Ribosomal protection represents an important tactic for promoting tetracycline resistance in both gram-positive and -negative species. Tet(O) and Tet(M) are the best studied of these determinants and were originally isolated from Campylobacter jejuni and Streptococcus spp., respectively, although both are widely distributed (10). These are the only two ribosomal protection proteins (RPPs) that have been studied in detail, and therefore, they have been dealt with extensively in this review. It is assumed, however, that the other members of this class of RPPs [Tet(S), Tet(T), Tet(Q), TetB(P), Tet(W), and OtrA] function through similar mechanisms. The distribution of these determinants in the eubacteria has been extensively reviewed by Chopra and Roberts (10) and more recent information can also be found at http://faculty.washington.edu/marilynr/.
Although this review focuses primarily on RPPs, it should be noted that a great variety of tetracycline resistance mechanisms exist (for a review, see reference 10). These determinants include (i) the efflux-based mechanisms found in gram-positive and gram-negative bacteria (10), (ii) the enzymatic degradation of tetracyclines found in Bacteroides (46), (iii) the rRNA mutations found in Propionibacterium acnes and Helicobacter pylori (19, 40, 55), and (iv) a host of undetermined mechanisms which bear little resemblance to the well-documented determinants mentioned above (10).
In this review, we will survey recent advances in the study of the ribosome, tetracycline, and the RPPs that further the understanding of RPP activity. Earlier work dealing with Tet(M) and Tet(O) as well as the other RPPs has been reviewed previously (51, 52).
INHIBITORY ACTIONS OF TETRACYCLINES
Tetracycline antibiotics.
Upon their introduction into medicine in 1948, tetracyclines were quickly accepted because they offered a broad spectrum of activity, being active against gram-positive and -negative bacteria, and more recently, they have been shown to be active against chlamydia, mycoplasmas, rickettsia, and some protozoan parasites (10). The tetracyclines can be separated into two groups, the atypical tetracyclines (e.g., anhydrotetracycline and 6-thiatetracycline) and typical tetracyclines (e.g., tetracycline, chlortetracycline, and minocycline) (9, 10, 35, 36, 38). The atypical tetracyclines function by disrupting bacterial membranes (36, 38). Alternatively, the typical tetracyclines, which are the subject of RPP-mediated resistance, bind to the ribosome and inhibit the elongation phase of protein synthesis (8, 13). More precisely, they inhibit accommodation of aminoacyl-tRNA (aa-tRNA) into the ribosomal A site (Fig. 1, reactions a and b) and, therefore, prevent the addition of new amino acids to the growing polypeptide (22, 25, 29, 48).
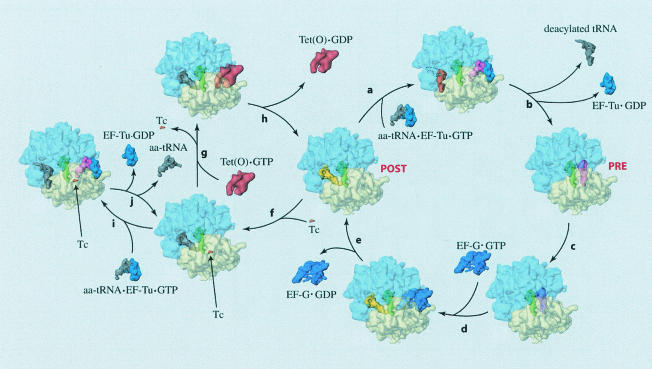
FIG. 1.
The pathway of Tet(O)-mediated tetracycline release is illustrated by cryo-EM reconstructions of ribosomes in various functional states (2, 45). The natural elongation cycle is represented by reactions a to e, such that if the ribosome is in the posttranslocational state (POST), a ternary complex of EF-Tu-aa-tRNA-GTP can decode the codon presented on the mRNA in the A site (reaction a). After correct codon-anticodon interaction, the GTPase activity of EF-Tu is triggered and the aa-tRNA is accommodated into the A site (reaction b), yielding a pretranslocational ribosome (PRE). After accommodation, the amino group of the A site-bound aa-tRNA attacks the ester bond of the P site-bound peptidyl-tRNA, thereby forming a peptide bond in a reaction called peptidyl transfer (reaction c). Following peptide bond formation, EF-G binds to the ribosome and promotes translocation of the tRNAs from the A and P sites to the P and E sites (reactions d and e), thus completing a single cycle and returning the ribosome to a POST state. Upon tetracycline binding (reaction f), the ribosome allegedly enters a nonproductive cycle illustrated by reactions i and j (4). In this cycle, the ternary complex repeatedly tries to bind aa-tRNA to the A site but fails. Tet(O) is able to rescue the ribosome from this nonproductive cycle by releasing tetracycline from its binding site on the 30S subunit (reaction g). After promoting the release of tetracycline, Tet(O) hydrolyzes its bound GTP and disassociates from the ribosome (reaction h), thereby returning the ribosome to the elongation cycle (reactions a to e). This figure has been reproduced from references 2 and 45 with permission of the publishers.
