamases, mainly types CTX-M-2 and CTX-M-3, were found to be widespread or even predominant ESBL types in several countries, including Argentina (31, 34; M. Galas, F. Pasteran, R. Melano, A. Petroni, G. Lopez, A. Corso, A. Rossi, et al., Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-109, 1998), Japan (41), and Poland (1).
Currently the CTX-M family includes more than 20 β-lactamases, which may be grouped on the basis of sequence similarity into four distinct clusters (subtypes) epitomized by CTX-M-1, CTX-M-2, CTX-M-8, and CTX-M-9 (26, 37; http://www.lahey.org/studies/webt.htm). Within each cluster there is a high degree of blaCTX-M gene sequence identity (>95%), although members of different subtypes share only 70 to 77% similarity at the nucleotide level. The recent finding of Kluyvera ascorbata species-specific β-lactamases (KluA) which share 98.6 to 100% identity with CTX-M-2 and CTX-M-5 and analysis of DNA sequences adjacent to the KluA- and CTX-M-coding genes provide strong evidence of the direct evolution of the CTX-M-2 cluster from the chromosomal enzyme of K. ascorbata (20, 36). Likewise, the chromosomal KLUG-1 enzyme of Kluyvera georgiana was found to be the most-probable progenitor of the CTX-M-8 that shares 99% identity with KLUG-1 (32). The origins of the other two CTX-M clusters are not so evident yet, although a number of class A chromosomal β-lactamases, including KLUC-1 of Kluyvera cryocrescens, FONA of Serratia fonticola, RAHN-1 of Rahnella aquatilis, OXY-1 of Klebsiella oxytoca, and SED-1 of Citrobacter sedlakii, display partial (72.9 to 85.9%) homology with different CTX-M enzymes (4, 6, 17, 29, 30).
In contrast with TEM- and SHV-type ESBLs, most of the CTX-M enzymes are much more active against cefotaxime and ceftriaxone than against ceftazidime. Thus, most of the CTX-M producers display levels of resistance to cefotaxime significantly higher than those to ceftazidime. However, the classical phenotype of resistance conferred by CTX-M β-lactamases is not universal among all CTX-M producers, since many factors, including production of additional β-lactamases (1) or mutations altering the substrate specificity of CTX-M enzymes (8, 21, 33), can mask their presence. Therefore, the phenotype of resistance to β-lactams may suggest the presence of CTX-M enzymes, but this is not a completely reliable approach. Isoelectric focusing is also inadequate, since the same isoelectric point can correspond to different β-lactamases. PCR has been used widely to detect blaCTX-M genes, but detection of all the known variants usually required multiple reactions with specific primers for different genes (12, 14). Consensus CTX-M primers have also been described (13, 36), but these primers were used for amplification of blaCTX-M genes from a limited number of isolates. Consequently, very few studies have assessed countrywide prevalence of ESBLs which belong to the distinct CTX-M clusters (N. Shibata, Y. Doi, K. Shibayama, T. Yagi, and Y. Arakawa, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-2235, 2001).
In the present study we explored the prevalence of various CTX-M ESBLs among nosocomial Escherichia coli and Klebsiella pneumoniae strains isolated in 28 Russian hospitals. CTX-M-coding genes were detected by PCR with consensus primers specifically recognizing all the known CTX-M variants, and the CTX-M clusters were subsequently differentiated by restriction analysis. We also analyzed the genetic relatedness of CTX-M-producing isolates and determined the partial sequences of the blaCTX-M genes for some isolates responsible for major outbreaks.
(The results of this work were presented in part at the 12th European Congress of Clinical Microbiology and Infectious Diseases [Milan, Italy] and the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy [San Diego, Calif.].)
MATERIALS AND METHODS
Bacterial isolates.
Consecutive nonduplicate nosocomial isolates of E. coli (n = 494) and K. pneumoniae (n = 410) were collected during the period from 1997 to 1998 in intensive care units of 28 hospitals located in 14 Russian cities: Ekaterinburg, Kazan, Krasnodar, Krasnoyarsk, Moscow, Novosibirsk, Omsk, Ryazan, Smolensk, St. Petersburg, Stavropol, Tomsk, Ufa, and Vladivostok. The geographic distribution of the surveyed hospitals is shown in Fig. 1. All the strains were reidentified in the laboratory of the Institute of Antimicrobial Chemotherapy using the API 20E system (bioMérieux, Marcy l'Etoile, France) and stored at −70°C.
FIG. 1.
Geographiclocations of hospitals surveyed in this study.
Susceptibility testing.
MICs of ceftazidime, cefotaxime, cefepime, cefoxitin, amoxicillin-clavulanic acid (2:1), piperacillin-tazobactam (tazobactam fixed at 4 μg/ml), imipenem, gentamicin, amikacin, and ciprofloxacin were determined using Etests (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar (Becton Dickinson, Sparks, Md.). The results of susceptibility testing were interpreted according to the current NCCLS standards (25). E. coli strains ATCC 25922 and ATCC 35218 were used as quality controls.
Phenotypic ESBL detection.
ESBL production was detected by a double-disk synergy test. Disks with cefotaxime (30 μg) and ceftazidime (30 μg) were placed 20 and 30 mm (center to center) from a disk with amoxicillin-clavulanic acid (20 and 10 μg, respectively). E. coli and K. pneumoniae strains producing the known enzymes TEM-12, SHV-2, and CTX-M-3 were used for quality control of ESBL detection.
Detection and subtyping of blaCTX-M genes by PCR-RFLP analysis.
