Abstract
Ten mutants of the yeast Saccharomyces cerevisiae resistant to the antimycotic terbinafine were isolated after chemical or UV mutagenesis. Molecular analysis of these mutants revealed single base pair exchanges in the ERG1 gene coding for squalene epoxidase, the target of terbinafine. The mutants did not show cross-resistance to any of the substrates of various pleiotropic drug resistance efflux pumps tested. The ERG1 mRNA levels in the mutants did not differ from those in the wild-type parent strains. Terbinafine resistance was transmitted with the mutated alleles in gene replacement experiments, proving that single amino acid substitutions in the Erg1 protein were sufficient to confer the resistance phenotype. The amino acid changes caused by the point mutations were clustered in two regions of the Erg1 protein. Seven mutants carried the amino acid substitutions F402L (one mutant), F420L (one mutant), and P430S (five mutants) in the C-terminal part of the protein; and three mutants carried an L251F exchange in the central part of the protein. Interestingly, all exchanges identified involved amino acids which are conserved in the squalene epoxidases of yeasts and mammals. Two mutations that were generated by PCR mutagenesis of the ERG1 gene and that conferred terbinafine resistance mapped in the same regions of the Erg1 protein, with one resulting in an L251F exchange and the other resulting in an F433S exchange. The results strongly indicate that these regions are responsible for the interaction of yeast squalene epoxidase with terbinafine.
Ergosterol is an essential and specific component of fungal membranes, in which it exhibits a variety of functions that modulate membrane fluidity, permeability, and the activities of membrane-bound enzymes (3). Therefore, the ergosterol biosynthesis pathway represents an important target for antifungal agents; and, indeed, some of the most effective compounds in the treatment of fungal infections, like azoles and allylamines, are inhibitors of essential steps in ergosterol biosynthesis. Azoles inhibit lanosterol 14α-demethylase (Erg11p) (14), and squalene epoxidase (Erg1p) is the target for allylamines (34), such as naftifine and terbinafine.
Azoles are widely used in the treatment of severe fungal infections, which has led to an increased occurrence of resistant clinical isolates of Candida albicans and other human fungal pathogens (25, 38, 46, 48). The elucidation of the molecular mechanisms by which C. albicans can exert resistance to azoles revealed that upregulation of the genes encoding efflux pumps leads to high levels of resistance to azoles and other antifungal drugs (27, 28, 40). This type of resistance is mediated by overexpression of transporters belonging to either the major facilitator superfamily (MFS) or the ATP binding cassette (ABC) transporter superfamily (27, 40, 42). Members of the C. albicans drug resistance gene family, CDR1 and CDR2, which code for ABC transporters, not only are involved in azole resistance but also confer resistance to terbinafine (28, 30, 41). In contrast, increased expression of the C. albicans MFS transporter Mdr1p leads to fluconazole resistance but not to terbinafine resistance (49).
In addition to efflux systems, resistance to metabolic inhibitors can also be mediated by overexpression of drug target proteins (38). Elevated levels of ERG11 mRNA were found in several azole-resistant clinical isolates of C. albicans and C. glabrata, suggesting that the amount of lanosterol 14α-demethylase (Erg11p) is higher and contributes to azole resistance in the isolates (8, 26, 28, 32).
Another mechanism of resistance to azoles involves modifications of the target enzyme Erg11p. A number of different mutations in the CaERG11 gene were identified in azole-resistant clinical C. albicans isolates, and the resulting amino acid substitutions were thought to alter the affinities of azole derivatives to Erg11p (7, 8, 15, 19, 20, 32, 39).
A second important target for antifungal drugs in the ergosterol biosynthesis pathway is represented by squalene epoxidase, which acts upstream of lanosterol 14α-demethylase. This essential enzyme is encoded by the ERG1 gene in Saccharomyces cerevisiae (13) and the CaERG1 gene in C. albicans (6) and catalyzes the epoxidation of squalene to (3S)-2,3-oxidosqualene (34, 43). In fungi this reaction is selectively inhibited by allylamines, such as naftifine and terbinafine (31, 34, 35). S. cerevisiae cells treated with these antifungal drugs accumulate squalene and are depleted of ergosterol, which finally results in growth inhibition (24, 31). Although terbinafine is widely used to treat infections caused by dermatophytes and other fungal pathogens, resistant mutants that appear as a consequence of terbinafine treatment have not been reported so far. However, increased use of terbinafine could lead to the development of resistant mutants and, consequently, to treatment failure. Therefore, the elucidation of the molecular mechanisms by which fungi can exert resistance to terbinafine is of great interest. We have chosen S. cerevisiae as a model organism to investigate the mechanisms by which a phenotype of terbinafine resistance can develop. We isolated a series of terbinafine-resistant S. cerevisiae mutants in which we identified mutations in the ERG1 gene. The mutants did not exhibit cross-resistance to other metabolic inhibitors, suggesting that active efflux of the drug is not involved in terbinafine resistance in these mutants. In this report we demonstrate for the first time that single mutations in the ERG1 gene can be the sole cause for terbinafine resistance in S. cerevisiae.
MATERIALS AND METHODS
Strains and growth conditions.