Location of tetracycline binding sites on the ribosome.
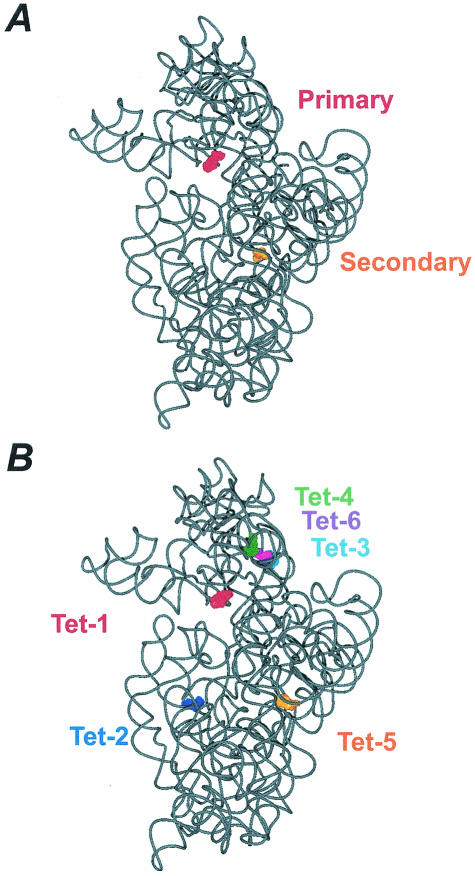
The inhibitory effect of tetracycline on A site occupation is presumed to be mediated by binding to a single high-affinity site (Kd = 1 to 20 μM) on the 30S ribosomal subunit (20). This is significant, as two independent 30S subunit-tetracycline crystal structures show tetracycline bound to either two (4) or six sites (37) on the 30S ribosomal subunit (Fig. 2). This, therefore, presents a problem in assigning one of the crystallographically determined sites to the biologically relevant inhibitory site. In the simplest case, one would expect that this single inhibitory site is in close proximity to the ribosomal A site, since it is aa-tRNA binding to the A site that is inhibited by the drug. In this respect, the Tet-1 and primary site in the Pioletti and Brodersen structures, respectively (Fig. 2), are located in the ribosomal A site, where tetracycline is bound by the irregular minor groove of helix 34 (h34) and the loop of helix 31 (h31) in the 16S rRNA (4, 37). In this position, it is believed that tetracycline would sterically interfere with aa-tRNA binding (4, 37). It is also interesting that, in the primary site, tetracycline makes interactions almost exclusively with the sugar phosphate backbone of the RNA (4, 37). In this sense, the lack of base-specific interactions may explain the broad specificity of the tetracyclines.
FIG. 2.
(A) The locations of the tetracycline binding sites determined by Brodersen et al. (4) are shown, where tetracycline bound in the primary site is red (surface representation) and tetracycline bound in the secondary site is orange. The structure shown is derived from the 3.4-Å model (PBD accession no. 1HNW). (B) The locations of the tetracycline binding sites determined by Pioletti et al. (37) are shown, where tetracycline bound to the Tet-1 site is red, tetracycline bound to the Tet-2 site is dark blue, tetracycline bound to the Tet-3 site is cyan, tetracycline bound to the Tet-4 site is green, tetracycline bound to the Tet-5 site is orange, and tetracycline bound to the Tet-6 site is purple. Note, the numbering of the tetracycline binding sites reflects their relative occupancy in the electron density map. The structure shown is derived from the 4.5-Å model (PBD accession no. 1I97). The tetracycline-ribosome interactions in the Tet-1 site are nearly identical to that in the primary site, whereas the Tet-5 and secondary site display distinct interactions. The figures were prepared with VMD (26) and PovRay (www.povray.org).
The secondary and Tet-5 tetracycline-binding sites (Fig. 2) are also likely candidates for the inhibitory binding site (4, 37). These binding sites are both associated with the so-called switch helix (h27) of the 16S rRNA (28), although the nature of their interactions with this helix are not exactly identical. In this position, tetracycline cannot directly interfere with tRNA binding, but rather, it could exert its inhibitory effect by interfering with the transition between the open and closed states of the 30S ribosomal subunit, which is important for the decoding reaction (34). The other four tetracycline-binding sites (Tet-2, -3, -4, and -6) (Fig. 2B) observed by Pioletti et al. (37) are not so easily correlated with the inhibitory action of tetracycline but do explain earlier data associated with photolabeling experiments (20, 33).
The inhibitory binding site.