A pair of primers (CTX-M/F' [5′-TTTGCGATGTGCAGTACCAGTAA-3′] and CTX-M/R' [5′-CGATATCGTTGGTGGTGCCATA-3′])matching the conserved sequences at positions 205 to 227 and positions 748 to 727 with respect to the CTX-M translational starting point was designed to amplify a 544-bp fragment of all the known blaCTX-M genes. Prior to PCR analysis bacterial strains were grown overnight at 35°C on MacConkey agar (Becton Dickinson, Franklin Lakes, N.J.) and DNA was extracted with Lyse-N-Go PCR reagent (Pierce, Rockford, Ill.) as recommended by the manufacturer. The 50-μl PCR mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 9), 0.1% Triton X-100, 2 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, one bead of TaqBead Hot Start Polymerase (Promega, Madison, Wis.), and 5 μl of template DNA. Amplification reactions were carried out in a PTC-200 thermocycler (MJ Research, Waltham, Mass.) under the following conditions: initial denaturation at 94°C for 2 min followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 51°C for 30 s, and elongation at 72°C for 30 s. The final elongation step was extended to 3 min. A computer analysis with DNASYS software (version 2.5; Hitachi Software Engineering Co., London, United Kingdom) was used to identify restriction endonucleases capable of distinguishing the subtypes of blaCTX-M genes. These restriction endonucleases and their predicted patterns are shown in Table 1. Ten-microliter aliquots of PCR products were directly subjected to digestion with 9 U of PstI and 4 U of PvuII enzymes (Amersham Pharmacia Biotech, Piscataway, N.J.) in One-Phor-All Plus Buffer (10 mM Tris acetate [pH 7.5], 10 mM magnesium acetate, 50 mM potassium acetate) for 3 h at 37°C. The PCR products and restriction fragments were analyzed by electrophoresis in 3.5% agarose (AmpliSize; Bio-Rad, Philadelphia, Pa.) gel and ethidium bromide staining. Bacterial strains producing the known β-lactamases—Salmonella enterica serovar Typhimurium CAS5 (CTX-M-2), Citrobacter freundii 2525 (CTX-M-3), E. coli (CTX-M-9), E. coli C600 (TEM-1), E. coli J53 (SHV-1), and K. ascorbata T861 (KluA)—were used for quality control and assessment of specificity of PCR-restriction fragment length polymorphism (RFLP) analysis.
TABLE 1.
Restriction patterns of PCR products showing differentiation between the CTX-M clusters
| CTX-M clustera | No. of restriction sites
|
Expected length(s) (bp) of PstI-PvuII digestion product(s) | |
|---|---|---|---|
| PstI | PvuII | ||
| CTX-M-1 (CTX-M-1, -3, -10, -11, -12, -15 [UOE-1], -22) | 0 | 2 | 267, 156, 121 |
| CTX-M-2 (CTX-M-2, -4, -5, -6, -7, and -20; Toho-1; KluA) | 1 | 0 | 355, 188 |
| CTX-M-8 | 0 | 0 | 544 |
| CTX-M-9 | |||
| CTX-M-9, -13, and -16 | 1 | 1 | 426, 72, 46 |
| CTX-M-14 (-18, UOE-2, Toho-3), -15, -17, -19, and -21 | 0 | 1 | 472, 72 |
| Toho-2 | 0 | 1 | 466, 72 |
Grouping is based on >95% sequence identity of the coding genes.
ERIC-PCR fingerprinting.
Molecular typing of all the CTX-M-positive strains was performed with primer ERIC1 (38). PCR mixtures were set up in Ready-To-Go PCR Bead (Amersham Pharmacia Biotech) format. PCR mixtures contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 1.5 U of Taq polymerase, a 2 μM concentration of ERIC1 primer, and 2 μl of template DNA in a final volume of 25 μl. The amplification was carried out in a PTC-200 thermocycler (MJ Research) under the following conditions: initial denaturation at 94°C for 2 min 30 s followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 47°C for 1 min, and elongation at 72°C for 1 min. The final elongation step was extended to 4 min. PCR products were separated by electrophoresis in 1.3% agarose gel and visualized by ethidium bromide staining. The gels were documented using a PhotoDoc-IT Link gel documentation system (UVP, Upland, Calif.). Cluster analysis of genomic fingerprints was done with the GelCompar software (version 4.1; Applied Maths BVBA, Sint-Martens-Latem, Belgium) using the Pearson correlation coefficient and unweighted pair-group method using an arithmetic averages algorithm. Isolates that produced similar fingerprints (correlation coefficient ≥ 90%) were considered to be related. The interday reproducibility of enterobacterial repetitive consensus PCR (ERIC-PCR) was ensured by repetitive testing of selected bacterial isolates (E. coli KZ-Ma 63, K. pneumoniae EK-Ro 9, and K. pneumoniae EK-Bo 246), generating ERIC-PCR profiles of relatively high complexity. Each of these isolates was tested with every new batch of analyzed strains and verified to produce identical fingerprints.
Transfer of resistance.
Nine CTX-M β-lactamase-producing clinical isolates were mated in broth with E. coli AB1456 (F− Rifr). The transconjugants were selected on agar containing rifampin (100 μg/ml) and cefotaxime (2 μg/ml).
Sequencing of PCR products.
Amplified 544-bp internal fragments of blaCTX-M genes were purified using QIAquick Spin Columns (QIAGEN Inc., Valencia, Calif.) and directly sequenced using the CTX-M/F' and CTX-M/R' primers on a CEQ-2000 automated sequencer (Beckman Coulter Inc., Fullerton, Calif.). Sequencing was done by the Eurogene company (Moscow, Russia).
RESULTS AND DISCUSSION
Assessment of specificity and efficiency of the PCR-RFLP method for detection and discrimination of blaCTX-M genes.
