Abstract
The elements conferring high-level gentamicin resistance in 64 clinical isolates of Enterococcus faecalis were characterized by PCR and by restriction enzyme hybridization analysis of genomic and plasmid DNA. There was a strong association between gentamicin resistance and the aac(6′)-aph(2") gene carried on IS256-based elements with different structures, locations, and transfer characteristics.
Enterococci exhibiting high-level gentamicin resistance (HLGR) have been reported widely as a cause of nosocomial infections in Europe, the United States, and other geographic locations (9, 11, 14). HLGR in enterococci is mediated by the aac(6′)-aph(2") gene, which encodes a bifunctional enzyme specifying both 6′ aminoglycoside acetyltransferase and 2" aminoglycoside phosphotransferase activities (5). This bifunctional enzyme gene mediates high-level resistance to virtually all commercially available aminoglycosides, except streptomycin, resulting in elimination of synergism between cell wall-active agents and aminoglycoside antibiotics (11, 14).
The aac(6′)-aph(2") gene is plasmid borne in most gentamicin-resistant isolates of Enterococcus faecalis and is located on elements related to the staphylococcal transposon Tn4001 (7). Transposon Tn4001 is composed of a central region containing the aac(6′)-aph(2") gene, flanked on each side by inverted copies of insertion sequence IS256 (1, 2, 10, 18). Several variants of transposon Tn4001 have been identified in enterococci from diverse geographic locations; the transposon Tn5281, in which there is a tandem duplication of IS256 at one terminus of Tn4001 (7), the Tn4001-IS257 hybrid (7), and the Tn4001-truncated elements (3, 8, 19). Moreover, new transposable elements involved in the dissemination of gentamicin resistance have been described (15, 16, 21).
The aim of the present study was to characterize the elements carrying the gentamicin resistance determinants involved in the dissemination of HLGR among E. faecalis isolates in Greece.
The bacterial isolates used in the present study were randomly selected from a collection of isolates that had been recovered from clinical specimens of individual patients at three different hospitals located in the Athens metropolitan area from 1996 to 2000. E. faecalis SF339 and SF350, which are highly resistant to gentamicin, were used as control strains (22). Susceptibilities to commonly used antimicrobial agents were determined by the broth microdilution method (13). High-level resistance to gentamicin, tobramycin, netilmicin, amikacin, and streptomycin was confirmed by measuring bacterial growth on brain heart infusion (BHI) agar containing 2,000 μg of each aminoglycoside/ml.
Transferability of resistance determinants was examined by using filter matings as described previously (6). E. faecalis FA2-2 (rifampin and fusidic acid resistant) was used as the susceptible recipient strain (4). Incubation was performed at 37°C for 6 to 8 h and for 24 h. The transfer frequencies were expressed as the number of transconjugants per recipient cell present at the time of plating on selective media.
The relatedness among the strains was examined by pulsed-field gel electrophoresis of the SmaI digest of genomic cell DNA as previously described (12). Gels were processed by using clamped homogeneous electric fields (CHEF-DRIII; Bio-Rad), stained in ethidium bromide, and photographed. Previously published guidelines were used for the interpretation of pulsed-field electrophoresis patterns (20).
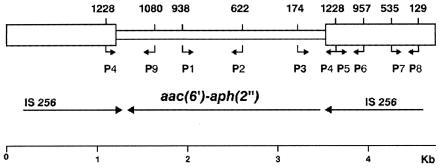
Bacterial cells were lysed, and DNA was extracted (23). Five microliters of DNA suspension was used as the template in 45-μl PCR mixtures. Five pairs of synthetic oligonucleotide primers were used to amplify five regions of transposon Tn4001 (Fig. 1): the first from the central region of the aac(6′)-aph(2") gene (primers P1 and P2), the second from the internal region of IS256 (primers P5 and P6), the third from the external region of IS256 (primers P7 and P8), the fourth from the right aac (6′)-aph (2")-IS256 junction region (primers P3 and P4), and the fifth from the left aac (6′)-aph (2")-IS256 junction region (primers P4 and P9). The sequences of oligonucleotide primers were derived from the known sequences of aac(6′)-aph(2") and IS256 (2, 5). A reaction master mixture containing 1 U of Taq polymerase, Taq polymerase buffer, 1.5 mM MgCl2, 200 μM (each) nucleotide, and 100 pmol of each primer per reaction (Roche Diagnostic Systems, Inc., Branchburg, N.J.) was prepared. The samples were initially denatured at 94°C for 3 min, and 30 amplification cycles were completed. Each cycle consisted of denaturation at 94°C for 30 s, annealing at 62°C for 45 s, and extension at 72°C for 30 s. All amplifications were performed on a Perkin-Elmer Cetus model 9600 DNA thermocycler. The PCR products were electrophoresed on 2% agarose gel, stained with ethidium bromide, and photographed.
FIG. 1.
Schematic representation of transposon Tn4001, composed of the aac(6′)-aph(2") gene (central box) and of two inverted copies of IS256 (side boxes). Arrows indicate orientation. P1 to P9, oligonucleotide primers used in the study (numbers indicate positions; bent arrows indicate transcriptional direction).
Plasmid and chromosomal DNAs were separated with a cesium chloride density gradient (4). Restriction enzyme analysis was performed with HindIII according to the manufacturer's recommendations. Samples were run on a 0.7% agarose gel, stained with ethidium bromide, and visualized under UV light. For localization of the gentamicin resistance determinant, hybridization experiments were carried out under stringent conditions. The probe used, pSF815A, contained the AluI fragment of the aac(6′)-aph(2") gene (5). The probe was biotin labeled by nick translation (Bethesda Research Laboratories). Hybridization and visualization were done with BlueGene (Bethesda Research Laboratories).