All strains and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source |
|---|---|---|
| Strains | ||
| E. coli XL1 | endA1 hsdR17 (hsdR hsdM+) supE44 thi-1 recA1 gyrA96 relA1 Δ(lac) [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| S. cerevisiae A2 | MATaleu2 his3 can1 | A. Hartig |
| S. cerevisiae W303-1B | MATα leu2 ura3 his3 ade2 trp1 can1 | J. Kolarov |
| S. cerevisiae W303/a | MATaleu2 ura3 his3 ade2 trp1 | S. D. Kohlwein |
| S. cerevisiae H1A, H1B, H2, H3, H4, H5, H6, H7, H8 | Terbinafine-resistant mutants of W303-1B after MNNG mutagenesis | This study |
| S. cerevisiae A2M8 | Terbinafine-resistant mutant of A2 after UV mutagenesis | Jandrositz et al. (13) |
| S. cerevisiae KLN1 | MATα, ERG1::URA3 leu2 ura3 trp1 | Landl et al. (21) |
| S. cerevisiae A2T8 | A2 with erg1 allele for terbinafine resistance of A2M8 | This study |
| S. cerevisiae A2H3 | A2 with erg1 allele for terbinafine resistance of H3 | This study |
| S. cerevisiae KLNH1B | KLN1 with erg1 allele for terbinafine resistance of H1B | This study |
| S. cerevisiae KLNH2 | KLN1 with erg1 allele for terbinafine resistance of H2 | This study |
| S. cerevisiae KPHJ1 | A2 MATα PDR1-12::HIS3 pet | Wendler et al. (47) |
| S. cerevisiae KPHJ2 | A2 MATα PDR3-33::HIS3 pet | Wendler et al. (47) |
| Plasmids | ||
| pYEp13 | Multicopy yeast-E. coli shuttle vector | A. Hartig |
| pYEp351 | Multicopy yeast-E. coli shuttle vector | A. Hartig |
| pRS315 | Centromere yeast-E. coli shuttle vector | K. Kuchler |
| pBIG1 | 2.3-kb PstI fragment with the wild-type ERG1 gene in pBluescript | This study |
| pKL1 | 3.9-kb SacI fragment with the erg1 allele for terbinafine resistance of A2M8 in pBluescript | This study |
| pKL2 | pKL1 with the LEU2 cassette in the PstI site | This study |
| pKLH3 | erg1 allele for terbinafine resistance of mutant H3 in pKL2 | This study |
| pAF22 | 4.8-kb Sau3A fragment with the erg1 allele for terbinafine resistance of A2M8 | Jandrositz et al. (13) |
| pML1 | 2.3-kb PstI fragment with the wild-type ERG1 gene in pRS315 | This study |
| pNS1 | 2.3-kb PstI fragment with the wild-type ERG1 gene in pRS315 (insert in the orientation opposite that in pML1) | This study |
| pML2 | 2.3-kb PstI fragment with the erg1 allele for terbinafine resistance of A2M8 in pRS315 | This study |
| pML3 | 2.3-kb PstI fragment with the erg1 allele ML3 for terbinafine resistance in pRS315 | This study |
| pNS2 | 2.3-kb PstI fragment with erg1 allele NS2 for terbinafine resistance in pRS315 | This study |
| pYSTS1 | STS1 (PDR5) gene in pYEp13 | Bissinger and Kuchler (2), K. Kuchler |
Escherichia coli XL1 carrying recombinant plasmids was grown in 2× TY medium (37) in the presence of 100 μg of ampicillin per ml at 37°C. Wild-type yeast strains W303-1B and A2 and terbinafine-resistant mutants were grown in yeast extract-peptone-dextrose (YPD) medium (44) at 30°C on a rotary shaker. Wild-type yeast strains carrying recombinant plasmids and A2 transformants after gene replacement were grown in yeast nitrogen base (YNB) minimal medium supplemented with all amino acids except leucine (11, 44). S. cerevisiae KLN1 was grown in YPD medium containing ergosterol and Tween 80 under oxygen-limiting conditions, as described previously (21). Strain KLN1 with ERG1-containing recombinant plasmids was grown in YPD medium under aerobic conditions. Tetrad analysis was performed as described previously (21). The components of the media were purchased from Difco, Gibco BRL, and Merck.
Mutagenesis and selection of terbinafine-resistant mutants.
Mutagenesis of S. cerevisiae W303-1B was carried out as described by Lawrence (23) with 40 μg of N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) per ml in 50 mM potassium phosphate buffer (pH 7.0) for 20 min at 30°C. Mutagenized cells were washed, incubated in YPD medium for 3 h at 30°C, and transferred to fresh YPD medium (pH 6.0) containing 50 μg of terbinafine per ml. After overnight incubation at 30°C the cells were plated on YPD agar plates. Colonies were tested for resistance to terbinafine on YPD plates containing 50 μg of terbinafine per ml, and nine resistant clones were selected for further analysis. The terbinafine-resistant mutant S. cerevisiae A2M8, isolated after UV mutagenesis, was described previously (13).
In another approach the wild-type ERG1 gene was randomly mutagenized in vitro by PCR amplification. Recombinant plasmid pBIG1, which carries the wild-type ERG1 gene on a 2.3-kb PstI fragment in pBluescript, served as the template for amplification of the ERG1 gene with the universal primer (5′-ACGACGTTGTAAAACGACGGCCAG-3′) and the reverse primer (5′-TTCACACAGGAAACAGCTATGACC-3′) (VBC Genomics, Vienna, Austria). The amplification was performed with DyNAzyme II DNA polymerase (Finnzymes) in the presence of 5 mM MgCl2 and 0.05 mM MnCl2. The PCR fragments were digested with PstI, cloned into the centromere vector pRS315, and transformed into E. coli XL1. Plasmid DNA was isolated from approximately 1,000 E. coli transformants by standard procedures (37) and transformed into S. cerevisiae KLN1 (21), which contains the chromosomal erg1::URA3 disruption, by the protocol of Gietz et al. (9). Transformants expressing a functional squalene epoxidase were selected by complementation of the aerobic growth defect of KLN1 on YPD agar plates. The transformants were tested for terbinafine resistance on YPD plates containing 100 μg of terbinafine per ml. Plasmid DNA was isolated from resistant S. cerevisiae KLN1 transformants (45), amplified in E. coli XL1, and characterized by DNA sequence determination.
Drug susceptibility testing.
For drug susceptibility determination in liquid medium, 2 ml of YPD medium containing 20, 50, or 100 μg of terbinafine per ml dissolved in ethanol was inoculated with aliquots of overnight cultures (ONCs) of the respective yeast mutants or wild-type strains to give an initial optical density at 600 nm (OD600) of 0.005. Control cultures of each strain were supplemented with the respective solvent. The cultures were incubated for 45 or 24 h at 30°C with shaking. Growth of the yeast strains was measured spectrophotometrically by determination of the OD600.
The susceptibilities of the yeast strains to terbinafine were also determined on agar plates. ONCs in YPD medium or YNB minimal medium were diluted to an OD600 of 0.1, and 5 μl of serial dilutions (100 to 10−3) was spotted on YPD agar plates containing terbinafine (10.0 to 100 μg/ml) and on control plates supplemented with the solvent. The plates were incubated for 2 days at 30°C. The sensitivities of the terbinafine-resistant yeast mutants to other metabolic inhibitors were determined with ONCs grown in YPD medium for 20 h at 30°C. The cells were washed and diluted to a cell density of 106/ml. Drops of 10 μl of these suspensions and serial dilutions (100 to 10−3) were spotted on YPD agar plates buffered with sodium succinate to pH 6.0 with and without the inhibitors. The inhibitors tested were cycloheximide (0.1 to 2.0 μg/ml), itraconazole (5.0 to 30 μg/ml), resazurin (50 to 200 μg/ml), 4-nitroquinoline-N-oxide (0.1 to 1.0 μg/ml), crystal violet (0.5 to 5.0 μg/ml), 6-amino-2n-pentylthiobenzothiazole (10 to 50 μg/ml), nystatin (0.5 to 2.5 μg/ml), and amorolfine (0.02 to 0.1 μg/ml). The susceptibilities to the mitochondrial inhibitors rhodamine 6G (1.0 to 5.0 μg/ml), ethidium bromide (0.5 to 5.0 μg/ml), and oligomycin (0.1 to 2.0 μg/ml) were tested on YP agar plates containing 1% (vol/vol) ethanol and 2% (vol/vol) glycerol instead of glucose. The plates were incubated for 3 days at 30°C. The inhibitors were obtained from Sigma (cycloheximide, oligomycin, 4-nitroquinoline-N-oxide), Serva (ethidium bromide, resazurin, nystatin), Loba Chemie Austria (crystal violet), and Lambda Physik Germany (rhodamine 6G). Terbinafine was a gift of A. Stütz, Novartis Research Institute, Vienna, Austria; and itraconazole, amorolfine, and 6-amino-2n-pentylthiobenzothiazole were obtained from T. Kuchta, Food Research Institute, Bratislava, Slovak Republic.
Nucleic acid preparation and analysis.