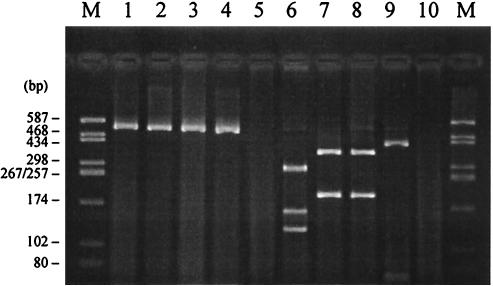
Using the primers and PCR conditions described here, a single DNA fragment of the expected size (544 bp) was amplified from the control E. coli strains producing β-lactamases CTX-M-2, -3, -4, and -9 and from the K. ascorbata type strain T861 carrying the species-specific β-lactamase KluA. These enzymes represent three major genetic clusters of the CTX-M family. Therefore, the proposed method was able to detect the blaCTX-M genes belonging to different CTX-M clusters. Further digestion of PCR products with selected restriction endonucleases allowed us to distinguish the subtypes of blaCTX-M genes as shown in Fig. 2.
FIG. 2.
Detection and differentiation of blaCTX-M genes by PCR-RFLP. Lanes 1 to 5, undigested PCR products; lanes 6 to 10, PstI-PvuII-digested PCR products; lanes 1 and 6, C. freundii 2525 (CTX-M-3); lanes 2 and 7, S. enterica serovar Typhimurium CAS5 (CTX-M-2); lanes 3 and 8, K. ascorbata T861 (KluA); lanes 4 and 9, E. coli (CTX-M-9); lanes 5 and 10, E. coli C600 (TEM-1); lanes M, molecular size marker (pUC18-HaeIII).
The specificity of PCR was confirmed by negative amplification results with bacteria possessing TEM- and SHV-type plasmid-mediated β-lactamases (only the data for the TEM-1 producer are shown in Fig. 2). However, the method described here is not applicable for direct detection of plasmid-coded CTX-M enzymes in K. oxytoca, Kluyvera spp., R. aquatilis, and C. sedlakii, which may produce false-positive results due to the high homology between primer binding sites in blaCTX-M and the chromosomal β-lactamase genes of these species (blaOXY-2, kluA, kluC, blaRAHN-1, and sed1).
Prevalence of ESBL production and proportion of isolates possessing CTX-M-type ESBLs.
The overall frequency of ESBL producers observed in this study was 15.8% in E. coli and 60.8% in K. pneumoniae, although the percentage of ESBL-producing E. coli and K. pneumoniae isolates ranged from 8.1 to 90% in different hospitals (Table 2). The rate of ESBL production did not depend on the geographic location of the hospital. For example, the frequency of ESBL-positive strains ranged between 10 and 90% in eight Moscow hospitals and between 26.1 and 56.3% in two Krasnoyarsk hospitals, thus reflecting a specific situation in each medical center.
TABLE 2.
Prevalence of ESBL-positive E. coli and K. pneumoniae isolates in different hospitals and multiplicity of strains producing different types of CTX-M β-lactamases
| Center |
E. coli
|
K. pneumoniae
|
Both species
|
CTX-M cluster (type)c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of isolates
|
No. of ERIC-PCR profilesa | No. of isolates
|
No. of ERIC-PCR profiles | % ESBL positive | % CTX-M positiveb | ||||||
| Total | ESBL positive | CTX-M positive | Total | ESBL positive | CTX-M positive | ||||||
| Ekaterinburg-1 | 1 | 1 | 0 | 14 | 12 | 6 | 5 | 86.7 | 46.2 | 1 | |
| Ekaterinburg-2 | 6 | 0 | 0 | 9 | 6 | 6 | 3 | 40.0 | 100.0 | 1 (CTX-M-3) | |
| Kazan | 19 | 8 | 8 | 1 | 21 | 10 | 0 | 45.0 | 44.4 | 2 (CTX-M-5) | |
| Krasnodar | 10 | 2 | 1 | 1 | 4 | 2 | 1 | 1 | 28.6 | 50.0 | 1 |
| Krasnoyarsk-1 | 13 | 3 | 2 | 2 | 19 | 15 | 2 | 2 | 56.3 | 22.2 | 1 |
| Krasnoyarsk-2 | 13 | 1 | 1 | 1 | 10 | 5 | 1 | 1 | 26.1 | 33.3 | 1 |
| Moscow-1 | 14 | 5 | 3 | 3 | 10 | 9 | 2 | 2 | 58.3 | 35.7 | 1 |
| Moscow-2 | 13 | 11 | 1 | 1 | 18 | 16 | 0 | 87.1 | 3.7 | 1 | |
| Moscow-3 | 14 | 12 | 5 | 2 | 26 | 24 | 7 | 4 | 90.0 | 33.3 | 1 (CTX-M-15) |
| Moscow-4 | 46 | 5 | 0 | 7 | 4 | 1 | 1 | 17.0 | 11.1 | 1 | |
| Moscow-5 | 40 | 1 | 0 | 30 | 6 | 1 | 1 | 10.0 | 14.3 | 1 | |
| Moscow-6 | 21 | 2 | 1 | 1 | 3 | 2 | 0 | 16.7 | 25.0 | 1 | |
| Moscow-7 | 27 | 5 | 0 | 11 | 4 | 1 | 1 | 23.7 | 11.1 | 1 | |
| Moscow-8 | 19 | 1 | 0 | 15 | 8 | 1 | 1 | 26.5 | 11.1 | 1 | |
| Novosibirsk | 5 | 1 | 1 | 1 | 34 | 30 | 20 | 2 | 79.5 | 67.7 | 1 (CTX-M-3) |
| Omsk | 29 | 0 | 0 | 8 | 3 | 1 | 1 | 8.1 | 33.3 | 1 | |
| Smolensk | 10 | 1 | 0 | 31 | 24 | 5 | 4 | 61.0 | 20.0 | 1 | |
| St. Petersburg | 6 | 3 | 2 | 1 | 14 | 13 | 2 | 1 | 80.0 | 25.0 | 1 (CTX-M-3) |
| Stavropol | 13 | 1 | 1 | 1 | 34 | 23 | 17 | 14 | 51.1 | 75.0 | 1 |
| Tomsk | 19 | 2 | 2 | 2 | 20 | 13 | 12 | 6 | 38.5 | 93.3 | 1 |
| Vladivostok | 8 | 1 | 0 | 8 | 2 | 1 | 1 | 18.8 | 33.3 | 1 | |
| Other hospitals | 148 | 12 | 0 | 64 | 17 | 0 | 13.7 | 0.0 | |||
| All | 494 | 78 | 28 | 17 | 410 | 248 | 87 | 48 | 36.1 | 35.3 | |
Numbers of distinct ERIC-PCR profiles among CTX-M producers.