Determining which of the above-mentioned tetracycline binding sites represents the inhibitory site is facilitated by data describing the nature of the inhibitory site, the interaction of tetracycline with the ribosome, and the mechanisms conferring resistance to tetracycline. For example, (i) the primary (and Tet-1) site is the most highly occupied site in both structures showing tetracycline bound to the 30S subunit (4, 37) consistent with the idea that the high-affinity site is the inhibitory site (17, 47, 56). (ii) In the primary (and Tet-1) site, several tetracycline-RNA interactions are mediated through a magnesium ion which is known to be important for tetracycline binding (4, 57). (iii) The crystal structures show that the face of tetracycline that interacts with the rRNA in the primary (and Tet-1) site is also the face where modifications result in a loss of biological activity (4, 39). (iv) Chemical probing showed that all tetracycline derivatives that bind the ribosome and inhibit protein synthesis enhance the dimethyl sulfate (DMS) reactivity of C1054 and U1052 in the 16S rRNA (associated with the primary tetracycline binding site). In contrast, only a subset of these derivatives were found to protect A892 (associated with the secondary tetracycline binding site) from DMS modification (38). (v) 16S rRNA mutations seen in H. pylori (19, 55) and P. acnes (40) that confer resistance to tetracycline are in close proximity to the primary binding site. (vi) Tet(O), an RPP which confers resistance to tetracycline, chases tetracyclines from the primary binding site but not the secondary binding site (12).
Proposed mechanism of tetracycline action.
Brodersen et al. (4) postulated that with tetracycline bound to the primary site, the ternary complex would be able to initiate decoding, such that the interaction between the codon and the anticodon of the EF-Tu-bound aa-tRNA would be unaffected by the presence of the drug (Fig. 1, reaction i). The subsequent step involving the release of the aa-tRNA from EF-Tu and its accommodation into the A site would, however, be inhibited (Fig. 1, reaction j), such that as the aa-tRNA rotates into the A site, the anticodon loop of the tRNA would clash with tetracycline (4, 37). Although the accommodation reaction is inhibited, EF-Tu-dependent GTP hydrolysis is not (22), and therefore, Brodersen et al. (4) speculate that a nonproductive cycle of ternary complex binding and GTP hydrolysis without A site occupation will ensue.
RIBOSOMAL PROTECTION PROTEINS
RPPs, such as the well-studied Tet(O) and Tet(M) (75% sequence similarity), are soluble cytoplasmic proteins (∼72 kDa) which mediate tetracycline resistance (51). Tet(O) was first cloned from a transferable plasmid pUA466 found in the food-borne pathogen C. jejuni (50). However, Tet(O), like the other RPPs, seems to have originated in the natural producer of oxytetracycline, Streptomyces rimosus, which harbors otrA, an RPP determinant (16, 44, 49). Accordingly, many of the RPP determinants are located on mobile genetic elements which may have facilitated their spread throughout the eubacteria via lateral gene transfer events (reviewed in reference 10).
Similarity between RPPs and elongation factors.
The RPPs display sequence similarity to the ribosomal elongation factors, EF-G and EF-Tu (41), and are grouped into the translation factor superfamily of GTPases (27). Accordingly, the RPPs bind and hydrolyze GTP in a ribosome-dependent manner (5, 6, 53), and maintenance of this activity is important for in vivo activity (11, 23). Sanchez-Pescador et al. interpreted this sequence similarity to indicate that the RPPs are functioning as tetracycline-resistant elongation factors (41); however, Burdett (5, 6) showed that Tet(M) cannot substitute for the elongation factors in vivo or in vitro. Nevertheless, the RPPs may be evolutionarily derived from the elongation factors, such that they lost their original function and have been adapted to function in tetracycline resistance.
Protection of the ribosome from tetracycline.
Tet(O) and Tet(M) can dislodge tetracycline from the ribosome (6, 54) and, in so doing, increase the apparent dissociation constant (Kd) of tetracycline binding to the ribosome from 5 to 30 μM. The ability of Tet(O) and Tet(M) to dislodge tetracycline is strictly dependent on the presence of GTP (6, 54), although there is some discrepancy concerning the role of GTP hydrolysis as a nonhydrolyzable GTP analogue was active with Tet(O) (54) but only partially active with Tet(M) (6). This, however, probably does not reflect a functional difference, but instead, it may result from the different methods used in each laboratory. In accordance with the ability of Tet(O) and Tet(M) to remove tetracycline, Burdett demonstrated that tRNA binding to the A site, which is normally inhibited by tetracycline, is, in fact, protected in the presence of Tet(M) (6). Thus, it appears that Tet(O) and Tet(M) confer tetracycline resistance by releasing tetracycline from the ribosome and thereby freeing the ribosome from the inhibitory effects of the drug, such that aa-tRNA can bind to the A site and protein synthesis can continue.
The ribosome-binding site for RPPs.
Much work was done to define the binding site by using biochemical assays (15), cryoelectron microscopy (cryo-EM) (45), and chemical probing (11, 12). The binding site was first localized to the elongation factor binding site when Dantley et al. (15) showed that Tet(M) and EF-G compete for a similar site on the ribosome. Additionally, the experiments of Dantley et al. (15) suggested that a component of this common site is the L11 region on the 50S subunit. This derives from the fact that the antibiotic thiostrepton, which binds this region and apparently locks in it a conformation unfavorable for EF-G binding (7), also inhibits the binding of Tet(M) (15).