Chromosomal and plasmid DNAs were isolated by standard procedures (37, 45). The ERG1 gene of the terbinafine-resistant mutants was amplified by PCR with High Fidelity DNA Polymerase (Roche Diagnostics), forward primer UE1 (5′-GTCCAGTATTGAACAATACAGGTT-3′), reverse primer DE1 (5′-TTGACGGTTCCTATCCTCTCTC-3′), and chromosomal DNA as the template. For DNA sequence determination, 1 μg of DNA was labeled by using the Thermo Sequenase fluorescently labeled primer cycle sequencing kit from Amersham (Little Chalfont, United Kingdom) and analyzed with the ALFexpress DNA sequencer from Amersham Pharmacia Biotech. The primers were purchased from VBC Genomics. The Genetics Computer Group program was applied for sequence alignments (4). RNA was isolated from lysed spheroplasts or mechanically disrupted cells with the RNeasy kits of Qiagen. Northern blot assays were performed with ERG1- and ACT1-specific probes as described previously (24).
Construction of recombinant plasmids pKL2 and pKLH3 for gene replacement.
Recombinant plasmid pAF22 carries a 4.8-kb insert with the erg1 allele from terbinafine-resistant mutant A2M8 in pYEp351 (13). pAF22 was restricted with SacI, followed by treatment with Klenow polymerase to generate blunt ends, and the 3.9-kb SacI fragment was isolated after agarose gel electrophoresis. The pBluescript vector was cleaved with SacI and KpnI to remove the multiple-cloning site and was also treated with Klenow polymerase. Ligation of the SacI fragment carrying the erg1 allele of pAF22 with the purified vector fragment and transformation into E. coli XL1 yielded recombinant plasmid pKL1. The unique PstI restriction site within the SacI fragment downstream of the ERG1 structural gene was used to insert a LEU2 cassette for selection purposes. The resulting plasmid was designated pKL2. The erg1 allele of terbinafine-resistant mutant H3 was amplified from the chromosomal DNA by PCR with forward primer UE1, reverse primer DE1, and High Fidelity DNA Polymerase (Roche Diagnostics). The PCR product was cleaved with HpaI, and the 1.4-kb fragment was used to replace the erg1 allele in pKL2 by ligation with the appropriately cleaved pKL2. The recombinant plasmid was designated pKLH3. The correct DNA sequences of the mutated erg1 alleles in pKL2 and pKLH3 were verified by DNA sequence determination.
Replacement of ERG1 gene by homologous recombination.
Recombinant plasmids pKL2 and pKLH3 were cleaved with SphI and SmaI, and the linear fragments carrying the erg1 alleles and the LEU2 marker were transformed into S. cerevisiae A2 by the protocol of Gietz et al. (9). Insertion of the fragments in the chromosome occurred at the ERG1 locus by homologous recombination. Transformants were selected on YNB minimal medium lacking leucine. Correct gene replacement was verified by PCR amplification and DNA sequence determination. The transformants carrying the chromosomally integrated erg1 alleles of terbinafine-resistant mutants A2M8 and H3 were designated A2T8 and A2H3, respectively.
The erg1 alleles of mutants H1B and H2 were integrated into the chromosome of KLN1 by a different approach. Briefly, the erg1 alleles were amplified from the chromosomal DNA of the mutants by PCR with primers UE1 and DE1, and the DNA sequences were verified by sequence analysis. Approximately 5 μg of the linear fragments was transformed into S. cerevisiae KLN1, in which the ERG1 gene was disrupted by a URA3 cassette (21). Successful integration of the fragments by homologous recombination at the chromosomal ERG1 locus allowed the transformants to grow aerobically and resulted in the loss of the URA3 cassette. Correct integration of the erg1 alleles was verified by PCR amplification and DNA sequencing. The transformants were designated KLNH1B and KLNH2, respectively.
Cloning of wild-type ERG1 gene and erg1 alleles for terbinafine resistance into centromere vector pRS315.
The wild-type ERG1 gene was isolated as a 2.3-kb PstI fragment from pBIG1 and cloned into the PstI site of the low-copy-number vector pRS315. The recombinant plasmid was designated pML1. A similar approach was taken to construct pML2, which carried the erg1 allele of A2M8, and pNS2, which contained the NS2 erg1 allele for terbinafine resistance. The plasmids were transformed into S. cerevisiae KLN1 without integration into the chromosome and were selected for aerobic growth due to complementation of the KLN1 phenotype.
RESULTS
Generation of terbinafine-resistant S. cerevisiae mutants.
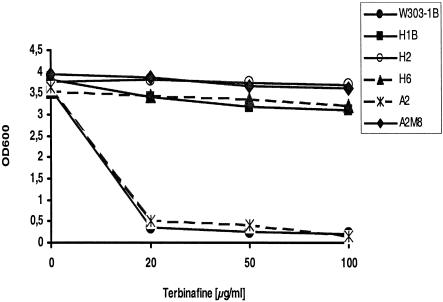
Terbinafine-resistant S. cerevisiae mutant A2M8 was isolated after UV mutagenesis of wild-type strain A2, as described previously (13). To isolate novel mutants, S. cerevisiae W303-1B cells were treated with MNNG, as described in Materials and Methods, and nine terbinafine-resistant clones growing on YPD agar plates containing 100 μg of terbinafine per ml were selected for further analysis. The sensitivities of the mutants to terbinafine were determined in liquid cultures after growth for 45 h at 30°C in the presence of terbinafine at concentrations of 20, 50, or 100 μg/ml in YPD medium. Figure 1 shows the growth yields of selected representatives (mutants H1B, H2, and H6 and the corresponding wild-type strain, W303-1B, as well as mutant A2M8 and its wild-type parent, A2) plotted as the OD600 against the terbinafine concentration. While the growth of wild-type strains W303-1B and A2 was completely inhibited by 20 μg of terbinafine per ml, the resistant S. cerevisiae mutants continued to grow even in the presence of 100 μg of terbinafine per ml. Similar results were obtained with resistant mutants H1A, H3, H4, H5, H6, H7, and H8 (data not shown).
FIG. 1.
Growth of wild-type S. cerevisiae strains A2 and W303-1B and terbinafine-resistant mutants in the presence and absence of terbinafine. The yeast strains were inoculated into 2 ml of YPD medium to an OD600 of 0.005 and grown at 30°C for 45 h. The medium was supplemented with 20, 50, and 100 μg of terbinafine per ml dissolved in ethanol. The control culture without terbinafine was supplemented with ethanol. Growth was monitored by measuring the OD600.
Since the mutants were originally isolated after chemical or UV mutagenesis, several mechanisms could be involved in the phenotypic expression of terbinafine resistance. Mutants A2M8 and H2 were crossed with corresponding wild-type yeast strains of the opposite mating type, the diploid cells were sporulated, the tetrads were dissected, and the spore cells were tested for terbinafine sensitivity on YPD agar plates containing 100 μg of terbinafine per ml. Regular 2:2 segregation of terbinafine resistance in both crosses (data not shown) indicated that a single nuclear gene locus is involved in the resistance phenotype of each of these mutants.
Mutations in the S. cerevisiae ERG1 gene are involved in terbinafine resistance.