Percentage of isolates carrying blaCTX-M genes among ESBL producers.
CTX-M clusters and types of enzymes were determined by RFLP analysis and sequencing of amplification products, respectively.
All the ESBL-producing strains (n = 326) were examined by PCR for the presence of blaCTX-M genes. Positive amplification results were observed for 28 (35.9%) E. coli and 87 (34.9%) K. pneumoniae isolates expressing the ESBL phenotype. These results demonstrate for the first time the high proportion of CTX-M enzymes among various ESBL-producing nosocomial strains in Russian hospitals.
CTX-M β-lactamases were detected in 21 (75%) surveyed hospitals from all the cities representing geographically distant areas of Russia, and in five (17.9%) hospitals this was the predominant ESBL. The highest rates of CTX-M β-lactamase-producing E. coli and K. pneumoniae were observed in the hospitals of Ekaterinburg, Tomsk, Krasnodar, and Stavropol, which represent the areas of Ural, Siberia, and South Russia (Fig. 1).
Distribution of CTX-M subtypes.
As was demonstrated by PCR-RFLP analysis, all CTX-M β-lactamases found in K. pneumoniae isolates from this study belonged to the CTX-M-1 cluster. Sequencing of PCR products from strains representing major clonal outbreaks (see below) showed that the internal fragments of blaCTX-M-1-related genes in three K. pneumoniae isolates (NOV37, EK-Ro171, and SP477.1) from Novosibirsk, Ekaterinburg, and St. Petersburg were identical to blaCTX-M-3 (GenBank accession no. Y10278). The CTX-M-3 enzyme is one of the most common and broadly disseminated β-lactamases of the CTX-M-1 cluster. Initially identified in Poland (19), it was later found in France (16, 18), Greece (23), and Taiwan (42, 44). The countrywide spread of CTX-M-3 β-lactamase-producing organisms was recently reported in Poland, where this type of ESBL was found in six species of the family Enterobacteriaceae, including K. pneumoniae (1).
As shown by PCR-RFLP analysis, the majority (71.4%) of CTX-M β-lactamases produced by E. coli isolates were also distributed to the CTX-M-1 cluster. The sequences similar to blaCTX-M-3 were obtained from two E. coli strains (BH6884 and BH3223/2) isolated in a single Moscow hospital. Both sequences contained the same point mutation leading to the Asp240Gly substitution (according to Ambler's numbering scheme [R. P. Ambler, A. F. W. Coulson, J.-M. Frère, J.-M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley, Letter, Biochem. J. 276:269-270, 1991]). This mutation is known to increase hydrolytic activity against ceftazidime (21, 33) and is present in the CTX-M variant described by different authors as UOE-1 (GenBank accession no. AY013478) or CTX-M-15 (GenBank accession no. AY044436). Enterobacterial strains producing CTX-M-15 were initially isolated in Japan and India (21) and recently found in Canada (M. P. Muller, D. Boyd, A. Ashi-Sulaiman, C. Larocque, M. Mulvey, D. Reynolds, A. McGeer, and B. M. Willey, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-233, 2001), Bulgaria (I. Schneider, E. Keuleyan, R. Markovska, and A. Bauernfeind, Abstr. 12th Eur. Congr. Clin. Microbiol. and Infect. Dis., abstr. P430, 2002), Poland (2), and France (27). This study therefore reports further dissemination of the CTX-M variant which confers significant levels of resistance to both cefotaxime and ceftazidime in strains that produce it.
The CTX-M-2-related enzymes were detected by PCR-RFLP analysis in only eight E. coli isolates from a single hospital located in Kazan. The sequence of the internal blaCTX-M gene fragment was determined for one isolate (KZ-Ma12) from this group and was found to be identical to that of blaCTX-M-5. The CTX-M-5 enzyme was initially found in isolates of S. enterica serovar Typhimurium from Latvia (10), and it had not been identified previously in other species of the family Enterobacteriaceae. Consequently we describe here the first occurrence of a CTX-M-5-like enzyme in E. coli. As shown below, all E. coli isolates from Kazan were clonally related according to the results of ERIC-PCR typing and did not transfer resistance to cefotaxime in conjugation experiments. This observation is in good agreement with the previous publication of Bradford et al. (10) reporting the location of genes encoding CTX-M-5 on small non-self-transferable plasmids. It may also explain the relatively low proportion of CTX-M-5-producing isolates compared to those bearing CTX-M-3 and other related enzymes which are usually encoded by conjugative plasmids.
Neither CTX-M-8- nor CTX-M-9-related enzymes were found in this study, although in recent years ESBLs of the latter genetic group were found in other European countries (Spain [9, 11, 35], France [18, 36], and the United Kingdom [Alobwede et al., letter]) as well as in Asian countries (Japan [41], China [14], Taiwan [22, 44], Korea [28], and Vietnam [13]) and Brazil (8).
Molecular epidemiology of CTX-M-producing clinical isolates.