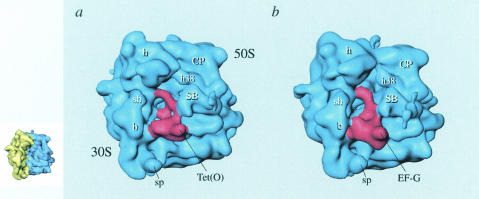
The interaction of Tet(O) with the ribosome was also studied by cryo-EM, a structural technique that is able to generate three-dimensional models of macromolecular complexes with a resolution between 10 and 30 Å (18). The final 16-Å three-dimensional reconstruction of Tet(O) bound to the ribosome can be seen in Fig. 3A, where a reconstruction of an EF-G-ribosome complex (Fig. 3B) is shown for comparison (1, 45). Noticeably, the density attributed to Tet(O) in the cryo-EM reconstruction has an overall shape similar to that of EF-G (Fig. 3), in agreement with the sequence similarity noted above. Also evident in the reconstruction is that Tet(O) and EF-G are binding to a common site located at the interface of the ribosomal subunit on the A site side, at the base of the L7/L12 stalk (Fig. 3), in agreement with Dantley et al. (15).
FIG. 3.
Cryo-EM reconstructions of Tet(O)-GTPγS (45) (a) and EF-G-GMPPCP (1) (b) ribosomal complexes. The ribosome (blue density) is shown in the same orientation as seen in the insert on the left, where the 30S subunit is colored yellow and the 50S subunit is colored blue. Tet(O) and EF-G are shown as red densities. Ribosomal landmarks are indicated. h, head; CP, central protuberance; h38, helix 38 of 23S rRNA; SB, stalk base; sp, spur; sh, shoulder; b, body. This figure has been reproduced from reference 45 with permission of the publisher.
In the cryo-EM study, Spahn et al. localize the sites of interaction between Tet(O) and the ribosome (Table 1), demonstrating that that the majority of the interactions are between Tet(O) and the rRNA (45). The only exception is a single interaction between domain III of Tet(O) and the ribosomal protein S12 (45). Furthermore, a comparison of the EF-G and Tet(O) ribosomal contacts indicates that they differ primarily in the vicinity of domain IV (Table 1), where EF-G contacts H69 of the 23S rRNA (21, 45) and Tet(O) interacts with h18/34 of the 16S rRNA (45). This is significant, as domain IV in EF-G has been implicated as an important determinant for promoting translocation of the tRNAs (30, 31, 42). In this case, these differences in domain IV may serve to distinguish Tet(O) and EF-G with respect to their activities; namely, domain IV of EF-G more intimately overlaps with the A site-bound tRNA, an idea that is consistent with the role of domain IV of EF-G in translocation. In contrast, the interaction of domain IV of Tet(O) and h34 of the 30S subunit is consistent with its role in tetracycline release (45) because h34 is a component of the primary tetracycline binding site (4, 37).
TABLE 1.
EF-G and Tet(O) interactions with the ribosomea
| Domain | Location of interaction with:
|
|
|---|---|---|
| EF-G | Tet(O) | |
| G | H95 | H95 |
| II | h5 | h5 |
| III | S12 | S12 |
| IV | H69 | h18/34 |
| V | H43/44 | H43/44 |
This table is an adaptation of one published previously (45) (reprinted with permission of the publisher). Helix is abbreviated with a lowercase h when referring to a helix within the 16S rRNA, whereas an uppercase H refers to a helix within the 23S rRNA.