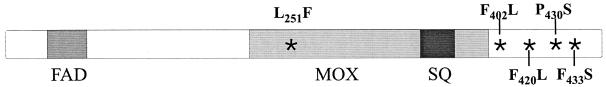
One possible mechanism leading to terbinafine resistance in S. cerevisiae is the expression of a mutated squalene epoxidase. Chromosomal DNA was isolated from the terbinafine-resistant mutants and served as the template to amplify the ERG1 gene by PCR. The DNA sequences of the PCR products were compared with the wild-type sequence of the ERG1 gene. The results showed that each of the erg1 alleles of the terbinafine-resistant mutants contained only 1 nucleotide exchange which led to single amino acid substitutions in the Erg1 protein (Fig. 2 and Table 2). Five of the 10 resistant mutants, i.e., H3, H4, H6, H7, and H8, carried an identical C-to-T transition in position 1288 of the open reading frame of the ERG1 gene, leading to the replacement of proline by serine at position 430 in Erg1p (P430S). Three of the mutants, i.e., A2M8, H1A, and H5, carried a C-to-T transition at position 751 which caused the change of amino acid 251 from leucine to phenylalanine (L251F). In addition, two other mutations resulted in the replacement of the phenylalanine residues by leucine at positions 402 (F402L in mutant H2) and 420 (F420L in mutant H1B). The high incidence of mutations leading to amino acid substitutions at positions 251 and 430 of the Erg1 protein points to an involvement of these amino acids in the interaction of squalene epoxidase with the inhibitor terbinafine. To gain a better insight into which region of the Erg1 protein is involved in terbinafine resistance, the ERG1 gene was randomly mutagenized by PCR. This approach led to the isolation of several terbinafine-resistant erg1 alleles, two of which carried single point mutations. One of them (ML3) carried the mutation leading to the same L251F substitution detected in A2M8, H1A, and H5, while another allele (NS2) carried a T-to-C transition at position 1298 of the open reading frame, resulting in an F433S exchange (Fig. 2 and Table 2).
FIG. 2.
Schematic presentation of the Erg1 protein of S. cerevisiae. The putative FAD, monooxygenase (MOX), and substrate binding (SQ) domains of squalene epoxidase are indicated together with the positions of the mutations identified in the terbinafine-resistant variants (*) and the respective amino acid exchanges.
TABLE 2.
Terbinafine-resistant mutants with single base pair exchanges leading to amino acid substitutions in the Erg1 protein
| Mutant | Base pair exchange in ERG1 gene | Amino acid substitution in Erg1 protein |
|---|---|---|
| A2M8, H1A, H5, ML3a | C751T | L251F |
| H2 | C1206A | F402L |
| H1B | C1260A | F420L |
| H3, H4, H6, H7, H8 | C1288T | P430S |
| NS2a | T1298C | F433S |
Alleles ML3 and NS2 for terbinafine resistance were isolated after in vitro PCR mutagenesis of the ERG1 gene.
Terbinafine resistance in the mutants is not linked to overexpression of ERG1 mRNA.
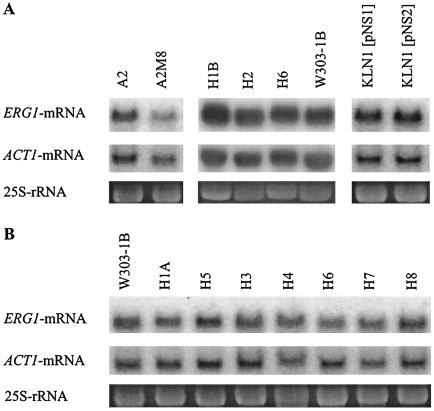
Since the mutations in the erg1 alleles may not be the only cause for the resistance phenotypes of the mutants and since the overexpression of target genes can lead to resistance to metabolic inhibitors (38), we determined the levels of ERG1 transcripts in the mutants by Northern analysis. Cultures were grown to early log phase in YPD medium, and total RNA was extracted and separated on agarose gels, as described in Materials and Methods. The RNA was probed with radiolabeled ERG1 DNA, and the amount of RNA loaded in each lane was monitored by use of ACT1 mRNA and 25S rRNA. The results are shown in Fig. 3. In terbinafine-resistant mutant A2M8, which was isolated after UV mutagenesis, the ERG1 mRNA level normalized to the amount of total RNA was approximately 20% lower than that in corresponding wild-type strain A2 (Fig. 3A). The ERG1 mRNA levels in mutants H1B, H2, and H6 also did not differ from that in parent strain W303-1B. Similarly, the NS2 allele for terbinafine resistance, which was isolated after PCR mutagenesis, was expressed at the same level as the wild-type NS1 allele in strain KLN1, in which erg1 was disrupted (Fig. 3A). In addition to the mutants shown in Fig. 3A, each of which had a different mutation in the ERG1 gene (Table 2), we determined the levels of expression of the erg1 alleles in the other mutants of W303-1B which had also been selected for terbinafine resistance after chemical mutagenesis (Fig. 3B). H1A and H5 carried the same mutation as mutant A2M8, while H3, H4, H6, H7, and H8 contained an identical mutation at position 1288 of the ERG1 gene (Table 2). The ERG1-specific mRNA levels in the mutants were quantitated and amounted to 72 to 101% of the ERG1 mRNA level of wild-type parent strain W303-1B. These data demonstrate that terbinafine resistance in our mutants is not caused by overexpression of the target gene.
FIG. 3.
Northern blot analysis of the ERG1 mRNA of terbinafine-resistant mutants. Cultures of terbinafine-resistant mutants, wild-type strains A2 and W303-1B, and strain KLN1 with an erg1 disruption with the recombinant plasmids pNS1 and pNS2 were grown in YPD medium to early log phase. Total RNA was extracted from either lysed spheroplasts (strains H1B, H2, H6, and W303-1B in panel A) or mechanically disrupted cells [strains A2, A2M8, KLN1(pNS1), KLN1(pNS2) in panel A and all strains in panel B], and 10 μg of the RNA was separated by agarose gel electrophoresis. Hybridization was performed as described in Materials and Methods by using an ERG1-specific DNA probe to detect the transcript from the chromosomal ERG1 locus and low-copy-number recombinant plasmids pNS1 and pNS2. The amounts of ACT1 mRNA and 25S rRNA were used to normalize the amount of RNA loaded on the gel.
Terbinafine-resistant erg1 mutants do not exhibit cross-resistance to other metabolic inhibitors.