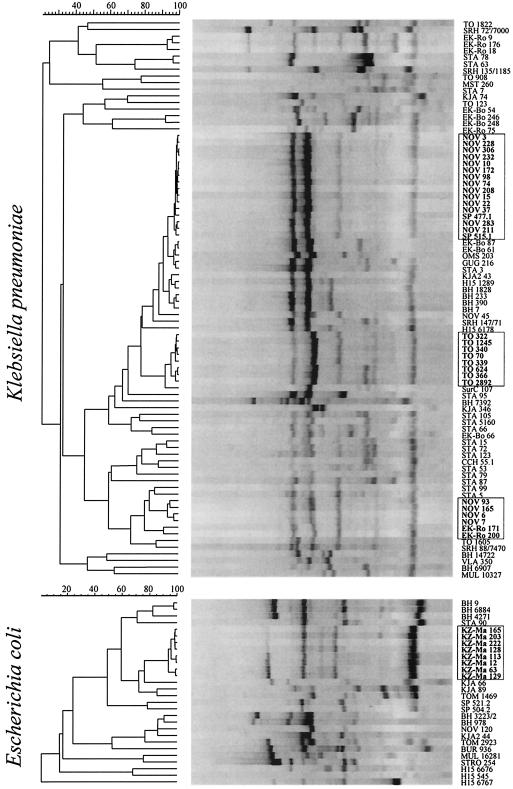
ERIC-PCR typing revealed considerable diversity of genetic types of CTX-M producers in Russia (Fig. 3). Distinct ERIC-PCR patterns were obtained for 17 and 48 E. coli and K. pneumoniae isolates, respectively. Nevertheless, several major clonal outbreaks caused by the strains of both species were recognized.
FIG. 3.
Unweighted pair-group method using arithmetic averages clustering of ERIC-PCR profiles of CTX-M-producing strains. The isolates representing major clonal outbreaks are outlined.
The largest outbreak occurred at Novosibirsk Regional Hospital, where 20 K. pneumoniae isolates producing a CTX-M-3-like β-lactamase were obtained from patients in surgical and intensive care units during a 2-year period. Out of these isolates 15 had identical ERIC-PCR patterns and 4 others were grouped into a distinct cluster, suggesting the dissemination of two major clones in a single hospital. It is noteworthy that isolates which apparently belonged to the Novosibirsk epidemic clones were also found in the hospitals of geographically distant cities; two strains were isolated in St. Petersburg, and the other two were isolated in Ekaterinburg. Random amplified polymorphic DNA typing with primer M13 (39) confirmed the relatedness of these isolates (data not shown); however, the means of their transmission remain unknown.
Another outbreak of CTX-M-producing K. pneumoniae occurred at Tomsk Regional Clinical Hospital. Eight of the twelve K. pneumoniae strains isolated in this hospital displayed very similar profiles (differing by not more than two bands).
As mentioned above, all the E. coli isolates from Kazan producing the CTX-M-5-like enzyme were also found to be clonally related. Smaller groups of two to four isolates with identical or similar ERIC-PCR profiles were found in many centers, but in almost all the surveyed hospitals multiple clones of CTX-M-producing organisms were detected.
Therefore, it is possible that in addition to the clonal spread of the strains, plasmid transfer played an important role in the global dissemination of CTX-M-encoding genes among nosocomial E. coli and K. pneumoniae strains in Russian hospitals. In support of this assumption, mating experiments with selected clinical isolates demonstrated that the genes for the CTX-M-1-cluster enzymes were readily transferred from all the isolates to the recipient strain.
Resistance phenotypes of ESBL-producing isolates.
The data on the activities of various antimicrobial agents against ESBL-producing E. coli and K. pneumoniae isolates are summarized in Table 3. β-Lactam-β-lactamase inhibitor combinations, carbapenems, aminoglycosides, and fluoroquinolones are considered to be potentially active drugs against ESBL-producing organisms. However, of the antimicrobial agents tested, only imipenem demonstrated universal activity against E. coli and K. pneumoniae isolates. The MICs of imipenem were not dependent on the type of ESBL produced. Among penicillin-β-lactamase inhibitor combinations, piperacillin-tazobactam was active against 70.5% of E. coli isolates and 55.2% of K. pneumoniae isolates, whereas only 14.1% of E. coli isolates and 19% of K. pneumoniae isolates were susceptible to amoxicillin-clavulanate. Interestingly, intermediate or high-level resistance to amoxicillin-clavulanate was more common among CTX-M producers (96.4% of E. coli isolates and 90.8% of K. pneumoniae isolates) than among strains producing non-CTX-M-type ESBLs (80.0% of E. coli isolates and 75.8% of K. pneumoniae isolates), whereas resistance to piperacillin-tazobactam was conversely less frequent among CTX-M producers (17.9% of E. coli isolates and 37.9% of K. pneumoniae isolates) than among strains producing non-CTX-M-type ESBLs (36.0% of E. coli isolates and 48.4% of K. pneumoniae isolates). The greater activity of piperacillin-tazobactam against CTX-M-positive strains is probably explained by the fact that the CTX-M enzymes are better inhibited by tazobactam than by clavulanate (10, 35, 37).
TABLE 3.