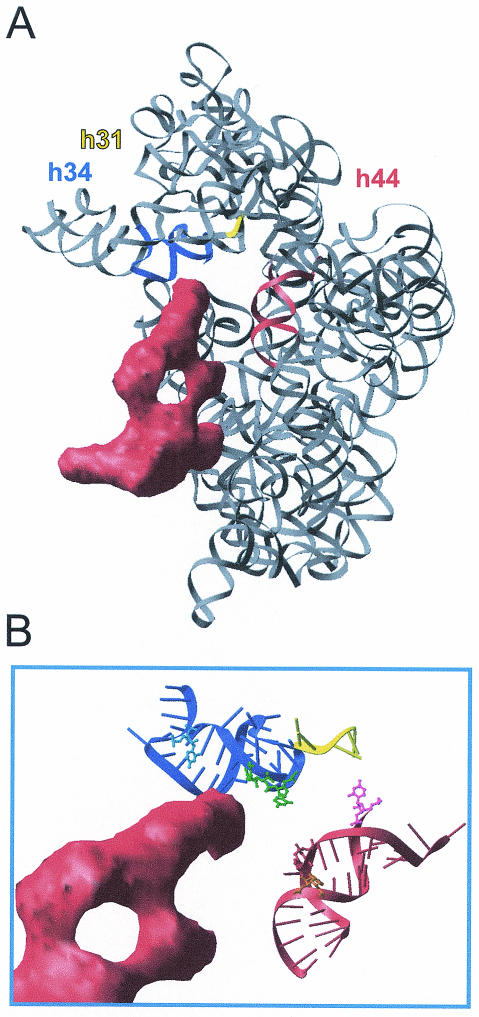
DMS, a chemical probe that modifies the N1 and N3 positions of adenosine and cytosine, respectively, has also been employed to define the interaction of Tet(O) with the ribosome (12). On the 30S subunit, sites of interaction were localized to h34 (C1214) and h44 (A1408), near the decoding site and the primary tetracycline-binding site (Fig. 4). The protection of C1214 from DMS modification would indicate that this base is directly shielded by Tet(O), and this conclusion is supported by the fact that the Tet(O) binding site observed by cryo-EM approaches C1214 (Fig. 4B). The close association of C1214 with the Tet(O) binding site contrasts with the enhancement of A1408 by DMS modification because, as illustrated in Fig. 4B, Tet(O) does not approach A1408 in h44 (45). Furthermore, an enhancement of chemical modification is clearly indicative of a conformational change; therefore, this result can be taken to indicate that Tet(O) is inducing long-range rearrangements in the ribosome. These changes could be mediated by S12, which is in close proximity to the top of h44 (43, 58) and also appears to interact with Tet(O) (45).
FIG. 4.
rRNA bases that are altered in DMS modification by the binding of Tet(O) cluster around the decoding center. (A) Tet(O) (red density) (45) bound to the 30S subunit (58) (PDB identification code 1FJF) in the same orientations as seen in panel B. Helices 31 (nucleotides 964 to 968), 34 (nucleotides 1199 to 1217 and 1058 to 1046) and 44 (nucleotides 1400 to 1414 and 1486 to 1503) are represented as yellow, blue, and red ribbons, respectively, and the remaining rRNA is represented as a grey ribbon. (B) Interaction of domain IV of Tet(O) (red density) with the region around the primary tetracycline binding site. Helices 31, 34, and 44 are represented as in panel A. The bases that experience changes in DMS accessibility upon tetracycline (U1052 and C1054, green), EF-G (A1408, orange; C1400, pink), or Tet(O) (C1214, blue; A1408, orange) binding are drawn in a ball and stick representation. This figure has been reproduced from reference 12 with permission of the publisher.
Proposed mechanism of Tet(O)-mediated tetracycline resistance.
A model describing Tet(O)-mediated tetracycline resistance (Fig. 1, reactions f to j) was presented by Spahn et al. (45) and summarizes most of the biochemical and structural work done on Tet(O) and Tet(M). In the absence of tetracycline, the 70S ribosome progresses through the various states of the elongation cycle (Fig. 1, reactions a to e) (see figure legend for a detailed description). In the presence of tetracycline, however, the ordered progression though the elongation cycle is interrupted and the ribosome becomes blocked in a posttranslocational state because subsequent A site occupation is inhibited. Although this blockage is likely due to a direct steric clash between tetracycline and the incoming aa-tRNA, it is possible that the binding of tetracycline to the ribosome (reaction f) is accompanied by a structural rearrangement. Although a gross conformational change is not observed in the crystal structure of tetracycline bound to the 30S subunit (4, 37), a conformational change can be inferred from several biochemical experiments (11, 14, 32, 56). For example, Noah et al. (32) presented evidence that tetracycline affects h44. They observed that a UV-dependent cross-link between C1402 and C1501, two bases located at the top of h44, is enhanced by the presence of tetracycline. These bases are distinct from the observed tetracycline binding sites and may indicate that tetracycline is promoting subtle structural rearrangement or fixing the ribosome in a conformation that is favorable for the establishment of the C1402-C1501 cross-link. The nature and role of this proposed conformational change is not known; however, the change might simply move the ribosome into a configuration compatible with stable tetracycline binding. Additionally, as h44 is a component of the decoding site, it may have a role in the inhibitory action of tetracycline. However, models presented in the papers of Brodersen et al. and Pioletti et al. (4, 37) suggest that tetracycline exerts it effect simply through a steric clash with an accommodating tRNA such that its binding is blocked, which presumably makes a conformational change unnecessary.