The potential participation of drug efflux pumps in the terbinafine resistance phenotype was investigated by determining the susceptibilities of mutants H1A, H1B, H2, H5, and H6 to the metabolic inhibitors cycloheximide, oligomycin, ethidium bromide, 6-amino-2n-phenylthiobenzothiazole, crystal violet, resazurin, itraconazole, nystatin, 4-nitroquinolin-N-oxide, rhodamine 6G, and amorolfine as representatives of substrates for various drug efflux pumps, such as Pdr5p, Snq2p, Yor1p, and Sge1p or unknown pumps (12, 16; V. Klobučníková and I. Hapala, unpublished results). Serial dilutions of ONCs of the resistant mutants were spotted on agar plates containing different concentrations of these inhibitors, as described in Materials and Methods. The results indicate that the terbinafine-resistant mutants in general behaved like the wild-type strain in the presence of the inhibitors tested, with the possible exception of mutant H5, which was slightly more sensitive to cycloheximide, 6-amino-2n-phenylthiobenzothiazole, crystal violet, and rhodamine 6G (data not shown). However, mutant H1A, which carries the same mutation in the squalene epoxidase gene as H5, did not exhibit altered sensitivity to the inhibitors tested. Thus, we assume that the sensitivity profile of mutant H5 is not related to the mutation in the ERG1 gene. The lack of cross-resistance indicates that terbinafine resistance in mutants H1A, H1B, H2, and H7 is not linked to increased drug efflux.
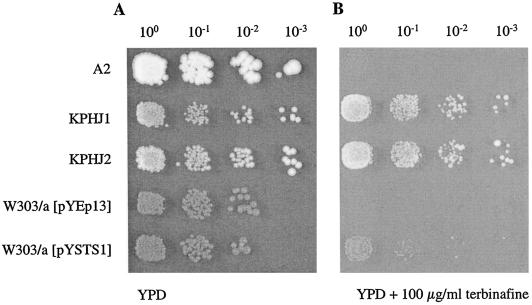
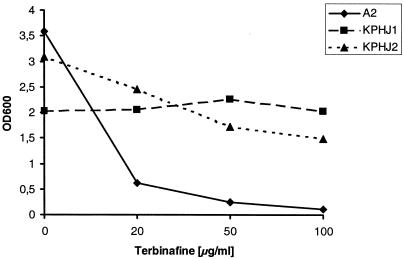
In order to investigate whether pleiotropic drug resistance can contribute to reduced terbinafine susceptibility, strains KPHJ1 and KPHJ2, which carry allelic forms of the genes PDR1 (PDR1-12) and PDR3 (PDR3-33), were tested for growth in the presence of terbinafine. The results in Fig. 4 show that these mutants grew on agar plates containing 100 μg of terbinafine per ml and are thus resistant to terbinafine, whereas wild-type strain A2 is sensitive. The susceptibilities of these mutants were also determined quantitatively in liquid medium, as described in Materials and Methods, and the results in Fig. 5 clearly demonstrate that both PDR mutants were resistant to terbinafine. The transcription factors Pdr1p and Pdr3p are key regulators in the pleiotropic drug resistance network of S. cerevisiae (1); and the PDR1-12 and PDR3-33 strains are known to show increased levels of expression of the efflux pump Pdr5p (47), indicating a contribution of Pdr5 in terbinafine resistance. Involvement of Pdr5p in terbinafine resistance was directly confirmed by overexpression of this efflux pump from recombinant plasmid pYSTS1 (2). S. cerevisiae W303/a with pYSTS1 could grow on agar plates containing 100 μg of terbinafine per ml, whereas the strain carrying the vector pYEp13 only remained sensitive (Fig. 4). In addition, deletion of the PDR5 gene in S. cerevisiae renders the strain significantly more sensitive to terbinafine than the isogenic wild-type parent (data not shown). Taken together, these results prove that terbinafine is a substrate for the efflux pump Pdr5 and that overexpression of Pdr5 leads to terbinafine resistance. Lack of cross-resistance to known substrates of Pdr5p, such as cycloheximide, itraconazole, 6-amino-2n-pentylthiobenzothiazole, and rhodamine 6G, therefore excludes the possibility that the pleiotropic drug resistance system is the cause of the terbinafine resistance phenotype in our series of mutants.
FIG. 4.
Resistance to terbinafine of mutants overexpressing Pdr5p. ONCs of wild-type strain S. cerevisiae A2 and two mutants containing allelic forms of PDR1 (PDR1-12 in KPHJ1) and PDR3 (PDR3-33 in KPHJ2) causing overexpression of Pdr5p were prepared at 30°C in YPD medium. Wild-type strain W303/a carrying the vector pYEp13 and wild-type strain W303/a carrying recombinant plasmid pYSTS1 (overexpressing Pdr5p) were grown in YNB minimal medium overnight at 30°C. The OD600s of the ONCs were adjusted to 0.1, 5 μl of the 100 to 10−3 dilutions was spotted on YPD agar plates without terbinafine (A) and with 100 μg of terbinafine per ml (B), and the strains were grown for 2 days at 30°C.
FIG. 5.
Growth of wild-type S. cerevisiae strain A2 and two mutants containing allelic forms of PDR1 (PDR1-12 in KPHJ1) and PDR3 (PDR3-33 in KPHJ2) causing overexpression of Pdr5p in the presence and absence of terbinafine. The yeast strains were inoculated into 2 ml of YPD medium to an OD600 of 0.005 and were grown at 30°C for 45 h. The medium was supplemented with 20, 50, or 100 μg of terbinafine per ml dissolved in ethanol. The control culture without terbinafine was supplemented with ethanol. Growth was monitored by measuring the OD600.
Replacement of the wild-type ERG1 gene by the mutated alleles renders the transformants terbinafine resistant.
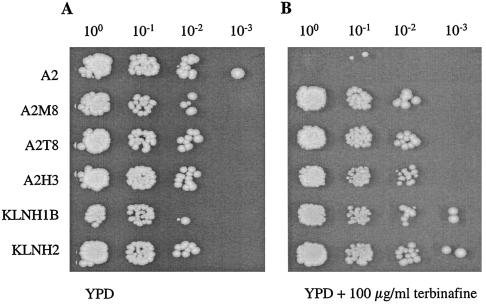
The terbinafine-resistant S. cerevisiae mutants contain single point mutations in the open reading frame of the ERG1 gene. In order to prove that each of the mutations is sufficient to confer terbinafine resistance, the wild-type ERG1 gene in S. cerevisiae A2 was replaced with the mutated erg1 alleles, isolated from A2M8 (leading to the L251F exchange) and H3 (leading to the P430S exchange), by homologous recombination, as described in Materials and Methods. The resulting transformants were designated A2T8 and A2H3, respectively. Correct gene replacement was verified by amplification of the erg1 alleles and DNA sequence determination. As shown in Fig. 6, chromosomal integration of the mutated erg1 alleles allows the transformants to grow in the presence of high terbinafine concentrations.
FIG. 6.
Effect of replacement of the wild-type ERG1 gene in A2 and the erg1::URA3 disruption in KLN1 with erg1 alleles of the resistant mutants on terbinafine susceptibility. The erg1 alleles for terbinafine resistance of mutants A2M8, H3, H1B, and H2 were integrated into the chromosome of wild-type strain A2 and strain KLN1 with the erg1 disruption at the ERG1 locus by homologous recombination. The resulting transformants (A2T8, A2H3, KLNH1B, and KLNH2, respectively) were grown overnight in YPD medium at 30°C, 5-μl aliquots of the ONCs were adjusted to an OD600 of 0.1, and serial dilutions (100 to 10−3) were spotted on YPD agar plates without terbinafine (A) and with 100 μg of terbinafine per ml (B). The plates were incubated for 2 days at 30°C.