In vitro activities of antimicrobial agents against E. coli and K. pneumoniae isolates that produce CTX-M and other types of ESBLs
| Antimicrobialc | No. (%) of resistant isolatesa | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| E. coli, CTX-M (n = 28) | ||||
| Ceftazidime | 28 (100) | 1-≥256 | 8 | ≥256 |
| Cefotaxime | 28 (100) | 8-≥256 | ≥256 | ≥256 |
| Cefepime | 28 (100) | 2-≥256 | 16 | 128 |
| AMC | 27 (96.4) | 4-≥256 | 32 | 64 |
| TZP | 5 (17.9) | 0.5-≥256 | 4 | ≥256 |
| Imipenem | 0 | 0.125-2 | 0.25 | 0.5 |
| Gentamicin | 25 (89.3) | 1-≥256 | 64 | ≥256 |
| Amikacin | 0 | 1-8 | 2 | 4 |
| Ciprofloxacin | 6 (21.4) | 0.004-≥32 | 0.03 | ≥32 |
| E. coli, non-CTX-M (n = 50) | ||||
| Ceftazidime | 50 (100) | 0.25-256 | 32 | ≥256 |
| Cefotaxime | 50 (100) | 0.06-≥256 | 16 | 128 |
| Cefepime | 50 (100) | 0.06-128 | 1 | 4 |
| AMC | 40 (80.0) | 4-≥256 | 32 | 128 |
| TZP | 18 (36.0) | 1-≥256 | 8 | ≥256 |
| Imipenem | 0 | 0.125-1 | 0.25 | 0.5 |
| Gentamicin | 38 (76.0) | 0.5-≥256 | 32 | ≥256 |
| Amikacin | 8 (16.0) | 0.5-≥256 | 2 | ≥256 |
| Ciprofloxacin | 25 (50.0) | 0.008-≥256 | 1 | ≥32 |
| K. pneumoniae, CTX-M (n = 87) | ||||
| Ceftazidime | 87 (100) | 0.5-≥256 | 8 | ≥256 |
| Cefotaxime | 87 (100) | 8-≥256 | 128 | ≥256 |
| Cefepime | 87 (100) | 1-≥256 | 16 | ≥256 |
| AMC | 79 (90.8) | 4-≥256 | 32 | 64 |
| TZP | 33 (37.9) | 0.5-≥256 | 8 | ≥256 |
| Imipenem | 0 | 0.125-2 | 0.5 | 0.5 |
| Gentamicin | 83 (95.4) | 1-≥256 | 128 | ≥256 |
| Amikacin | 17 (19.5) | 1-64 | 2 | 32 |
| Ciprofloxacin | 16 (18.4) | 0.016-≥32 | 0.06 | 8 |
| K. pneumoniae, non-CTX-M (n = 161) | ||||
| Ceftazidime | 161 (100) | 0.5-≥256 | 16 | ≥256 |
| Cefotaxime | 161 (100) | 0.06-≥256 | 4 | 64 |
| Cefepime | 161 (100) | 0.06-≥256 | 2 | 8 |
| AMC | 122 (75.8) | 1-≥256 | 32 | 64 |
| TZP | 78 (48.4) | 1-≥256 | 16 | ≥256 |
| Imipenem | 0 | 0.125-1 | 0.25 | 0.5 |
| Gentamicin | 128 (79.5) | 0.5-≥256 | 32 | ≥256 |
| Amikacin | 20 (12.4) | 1-≥256 | 2 | 32 |
| Ciprofloxacin | 38 (23.6) | 0.032-≥32 | 0.125 | 4 |
Refers to cumulative number (percent) of isolates categorized as intermediate or resistant according to the NCCLS breakpoint criteria. All ESBL producers were considered resistant to ceftazidime, cefotaxime, and cefepime independent of MICs.
50% and 90%, MIC50 and MIC90, respectively.
AMC, amoxicillin-clavulanic acid (2:1). The concentrations listed refer to amoxicillin. TZP, piperacillin-tazobactam (tazobactam fixed at 4 μg/ml). The concentrations listed refer to piperacillin.
Associated resistance to aminoglycosides or ciprofloxacin was frequently observed among ESBL-positive isolates of both species. In total, 80.8% of E. coli isolates and 85.1% of K. pneumoniae isolates were nonsusceptible to gentamicin. The resistance rates were about 15% higher among CTX-M producers than among isolates expressing other types of ESBLs. Amikacin, however, remained active against 89.7% of E. coli isolates, including all CTX-M-positive strains, and against 85.1% of K. pneumoniae isolates producing various types of ESBLs. Overall, as many as 39.7% of E. coli isolates and 21.8% of K. pneumoniae isolates producing ESBLs were nonsusceptible to ciprofloxacin. The percentages of CTX-M-positive E. coli and K. pneumoniae strains that appeared intermediate or resistant to ciprofloxacin were lower (21.4 and 18.4%, respectively).
Similar to our study, a very high rate of resistance to gentamicin (97%) has been reported by Winokur et al., who analyzed a relatively smaller number of ESBL-producing K. pneumoniae strains isolated in six Russian medical centers during the period from 1994 to 1996 (40). However, in our study, the incidence of fluoroquinolone resistance among ESBL-producing K. pneumoniae isolates was approximately seven times higher than that reported by Winokur et al.
Although, according to the NCCLS (24), all ESBL producers should be considered resistant to all cephalosporins, independent of MICs, it was interesting to compare the levels of resistance to some oxyimino-cephalosporins—especially ceftazidime, cefotaxime, and cefepime—among isolates producing CTX-M- and non-CTX-M-type ESBLs.
Cefotaxime MICs were expectedly higher for CTX-M-producing E. coli (MIC at which 50% of strains tested were resistant [MIC50], ≥256 μg/ml) and K. pneumoniae strains (MIC50, 128 μg/ml) than for those expressing other types of ESBLs (MIC50, 16 and 4 μg/ml, respectively). Nevertheless, a high degree of diversity of the levels of resistance to cefotaxime was observed for CTX-M-positive strains of both species, as illustrated by the broad range of MICs (8 to ≥256 μg/ml).
Cefepime also demonstrated significantly lower activity against CTX-M producers in terms of elevated MIC50 and MIC90 (Table 3). Yet, the levels of resistance to cefepime varied widely within the group of CTX-M producers (MIC range, 1 to 256 μg/ml) as well as within the group of strains producing other types of ESBLs (MIC range, 0.06 to 256 μg/ml). It is interesting that the three isolates of K. pneumoniae (EK-Ro171, NOV37, and SP477.1) which were found to produce the same type of CTX-M β-lactamase differed considerably in the levels of resistance to cefotaxime (MICs, 8, 64, and ≥256 μg/ml, respectively) and cefepime (MICs, 4, 16, and ≥256 μg/ml, respectively). At the same time they were equally susceptible to cefoxitin (MICs, 2 to 4 μg/ml), suggesting that altered permeability was not likely contributing to the increased resistance. This observation supports the findings of Baraniak et al., who found that resistance phenotype conferred by the CTX-M-3 enzyme may be variable, possibly reflecting the fluctuations in the level of its expression (1). Although in the study of Yu et al. high-level resistance to cefepime was strongly associated with the CTX-M-type ESBLs (43), our data indicate that the MICs of cefepime alone are not always predictive of a particular ESBL type.