In any case, tetracycline binding to the ribosome presumably does not interfere with initial decoding and EF-Tu-dependent GTPase activity but rather prevents stable occupation of the A-site by incoming aa-tRNA (4), a step that is termed accommodation. This may lock the ribosome in a nonproductive cycle of ternary complex binding and release (Fig. 1, reactions i and j) (4). In the presence of Tet(O), this nonproductive cycle would be averted, as Tet(O) would bind the tetracycline-blocked ribosome, release tetracycline, and return the ribosome to the elongation cycle (Fig. 1, reactions g and h). The mechanism by which Tet(O) distinguishes the tetracycline-blocked ribosome has not been conclusively established, but we suggest that it could involve two mechanisms. First, a tetracycline-induced conformational change in the ribosome may promote Tet(O) binding (11). Second, tetracycline blocks the ribosome in a state with an open A site, and a ribosome in this condition seems to be the preferred substrate for Tet(O) because Tet(O) cannot bind a ribosome with an occupied A site (11). In the presence of tetracycline, the ribosome is blocked with an open A site and this could provide a kinetic window for Tet(O) to act, thus distinguishing the tetracycline-blocked ribosome from a translating ribosome (11).
After Tet(O) has bound the tetracycline-blocked ribosome, it must free the ribosome from tetracycline (Fig. 1, reaction g). Trieber et al. (54) demonstrated that the binding of Tet(O) in the GTP state is sufficient to trigger the release of tetracycline, whereas DMS-probing experiments showed that Tet(O) specifically triggers the release of tetracycline from the primary tetracycline binding site (12). Additionally, cryo-EM studies demonstrated that when Tet(O) binds the ribosome, it does not directly overlap the primary tetracycline binding site, and therefore, Spahn et al. (45) proposed that Tet(O) triggers the release of tetracycline through an allosteric mechanism. It should be noted, however, that a direct interaction between Tet(O) and tetracycline bound to the primary site cannot be absolutely discounted, although both chemical probing and cryo-EM suggest this is not the case (12, 45). The proposed conformational change resulting in tetracycline release probably involves h34 as (i) h34 forms an integral part of the primary tetracycline binding site (4, 37), (ii) cryo-EM reconstructions show that domain IV of Tet(O) contacts the base of h34 (45), and (iii) Tet(O) protects C1214 at the base of h34 from chemical modification by DMS (12). As such, Spahn et al. (45) hypothesize that, upon binding, Tet(O) contacts the base of h34, which in turn causes a disturbance in this helix, which is propagated to the primary tetracycline-binding site, releasing the drug.
In addition to the proposed conformational change in h34 that results in tetracycline release, Tet(O) invokes structural rearrangements in h44 (12), a site distinct from both the primary tetracycline binding site (4, 37) and the Tet(O) binding site observed in the cryo-EM reconstruction (45). The reasons for this long-range rearrangement are not yet understood, but the following points should be considered: (i) Tet(O) may reverse a tetracycline-induced rearrangement in h44 (discussed above), (ii) the effect on h44 may be a consequence of Tet(O) being derived from EF-G and may not be related to Tet(O) activity (12), and (iii) Tet(O) may induce an altered conformation in the ribosome to prevent tetracycline rebinding and/or promote ternary complex binding (11). With respect to the last point, the fact that Tet(O) can stimulate the GTPase activity of EF-Tu suggests that Tet(O) can induce conformational changes in the ribosome that persist after it has dissociated (11).
Nevertheless, after removing tetracycline from the ribosome, Tet(O) must disassociate from the ribosome (Fig. 1, reaction h) so that the ternary complex (EF-Tu-aa-tRNA-GTP) can bind and protein synthesis can continue (Fig. 1, reactions a to e).
CONCLUDING REMARKS
Research in the last several years has contributed greatly to our understanding of RPP activity. For example, structural studies on the ribosome (3, 24, 43, 58) and the ribosome-tetracycline complex (4, 37) have greatly expanded our understanding of protein synthesis and the molecular mechanism of tetracycline action. When the biochemical (11, 12, 15) and structural (45) data describing the RPP's ribosomal binding site are combined, the interaction of the RPP with the ribosome can be modeled with high precision (Table 1). Furthermore, the combination of these data has provided a plausible mechanism that explains the mode of RPP action in molecular detail, namely that the RPPs interact with the base of h34, resulting in an allosteric disruption of primary tetracycline binding and consequently releasing the drug (45).
One puzzling aspect of RPP-mediated tetracycline resistance that remains unanswered is the question of whether or not the RPPs actively function to prevent tetracycline rebinding after triggering tetracycline release. This is an important question because, after being released, tetracycline is free to rebind the ribosome and again inhibit protein synthesis. In this sense, if an active mechanism does not exist, Tet(O) might be required to work successively before an aa-tRNA successfully competes with tetracycline for the A site. Alternatively, an attractive possibility is that Tet(O) may promote subtle rearrangements in the ribosomal architecture that slow tetracycline rebinding and actively enhance the ability of the aa-tRNA complex to bind to the A site.
Acknowledgments
We thank Daniel Wilson for critical reading of the manuscript and James Gunton for help during preparation of the manuscript.
This work was funded by the Alberta Heritage Foundation for Medical Research (AHFMR) through an AHFMR Studentship to S.R.C. and D.M.T, an AHFMR Scientist Award to D.E.T., a grant from the National Science and Engineering Research Council of Canada (NSERC) to D.E.T., and a grant from the Deutsche Forschungsgemeinschaft to K.H.N. (Ni174/8-3).