In another approach, the mutated erg1 alleles of H1B (leading to the F420L exchange) and H2 (leading to the F402L exchange) were amplified by PCR, and the linear DNA fragments were transformed into strain KLN1 with the erg1::URA3 disruption. This strain lacks a functional squalene epoxidase and cannot grow under aerobic conditions, even in the presence of ergosterol (21). Replacement of the disrupted erg1 gene in KLN1 with the resistant erg1 alleles by homologous recombination resulted in the complementation of the lethal phenotype, and thus, transformants could be selected for growth under aerobic conditions. The correct gene replacement was verified by PCR amplification and DNA sequence determination of the mutated erg1 alleles, and the transformants were designated KLNH1B and KLNH2, respectively. As shown in Fig. 6, the mutations leading to the F420L and F402L exchanges in the Erg1 protein also confer high levels of terbinafine resistance to yeast cells, as demonstrated by growth of the transformants on agar plates containing 100 μg of terbinafine per ml.
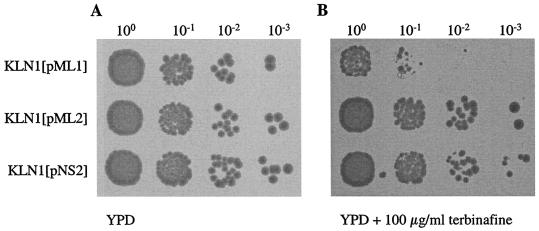
When S. cerevisiae KLN1 was transformed with pNS2, which carries the PCR-generated erg1 allele NS2 for terbinafine resistance on low-copy-number plasmid pRS315 without integration into the chromosome, the transformant became resistant to terbinafine (Fig. 7). Similar resistance was observed in KLN1 transformed with plasmid pML2 containing the erg1 allele of mutant A2M8, whereas KLN1 carrying pML1 with the wild-type ERG1 gene on the same plasmid remained sensitive. The growth of these strains was also monitored in liquid medium after incubation for 24 h at 30°C. KLN1(pNS2) and KLN1(pML2) continued to grow in the presence of up to 100 μg of terbinafine per ml, while the growth of KLN1(pML1) or KLN1(pNS1), each of which carried the wild-type ERG1 gene, was strongly inhibited in the presence of 20 μg of terbinafine per ml (data not shown). Taken together, the results of these experiments prove that single point mutations in the squalene epoxidase gene of S. cerevisiae, which result in an L251F, F402L, F420L, P430S, or F433S exchange, are sufficient to confer high-level resistance to terbinafine.
FIG. 7.
Effects of erg1 alleles expressed from low-copy-number plasmids on terbinafine resistance. S. cerevisiae KLN1 (erg1::URA3) was transformed with the low-copy-number plasmids pML1 carrying the wild-type ERG1 gene, pML2 carrying the erg1 allele for terbinafine resistance of A2M8, and pNS2 carrying erg1 allele NS2 for terbinafine resistance on pRS315. The strains were grown overnight in YPD medium, 5-μl aliquots of the ONCs were diluted to an OD600 of 0.1, and serial dilutions (100 to 10−3) were spotted on YPD plates without terbinafine (A) and with 100 μg of terbinafine per ml (B) and incubated for 2 days at 30°C.
DISCUSSION
Despite the extensive use of terbinafine for the treatment of infections caused by dermatophytes and other fungal pathogens (10, 33, 50), there are no reports in the literature on the isolation of terbinafine-resistant variants directly related to application of the drug. This is in striking contrast to the increasing incidence of clinical isolates of fungal pathogens resistant to azoles, another group of commonly used antimycotics (25, 27, 28, 40). The only terbinafine-resistant clinical isolate to have been recovered was recently reported by Mukherjee et al. (29). That Trichophyton rubrum strain, which was isolated from a patient before and during therapy with terbinafine, was shown to exhibit primary resistance to the drug. The clinical isolate was susceptible to azoles and griseofulvin but was fully cross-resistant to other known squalene epoxidase inhibitors, suggesting a target-specific resistance mechanism which, however, has not been elucidated at the molecular level (29). Terbinafine-resistant clones of the phytopathogenic fungus Nectria haematococca have been isolated after UV mutagenesis by Lasseron-De Falandre et al. (22). Although the pattern of sterols in these mutants indicated altered squalene epoxidase activity, the nature of these alterations has not been analyzed in detail, but a reduced affinity of the squalene epoxidase for terbinafine has been proposed. We have chosen S. cerevisiae as a model organism to investigate the mechanisms by which yeast can develop resistance to terbinafine, in particular, whether modifications in the target enzyme, squalene epoxidase, can alter the sensitivity to the inhibitory effect of terbinafine. The results presented in this report were obtained with a set of 10 terbinafine-resistant mutants isolated after chemical or UV mutagenesis and two mutated erg1 alleles generated by PCR mutagenesis. To our knowledge, this report provides the first detailed analyses of terbinafine resistance at the molecular level.
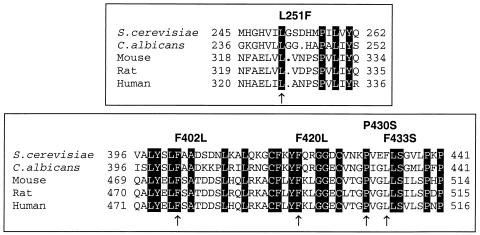
Single point mutations in the ERG1 gene from the terbinafine-resistant mutants leading to amino acid exchanges in the Erg1 protein were identified, with each of them being sufficient to confer terbinafine resistance, as shown by gene replacement experiments and expression of the mutated erg1 allele from a low-copy-number plasmid. The nucleotide exchanges were localized at five different positions of the ERG1 gene. Five of 10 mutants contained a single mutation at position 1288 of the open reading frame, leading to a P430S substitution in the C-terminal portion of the Erg1 protein. Three mutants carried erg1 alleles with a single point mutation at position 751 of the gene, resulting in an L251F substitution in the squalene epoxidase. Two other mutations were found in individual mutants, and they led to F402L and F420L amino acid exchanges, respectively. Investigation of alleles for terbinafine resistance isolated after in vitro mutagenesis of the ERG1 gene led to the identification of the already characterized L251F exchange and a new amino acid substitution, F433S, in the Erg1 protein. There are no structural data for the S. cerevisiae squalene epoxidase that would enable us to identify the amino acids participating in the interaction of squalene epoxidase with terbinafine. Since we have characterized five amino acid substitutions in the Erg1 protein that result in terbinafine-resistant variants of squalene epoxidase, we can speculate that all of them are involved in the enzyme-inhibitor interaction. The amino acid exchanges are clustered in two regions of the Erg1 protein; one is the L251F substitution, and the other exchanges are localized in the C-terminal portion of Erg1p. The overall amino acid sequence homologies between the Erg1 protein of S. cerevisiae and C. albicans, rat, mouse, and human squalene epoxidases are between 71 and 32% (6, 17, 18, 36). Alignment of the amino acid sequences of squalene epoxidases reveals that some regions are highly conserved from fungi to mammals. A putative FAD binding domain (FAD) is proposed to include amino acid residues 21 to 48 of Erg1p of S. cerevisiae (6), in which the conserved motif GXGXXG is present in all squalene epoxidases. A profile scan of the SwissProt database proposes a large monooxygenase domain that includes amino acid residues 201 to 396 in the Erg1 protein of S. cerevisiae. Within this domain, a fully conserved region of 11 identical amino acids could serve as a potential binding site for the substrate squalene (6). None of the amino acid exchanges detected in the Erg1 protein of terbinafine-resistant mutants are localized in the putative domains involved in FAD or substrate binding. The mutation leading to the L251F exchange was found in four mutant alleles and is located within the putative monooxygenase domain. Since this mutation could alter the enzymatic activity of the Erg1 protein, we assayed cell extracts of wild-type strain A2 and mutant A2M8 for squalene epoxidase activity. No difference in the specific activity of the mutant protein has been found; the in vitro susceptibility to terbinafine, however, was significantly reduced compared to that of the wild-type protein, which corresponds to the resistance phenotype of the mutant (13; E. Pitters and F. Turnowsky, unpublished results). The high incidence of mutations in the C-terminal region of the squalene epoxidase leading to terbinafine resistance indicates that this portion of the protein, and in particular, the proline residue at position 430 of Erg1p, may be crucial in the enzyme-inhibitor interaction.