More surprisingly, no statistically significant difference was observed in ceftazidime MICs between the groups of E. coli and K. pneumoniae isolates producing CTX-M- and non-CTX-M-type ESBLs (P = 0.1619 [Wilcoxon test]). The resistance phenotype of the majority of CTX-M-positive isolates was consistent with production of CTX-M ESBL. The cefotaxime MICs for such strains were two to nine twofold dilutions higher than those of ceftazidime (Table 4). Nevertheless, MICs of cefotaxime and ceftazidime were equal or differed by only one twofold dilution in 6 (21.4%) E. coli and 26 (29.9%) K. pneumoniae isolates producing CTX-M enzymes. All isolates equally resistant to cefotaxime and ceftazidime had the highest detectable MICs of these drugs (≥256 μg/ml). In some cases, high-level resistance to ceftazidime was possibly associated with production of mutant CTX-M enzymes. In fact, the Asp240Gly substitution increasing the ceftazidime-hydrolyzing activity of CTX-M-15 was found in two ceftazidime-resistant (MICs, ≥256 μg/ml) E. coli clones isolated in the same hospital in Moscow. It is also possible that several CTX-M-positive isolates which appeared highly resistant to ceftazidime (MICs, 128 to ≥256 μg/ml) but susceptible to cefoxitin (MICs, 2 to 8 μg/ml) coproduced additional ceftazidime-hydrolyzing β-lactamases. This was proved by conjugation experiments with two E. coli and three K. pneumoniae isolates, which transferred cefotaxime resistance to the recipient strain independently of ceftazidime resistance. Finally, it is likely that the permeability-based mechanisms contributed to the ceftazidime resistance in at least four CTX-M-producing E. coli and 12 CTX-M-producing K. pneumoniae isolates which had decreased susceptibility to cefoxitin (MICs, 16 to ≥256 μg/ml).
TABLE 4.
Frequency distribution of cefotaxime and ceftazidime MIC ratios among E. coli and K. pneumoniae isolates producing CTX-M and other types of ESBLs
| MIC ratio (cefotaxime/ceftazidime) | No. (%) of E. coli isolates producing:
|
No. (%) of K. pneumoniae isolates producing:
|
||
|---|---|---|---|---|
| CTX-M ESBLs (n = 28) | Non-CTX-M ESBLs (n = 50) | CTX-M ESBLs (n = 87) | Non-CTX-M ESBLs (n = 161) | |
| 1/64 | 0 | 0 | 0 | 3 (1.9) |
| 1/32 | 0 | 1 (2.0) | 0 | 13 (8.1) |
| 1/16 | 0 | 7 (14.0) | 0 | 20 (12.4) |
| 1/8 | 0 | 8 (16.0) | 0 | 25 (15.5) |
| 1/4 | 0 | 9 (18.0) | 0 | 15 (9.3) |
| 1/2 | 0 | 7 (14.0) | 0 | 25 (15.5) |
| 1 | 4 (14.3) | 8 (16.0) | 14 (16.1) | 37 (23.0) |
| 2 | 2 (7.1) | 4 (8.0) | 12 (13.8) | 14 (8.7) |
| 4 | 4 (14.3) | 5 (10.0) | 16 (18.4) | 7 (4.3) |
| 8 | 4 (14.3) | 0 | 17 (19.5) | 1 (0.6) |
| 16 | 5 (17.9) | 1 (2.0) | 18 (20.7) | 1 (0.6) |
| 32 | 6 (21.4) | 0 | 2 (2.3) | 0 |
| 64 | 2 (7.1) | 0 | 7 (8) | 0 |
| 128 | 1 (3.6) | 0 | 1 (1.1) | 0 |
On the other hand, the MICs of cefotaxime for 10 E. coli and 23 K. pneumoniae isolates producing non-CTX-M-type ESBLs were one to three twofold dilutions higher than those of ceftazidime. All the above data suggest that a phenotypic approach based on the comparison of cefotaxime and ceftazidime MICs has limited value in predicting the presence of the CTX-M-type ESBLs in clinical isolates.
Conclusions.
The present study demonstrated the usefulness of our PCR-RFLP method for detection and characterization of the CTX-M-type ESBLs. Using this method we have shown the high prevalence and broad geographic distribution of CTX-M-producing E. coli and K. pneumoniae nosocomial strains in Russia. β-Lactamases of the CTX-M-1 cluster were the predominant CTX-M enzymes in both species. Out of this group only CTX-M-3- and CTX-M-15-like enzymes were identified among selected isolates. In addition to the clonal spread of the strains, plasmid transfer played an important role in the dissemination of the CTX-M-1-cluster enzymes among nosocomial E. coli and K. pneumoniae strains in Russian hospitals. By contrast, the outbreak of CTX-M-5-producing E. coli that occurred in a single hospital was attributed only to clonal spread.
The alarming situation with global dissemination of CTX-M-producing strains highlights the need for their epidemiological monitoring and prudent use of antimicrobial agents.
Acknowledgments
We are grateful to the following people throughout Russia who supplied the clinical isolates used in this study: Yury Tikhonov, Svetlana Polikarpova, Valery Stroganov, Vladimir Kurchavov, Tatiana Vostrikova, Elena Gugucidze, Natalya Furletova, Natalya Bogomolova, Leonid Bolshakov, Irina Aleksandrova, and Larisa Ritchik (Moscow); Viktor Tec, Gennady Afinogenov, and Tatiana Suborova (St. Petersburg); Olga Krechikova and Marina Sukhorukova (Smolensk); Vladimir Biryukov (Riazan); Lyudmila Akhmetova and Lyubov Boronina (Ekaterinburg); Valentina Taraban and Irina Multih (Krasnodar); Evgeny Schetinin (Stavropol); Natalya Marusina and Olga Galeeva (Kazan); Svetlana Ivanova (Omsk); Sofya Khasanova (Ufa); Vera Ilyina (Novosibirsk); Lyubov Gudkova (Tomsk); Dmitry Zdzitovecky; Olga Peryanova (Krasnoyarsk); and Lyubov Karpukhina (Vladivostok). We thank Marek Gniadkowski (Warsaw, Poland), Rafael Canton (Madrid, Spain), Adolf Bauernfeind (Munich, Germany), and Federico Uruburu (Valencia, Spain) for the reference strains producing the known CTX-M-type β-lactamases.