REFERENCES
- 1.Agrawal, R. K., A. B. Heagle, P. Penczek, R. A. Grassucci, and J. Frank. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6:643-647. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, R. K., C. M. Spahn, P. Penczek, R. A. Grassucci, K. H. Nierhaus, and J. Frank. 2000. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J. Cell Biol. 150:447-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., W. M. Clemons, A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 5.Burdett, V. 1991. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J. Biol. Chem. 266:2872-2877. [PubMed] [Google Scholar]
- 6.Burdett, V. 1996. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J. Bacteriol. 178:3246-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, D. M., J. Thompson, P. E. March, and A. E. Dahlberg. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 319:27-35. [DOI] [PubMed] [Google Scholar]
- 8.Chopra, I. 1985. Mode of action of the tetracyclines and the nature of bacterial resistance to them, p. 317-392. In J. H. Boothe and J. J. Hlavka (ed.), The tetracyclines (handbook of experimental pharmacology), vol. 78. Springer-Verlag, Berlin, Germany.
- 9.Chopra, I. 1994. Tetracycline analogs whose primary target is not the bacterial ribosome. Antimicrob. Agents Chemother. 38:637-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell, S. R., C. A. Trieber, E. Einfeldt, G. P. Dinos, D. E. Taylor, and K. Nierhaus. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connell, S. R., C. A. Trieber, U. Stelzl, E. Einfeldt, D. E. Taylor, and K. H. Nierhaus. 2002. The tetracycline resistance protein, Tet(O), perturbs the conformation of the ribosomal decoding center. Mol. Microbiol. 45:1463-1472. [DOI] [PubMed] [Google Scholar]
- 13.Cundliffe, E. 1967. Antibiotics and polyribosomes. Chlortetracycline and polyribosomes of Bacillus megaterium. Mol. Pharmacol. 3:401-411. [PubMed] [Google Scholar]
- 14.Dahlberg, A. E., E. Lund, N. O. Kjeldgaard, C. M. Bowman, and M. Nomura. 1973. Colicin E3 induced cleavage of 16S ribosomal ribonucleic acid; blocking effects of certain antibiotics. Biochemistry 12:948-950. [DOI] [PubMed] [Google Scholar]
- 15.Dantley, K. A., H. K. Dannelly, and V. Burdett. 1998. Binding interaction between Tet(M) and the ribosome: requirements for binding. J. Bacteriol. 180:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle, D., K. J. McDowall, M. J. Butler, and I. S. Hunter. 1991. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol. Microbiol. 5:2923-2933. [DOI] [PubMed] [Google Scholar]
- 17.Epe, B., and P. Woolley. 1984. The binding of 6-demethylchlortetracycline to 70S, 50S and 30S ribosomal particles: a quantitative study by fluorescence anisotropy. EMBO J. 3:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, J. 2001. Cryo-electron microscopy as an investigative tool: the ribosome as an example. BioEssays 23:725-732. [DOI] [PubMed] [Google Scholar]
- 19.Gerrits, M. M., M. R. De Zoete, N. L. Arents, E. J. Kuipers, and J. G. Kusters. 2002. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2996-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman, R. A., T. Hasan, C. C. Hall, W. A. Strycharz, and B. S. Cooperman. 1983. Photoincorporation of tetracycline into Escherichia coli ribosomes. Identification of the major proteins photolabeled by native tetracycline and tetracycline photoproducts and implications for the inhibitory action of tetracycline on protein synthesis. Biochemistry 22:359-368. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Lorenzo, M. G., C. M. Spahn, R. K. Agrawal, R. A. Grassucci, P. Penczek, K. Chakraburtty, J. P. Ballesta, J. L. Lavandera, J. F. Garcia-Bustos, and J. Frank. 2000. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 19:2710-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon, J. 1969. Hydrolysis of guanosine 5′-triphosphate associated with binding of aminoacyl transfer ribonucleic acid to ribosomes. J. Biol. Chem. 244:5680-5686. [PubMed] [Google Scholar]
- 23.Grewal, J., E. K. Manavathu, and D. E. Taylor. 1993. Effect of mutational alteration of Asn-128 in the putative GTP-binding domain of tetracycline resistance determinant Tet(O) from Campylobacter jejuni. Antimicrob. Agents Chemother. 37:2645-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 25.Hierowski, M. 1965. Inhibition of protein synthesis by chlorotetracycline in the Escherichia coli in vitro system. Proc. Natl. Acad. Sci. USA 53:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 27.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 28.Lodmell, J. S., and A. E. Dahlberg. 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277:1262-1267. [DOI] [PubMed] [Google Scholar]
- 29.Lucas-Lenard, J., and A. L. Haenni. 1967. Requirement of guanosine 5′-triphosphate for ribosomal binding of aminoacyl-sRNA. Biochemistry 59:554-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martemyanov, K. A., and A. T. Gudkov. 1999. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 452:155-159. [DOI] [PubMed] [Google Scholar]