Terbinafine inhibits the enzymatic activity of fungal squalene epoxidases at much lower concentrations than it inhibits the activity of the mammalian enzyme (5, 35). It is therefore interesting that all but one (F433S) of the amino acid exchanges identified to confer resistance to terbinafine in S. cerevisiae involve amino acid residues which are conserved in all squalene epoxidases (Fig. 8). At present we have no reasonable explanation for this surprising result. In the squalene epoxidases of S. cerevisiae and C. albicans, but not in the mammalian enzymes, most of the amino acid residues surrounding the exchanges in the resistant Erg1 proteins are identical, suggesting that the highly homologous regions of the two fungal squalene epoxidases and not so much single amino acids are responsible for the selectivity of terbinafine inhibition. Similar observations were reported for azole resistance in C. albicans caused by mutations in lanosterol 14α-demethylase. Despite the high selectivity of azole inhibition, many amino acid substitutions affected positions that are conserved in fungal and mammalian demethylases (7, 15, 19, 20, 39). In most reported cases the amino acid exchanges led to a decreased affinity of the enzyme for azoles and a combination of mutations showed additive effects on the resistance level. In the case of squalene epoxidase, however, only single mutations were identified in the terbinafine-resistant mutants, and each of the mutations was sufficient to confer high-level resistance to the inhibitor.
FIG. 8.
Multiple-sequence alignment of S. cerevisiae Erg1 protein regions carrying the mutations with the corresponding regions of C. albicans, rat, mouse, and human squalene epoxidases. The amino acids conserved in all squalene epoxidases are shaded. Mutations leading to terbinafine resistance are indicated by arrows, and the corresponding amino acid exchanges are shown above the sequence.
The majority of reports on antifungal drug resistance deal with the pathogenic yeast C. albicans, but many resistance mechanisms will be applicable to other fungi as well (38, 48). Since multiple mechanisms can lead to drug resistance and often contribute simultaneously to the development of high-level resistance after long-term treatment with antimycotics (8, 46), we had to consider the potential involvement of additional mechanisms in the terbinafine resistance phenotypes of our mutants. The intracellular concentrations of drugs and the sensitivities of the targets determine the effectiveness of antifungal drugs. The much higher susceptibility of T. rubrum to terbinafine compared to that of S. cerevisiae can be explained by both the higher sensitivity of the squalene epoxidase and the possible accumulation of terbinafine in T. rubrum cells (5). On the other hand, reduced intracellular concentrations of inhibitors mediated by either reduced uptake into the cells or increased efflux out of the cells are clinically relevant causes of intrinsic or acquired resistance to antimycotics. Enhanced expression of drug efflux pumps belonging to the ABC transporter superfamily or the major facilitator superfamily is therefore a very important mechanism of resistance to antifungal drugs (27, 40, 42). Although the number of terbinafine-resistant mutants isolated during our studies is far from representative, it is nevertheless interesting that we have not identified clones with phenotypes linked to alterations in the pleiotropic drug resistance system. Many C. albicans isolates resistant to azoles were shown to overexpress the ABC transporters Cdr1p and Cdr2p, which are the Candida homologues of the ABC transporter Pdr5p of S. cerevisiae (42). These strains frequently show cross-resistance to terbinafine and other metabolic inhibitors (28, 41). Our results demonstrate the association of increased levels of the efflux pump Pdr5p with terbinafine resistance in S. cerevisiae mutants carrying mutations in the transcription factors Pdr1p and Pdr3p, both of which lead to the overexpression of Pdr5 (47). These mutants can grow in the presence of approximately 10 times higher concentrations of terbinafine than wild-type yeast cells. Similar levels of resistance to a number of other inhibitors have been reported for these mutants by Wendler et al. (47). Although we could show that overexpression of Pdr5 leads to terbinafine resistance, the involvement of efflux pumps in our set of mutants can be ruled out since they did not exhibit cross-resistance to other antifungal drugs.
The low incidence of terbinafine-resistant mutants even after chemical or UV mutagenesis could resemble the clinical situation, in which no fungal pathogen has been reported to develop terbinafine resistance during treatment. The T. rubrum strain that has been isolated from a patient who failed terbinafine treatment was later identified to be a strain with primary resistance that was present even before the onset of terbinafine therapy (29). Other failures of therapy for fungal infections with terbinafine could have been caused by the resistance of the pathogens to terbinafine; however, this has not been checked routinely (29). Our results with S. cerevisiae show that the potential for the development of terbinafine resistance mediated by changes in the target enzyme squalene epoxidases does exist. The clustering of these changes in specific regions of the Erg1 protein indicates their possible involvement in a specific interaction between the target enzyme and terbinafine. Mutant variants of the squalene epoxidase may thus help to elucidate the structure of the enzyme and to develop novel inhibitors for this essential enzyme in the ergosterol biosynthesis pathway.
Acknowledgments
We are grateful to H. Bergler for critical reading of the manuscript and many helpful discussions and comments. We thank A. Stütz, Novartis Forschungsinstitut Vienna, Austria, for terbinafine; T. Kuchta, Food Research Institute, Bratislava, Slovak Republic, for itraconazole, amorolfine, and 6-amino-2n-pentylthiobenzothiazole; and H. Jungwirth, Graz, Austria, K. Kuchler, Vienna, Austria, A. Hartig, Vienna, Austria, and S. D. Kohlwein, Graz, Austria, for supplying strains and plasmids. The construction of strains by M. Heiser and N. Scheer is gratefully acknowledged.
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich, project P14415 (to F.T.) and projects VEGA-2/1016/21 and APVT-20-016502 (to I.H.), and the Aktion Österreich-Slowakei, project 39s9 (to F.T. and I.H.).