REFERENCES
- 1.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., H. Grimm, and S. Schweighart. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294-298. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., J. M. Casellas, and M. Goldberg. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 6.Bellais, S., L. Poirel, N. Fortineau, J. W. Decousser, and P. Nordmann. 2001. Biochemical-genetic characterization of the chromosomally encoded extended-spectrum class A β-lactamase from Rahnella aquatilis. Antimicrob. Agents Chemother. 45:2965-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard, H., C. Tancrede, V. Livrelli, A. Morand, M. Barthélémy, and R. Labia. 1992. A novel plasmid-mediated extended-spectrum β-lactamase not derived from TEM- or SHV-type enzymes. J. Antimicrob. Chemother. 29:590-592. [DOI] [PubMed] [Google Scholar]
- 8.Bonnet, R., C. Dutour, J. L. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou, G., M. Cartelle, M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 β-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing beta-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinas, L., M. A. Moreno, M. Zarazaga, C. Porrero, Y. Saenz, M. Garcia, L. Dominguez, and C. Torres. 2003. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 47:2056-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantón, R., A. Oliver, T. M. Coque, M. del Carmen Varela, J. C. Pérez-Díaz, and F. Baquero. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanawong, A., F. H. M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coque, T. M., A. Oliver, J. C. Pérez-Díaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, et al. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, Probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 22.Ma, L., Y. Ishii, F. Y. Chang, K. Yamaguchi, M. Ho, and L. K. Siu. 2002. CTX-M-14, a plasmid-mediated CTX-M type extended-spectrum β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 46:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavroidi, A., E. Tzelepi, V. Miriagou, D. Gianneli, N. J. Legakis, and L. S. Tzouvelekis. 2002. CTX-M-3 β-lactamase-producing Escherichia coli from Greece. Microb. Drug Resist. 8:35-37. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, sixth edition. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing: 13th informational supplement. M100-S13 (M7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Navarro, F., and E. Miró. 2002. Update on CTX-M-type β-lactamases. Rev. Med. Microbiol. 13:63-73. [Google Scholar]
- 27.Neuwirth, C., E. Siebor, M. Pruneaux, M. Zarnayova, C. Simonin, J. P. Kisterman, and R. Labia. 2003. First isolation of CTX-M15-producing Escherichia coli from two French patients. J. Antimicrob. Chemother. 51:471-473. [DOI] [PubMed] [Google Scholar]
- 28.Pai, H., E. H. Choi, H. J. Lee, J. Y. Hong, and G. A. Jacoby. 2001. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39:3747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peduzzi, J., S. Farzaneh, A. Reynaud, M. Barthélémy, and R. Labia. 1997. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A β-lactamase from Serratia fonticola CUV. Biochim. Biophys. Acta 1341:58-70. [DOI] [PubMed] [Google Scholar]
- 30.Petrella, S., D. Clermont, I. Casin, V. Jarlier, and W. Sougakoff. 2001. Novel class A β-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of β-lactamases within the Citrobacter genus. Antimicrob. Agents Chemother. 45:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petroni, A., A. Corso, R. Melano, M. L. Cacace, A. M. Bru, A. Rossi, and M. Galas. 2002. Plasmidic extended-spectrum β-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob. Agents Chemother. 46:1462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirel, L., P. Kämpfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power, P., M. Radice, C. Barberis, C. de Mier, M. Mollerach, M. Maltagliatti, C. Vay, A. Famiglietti, and G. Gutkind. 1999. Cefotaxime-hydrolysing β lactamases in Morganella morganii. Eur. J. Clin. Microbiol. Infect. Dis. 18:743-747. [DOI] [PubMed] [Google Scholar]
- 35.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saladin, M., V. T. B. Cao, T. Lambert, J.-L. Donay, J.-L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 37.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 38.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel, L., E. van Oorschot, H. M. Maas, B. Minderhoud, and L. Dijkshoorn. 2000. Epidemiologic typing of Escherichia coli using RAPD analysis, ribotyping and serotyping. Clin. Microbiol. Infect. 6:82-87. [DOI] [PubMed] [Google Scholar]
- 40.Winokur, P. L., M. V. Eidelstain, O. Stetsiouk, L. Stratchounski, J. Blahova, G. K. Reshedko, M. A. Croco, R. J. Hollis, M. A. Pfaller, and R. N. Jones. 2000. Russian Klebsiella pneumoniae isolates that express extended-spectrum β-lactamases. Clin. Microbiol. Infect. 6:103-108. [DOI] [PubMed] [Google Scholar]
- 41.Yagi, T., H. Kurokawa, N. Shibata, K. Shibayama, and Y. Arakawa. 2000. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol. Lett. 184:53-56. [DOI] [PubMed] [Google Scholar]
- 42.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, W. L., M. A. Pfaller, P. L. Winokur, and R. N. Jones. 2002. Cefepime MIC as a predictor of the extended-spectrum β-lactamase type in Klebsiella pneumoniae, Taiwan. Emerg. Infect. Dis. 8:522-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, W. L., P. L. Winokur, D. L. Von Stein, M. A. Pfaller, J. H. Wang, and R. N. Jones. 2002. First description of Klebsiella pneumoniae harboring CTX-M β-lactamases (CTX-M-14 and CTX-M-3) in Taiwan. Antimicrob. Agents Chemother. 46:1098-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]