- 31.Martemyanov, K. A., A. S. Yarunin, A. Liljas, and A. T. Gudkov. 1998. An intact conformation at the tip of elongation factor G domain IV is functionally important. FEBS Lett. 434:205-208. [DOI] [PubMed] [Google Scholar]
- 32.Noah, J. W., M. A. Dolan, P. Babin, and P. Wollenzien. 1999. Effects of tetracycline on the tertiary structure of ribosomal RNA in the Escherichia coli 30S ribosomal subunit. J. Biol. Chem. 274:16576-16581. [DOI] [PubMed] [Google Scholar]
- 33.Oehler, R., N. Polacek, G. Steiner, and A. Barta. 1997. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia coli. Nucleic Acids Res. 25:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 35.Oliva, B., and I. Chopra. 1992. Tet determinants provide poor protection against some tetracyclines: further evidence for division of tetracyclines into two classes. Antimicrob. Agents Chemother. 36:876-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliva, B., G. Gordon, P. McNicholas, G. Ellestad, and I. Chopra. 1992. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob. Agents Chemother. 36:913-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pioletti, M., F. Schlunzen, J. Harms, R. Zarivach, M. Gluhmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, and F. Franceschi. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen, B., H. F. Noller, G. Daubresse, B. Oliva, Z. Misulovin, D. M. Rothstein, G. A. Ellestad, Y. Gluzman, F. P. Tally, and I. Chopra. 1991. Molecular basis of tetracycline action: identification of analogs whose primary target is not the bacterial ribosome. Antimicrob. Agents Chemother. 35:2306-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogalski, W. 1985. Chemical modification of the tetracyclines, p. 179-316. In J. H. Boothe and J. J. Hlavka (ed.), The tetracyclines (handbook of experimental pharmacology), vol. 78. Springer-Verlag, Berlin, Germany.
- 40.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Pescador, R., J. T. Brown, M. Roberts, and M. S. Urdea. 1988. Homology of the TetM with translational elongation factors: implications for potential modes of tetM-conferred tetracycline resistance. Nucleic Acids Res. 16:1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savelsbergh, A., N. B. Matassova, M. V. Rodnina, and W. Wintermeyer. 2000. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 300:951-961. [DOI] [PubMed] [Google Scholar]
- 43.Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102:615-623. [DOI] [PubMed] [Google Scholar]
- 44.Sougakoff, W., B. Papadopoulou, P. Nordmann, and P. Courvalin. 1987. Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol. Lett. 44:153-159. [Google Scholar]
- 45.Spahn, C. M. T., G. Blaha, R. K. Agrawal, P. Penczek, R. A. Grassucci, C. A. Trieber, S. R. Connell, D. E. Taylor, K. H. Nierhaus, and J. Frank. 2001. Localization of the ribosomal protection protein Tet(O) on the ribosome and the mechanism of tetracycline resistance. Mol. Cell 7:1037-1045. [DOI] [PubMed] [Google Scholar]
- 46.Speer, B. S., and A. A. Salyers. 1989. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J. Bacteriol. 171:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strel'tsov, S. A., M. K. Kukhanova, G. V. Gurskii, A. A. Kraevskii, and I. V. Beliavskaia. 1975. Oxytetracycline binding to E. coli ribosomes. Mol. Biol. (Moskow) 9:910-921. [PubMed] [Google Scholar]
- 48.Suarez, G., and D. Nathans. 1965. Inhibition of aminoacyl tRNA binding to ribosomes by tetracycline. Biochem. Biophys. Res. Commun. 18:743-750. [Google Scholar]
- 49.Taylor, D. E. 1992. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli to tetracycline, chloramphenicol, and erythromycin, p. 74-86. In I. Nachaminkin (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 50.Taylor, D. E. 1986. Plasmid-mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J. Bacteriol. 165:1037-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor, D. E., and P. Courvalin. 1988. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob. Agents Chemother. 32:1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor, D. E., L. J. Jerome, J. Grewal, and N. Chang. 1995. Tet(O), a protein that mediates ribosomal protection to tetracycline, binds, and hydrolyses GTP. Can. J. Microbiol. 41:965-970. [Google Scholar]
- 54.Trieber, C. A., N. Burkhardt, K. H. Nierhaus, and D. E. Taylor. 1998. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 379:847-855. [DOI] [PubMed] [Google Scholar]
- 55.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S ribosomal RNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tritton, T. R. 1977. Ribosome-tetracycline interactions. Biochemistry 16:4133-4138. [DOI] [PubMed] [Google Scholar]
- 57.White, J. P., and C. R. Cantor. 1971. Role of magnesium in the binding of tetracycline to Escherichia coli ribosomes. J. Mol. Biol. 58:397-400. [DOI] [PubMed] [Google Scholar]
- 58.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]