REFERENCES
- 1.Bauer, B. E., H. Wolfger, and K. Kuchler. 1999. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta 1461:217-236. [DOI] [PubMed] [Google Scholar]
- 2.Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 269:4180-4186. [PubMed] [Google Scholar]
- 3.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 4.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favre, B., and N. S. Ryder. 1996. Characterization of squalene epoxidase activity from the dermatophyte Trichophyton rubrum and its inhibition by terbinafine and other antimycotic agents. Antimicrob. Agents Chemother. 40:443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favre, B., and N. S. Ryder. 1997. Cloning and expression of squalene epoxidase from the pathogenic yeast Candida albicans. Gene 189:119-126. [DOI] [PubMed] [Google Scholar]
- 7.Favre, B., M. Didmon, and N. S. Ryder. 1999. Multiple amino acid substitutions in lanosterol 14α-demethylase contribute to azole resistance in Candida albicans. Microbiology 145:2715-2725. [DOI] [PubMed] [Google Scholar]
- 8.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz, R. D., R. H. Schiestl, A. R. Willems, and A. R. Woods. 1995. Studies on the transformation on intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 10.Hay, R. J. 1999. Therapeutic potential of terbinafine in subcutaneous and systemic mycoses. Br. J. Dermatol. 141(Suppl. 56):36-40. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch, P. J., and S. A. Henry. 1986. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 6:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquot, C., R. Julien, and M. Guilloton. 1997. The Saccharomyces cerevisiae MFS superfamily SGE1 gene confers resistance to cationic dyes. Yeast 13:891-902. [DOI] [PubMed] [Google Scholar]
- 13.Jandrositz, A., F. Turnowsky, and G. Högenauer. 1991. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene 107:155-160. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, S. L., A. Arnoldi, and D. E. Kelly. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, S. L., D. C. Lamb, J. Loeffler, H. Einsele, and D. E. Kelly. 1999. The G464S amino acid substitution in Candida albicans sterol 14α-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174-179. [DOI] [PubMed] [Google Scholar]
- 16.Kolaczkowski, M., A. Kolaczkowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 17.Kosuga, K., S. Hata, T. Osumi, J. Sakakibara, and T. Ono. 1995. Nucleotide sequence of a cDNA for mouse squalene epoxidase. Biochim. Biophys. Acta 1260:345-348. [DOI] [PubMed] [Google Scholar]
- 18.Laden, B. P., Y. Tang, and T. D. Porter. 2000. Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch. Biochem. Biophys. 374:381-388. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, D. C., D. E. Kelly, T. C. White, and S. L. Kelly. 2000. The R467K amino acid substitution in Candida albicans sterol 14α-demethylase causes drug resistance through reduced affinity. Antimicrob. Agents Chemother. 44:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb, D. C., D. E. Kelly, W.-H. Schunck, A. Z. Shyadehi, M. Akhtar, D. J. Lowe, B. C. Baldwin, and S. L. Kelly. 1997. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682-5688. [DOI] [PubMed] [Google Scholar]
- 21.Landl, K., B. Klösch, and F. Turnowsky. 1996. ERG1, encoding squalene epoxidase, is located on the right arm of chromosome VII of Saccharomyces cerevisiae. Yeast 12:609-613. [DOI] [PubMed] [Google Scholar]
- 22.Lasseron-De Falandre, A., D. Debieu, J. Bach, C. Malosse, and P. Leroux. 1999. Mechanisms of resistance to fenpropimorph and terbinafine, two sterol biosynthesis inhibitors, in Nectria haematococca, a phytopathogenic fungus. Pestic. Biochem. Physiol. 64:167-184. [Google Scholar]
- 23.Lawrence, C. W. 1991. Classical mutagenesis techniques. Methods Enzymol. 194:273-281. [DOI] [PubMed] [Google Scholar]
- 24.Leber, R., R. Zenz, K. Schröttner, S. Fuchsbichler, B. Pühringer, and F. Turnowsky. 2001. A novel sequence element is involved in the transcriptional regulation of expression of the ERG1 (squalene epoxidase) gene in Saccharomyces cerevisiae. Eur. J. Biochem. 268:914-924. [DOI] [PubMed] [Google Scholar]
- 25.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 26.Marichal, P., H. Vanden Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marr, K. A., C. N. Lyons, T. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morschhäuser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee, P. K., S. D. Leidich, N. Isham, I. Leitner, N. S. Ryder, and M. A. Ghannoum. 2003. Clinical Trichophyton rubrum strain exhibiting primary resistance to terbinafine. Antimicrob. Agents Chemother. 47:82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura, K., M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau, and R. D. Cannon. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 45:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paltauf, F., G. Daum, G. Zuder, G. Högenauer, G. Schulz, and G. Seidl. 1982. Squalene and ergosterol biosynthesis in fungi treated with naftifine, a new antimycotic agent. Biochim. Biophys. Acta 712:268-273. [Google Scholar]
- 32.Perea, S., J. L. López-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillán, M. Martínez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez, A. 1999. Terbinafine: broad new spectrum of indications in several subcutaneous and systemic and parasitic diseases. Mycoses 42(Suppl. 2):111-114. [PubMed] [Google Scholar]
- 34.Ryder, N. S. 1992. Terbinafine: mode of action and properties of the squalene epoxidase inhibiton. Br. J. Dermatol. 126(Suppl. 39):2-7. [DOI] [PubMed] [Google Scholar]
- 35.Ryder, N. S., and M. C. Dupont. 1985. Inhibition of squalene epoxidase by allylamine antimycotic compounds. A comparative study of the fungal and mammalian enzymes. Biochem. J. 230:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakakibara, J., R. Watanabe, Y. Kanai, and T. Ono. 1995. Molecular cloning and expression of rat squalene epoxidase. J. Biol. Chem. 270:17-20. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sanglard, D. 2002. Clinical relevance of mechanisms of antifungal drug resistance in yeasts. Enferm. Infecc. Microbiol. Clin. 20:462-470. [DOI] [PubMed] [Google Scholar]
- 39.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 42.Sanglard, D., K. Kuchler, F. Ischer, J.-L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satoh, T., M. Horie, H. Watanabe, Y. Tsuchiya, and T. Kamei. 1993. Enzymatic properties of squalene epoxidase from Saccharomyces cerevisiae. Biol. Pharm. Bull. 16:349-352. [DOI] [PubMed] [Google Scholar]
- 44.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 45.Strathern, J. N., and D. R. Higgins. 1991. Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol. 194:319-329. [DOI] [PubMed] [Google Scholar]
- 46.Vanden Bossche, H., F. Dromer, I. Improvisi, M. Lozano-Chiu, J. H. Rex, and D. Sanglard. 1998. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 36(Suppl. 1):119-128. [PubMed] [Google Scholar]
- 47.Wendler, F., H. Bergler, K. Prutej, G. Zisser, K. Kuchler, and G. Högenauer. 1997. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 272:27091-27098. [DOI] [PubMed] [Google Scholar]
- 48.White, C. W., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirsching, S., G. P. Moran, D. J. Sullivan, D. C. Coleman, and J. Morschhäuser. 2001. MDR1-mediated drug resistance in Candida dubliensis. Antimicrob. Agents Chemother. 45:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaias, N. 1997. Candida: a review of clinical experience with Lamisil. Dermatology 194(Suppl. 1):10-13. [DOI] [PubMed] [Google Scholar]