Eighty-four clinical isolates of E. faecalis were randomly selected for the study, 64 isolates for which gentamicin MICs were >128 μg/ml and 20 isolates for which gentamicin MICs were ≤128 μg/ml. All 64 isolates for which gentamicin MICs were >128 μg/ml exhibited high-level resistance to gentamicin, tobramycin, netilmicin, and amikacin (MIC > 2,000 μg/ml), and 62 isolates were also highly resistant to streptomycin (2 of 64 isolates with high-level gentamicin resistance were susceptible to streptomycin). Of 64 gentamicin-resistant isolates, 14 were beta-hemolytic on horse blood agar. Analysis of the SmaI restriction digests of genomic DNA of gentamicin-resistant isolates resulted in 42 different pulsed-field gel electrophoresis patterns; 31 of the isolates fell into nine groups (groups A to I) as follows: A, 6; B, 3; C, 5; D, 2; E, 3; F, 2; G, 3; H, 3; I, 4. The remaining 33 isolates had distinct patterns not related to each other and to any of the groups.
All 64 gentamicin-resistant wild-type isolates generated a 317-bp PCR product corresponding to the central region of the aac(6′)-aph(2") gene and two PCR products, 272 and 407 bp, corresponding to the internal and external regions of IS256, respectively. By amplifying the aac(6′)-aph(2")-IS256 left and right junction regions, it was found that all 64 gentamicin-resistant isolates yielded a 559-bp amplicon corresponding to the left junction region whereas only 34 of 64 isolates yielded a 691-bp amplicon corresponding to the right junction region. None of the gentamicin-susceptible isolates yielded any PCR product corresponding to the bifunctional enzyme gene, whereas five generated amplicons corresponding to IS256. The last finding is in agreement with previous reports, where the presence of IS256 was not always related to gentamicin resistance in enterococci (17). No structure of the IS256-based elements carrying the aac(6′)-aph(2") gene corresponded to a particular pulsed-field gel electrophoresis strain type.
Gentamicin resistance was transferable into E. faecalis FA2-2 for 52 of 64 isolates after a 6-h mating. No additional isolates transferred aminoglycoside resistance into the recipient strain after prolonged mating of up to 24 h. Sequences from the aac(6′)-aph(2") gene and IS256 were detected by PCR in 52 of 52 transconjugants. Transfer frequencies ranged from 10−2 to 10−8 transconjugants per recipient cell; 10−2 to 10−4 transconjugants per recipient cell for 30 isolates, <10−4 to 10−6 transconjugants per recipient cell for 10 isolates, and 10−6 to 10−8 transconjugants per recipient cell for 12 isolates.
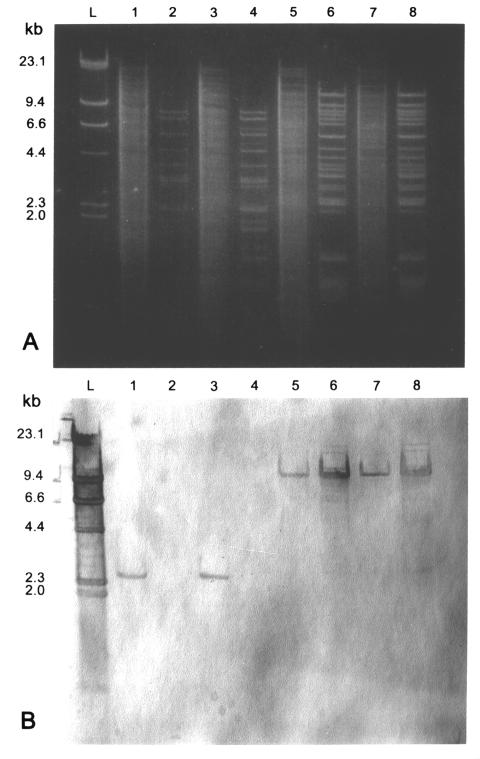
To determine the location of the elements conferring HLGR, 17 isolates were examined further by restriction enzyme and hybridization analysis of plasmid and genomic DNA preparations (Fig. 2; Table 1). Three groups of isolates were randomly selected according to frequency of transfer of gentamicin resistance. In the first group (5 isolates), the gentamicin resistance was not transferable at a detectable level (frequency < 10−9 transconjugants per recipient cell); in the second group (2 isolates), the transfer frequency was intermediate between 10−4 and 10−9 transconjugants per recipient cell; in the third group (10 isolates), the transfer frequency was >10−4 transconjugants per recipient cell. In the first group of isolates the aac(6′)-aph(2") gene probe hybridized to a 2.4-kb HindIII fragment located on chromosomal DNA in four isolates and on plasmid DNA in one. In the second group, the aac(6′)-aph(2") gene probe hybridized to a 9.4- or 6.6-kb HindIII fragment located on plasmid DNA and to a 2.4-kb HindIII fragment located on chromosomal DNA. In the third group of isolates, the probe hybridized to a 9.4-kb HindIII fragment located on plasmid DNA.
FIG. 2.
(A) Agarose gel electrophoresis (0.7% agarose) of HindIII digests of genomic and plasmid DNAs from HLGR E. faecalis isolates. (B) Southern blot of gel shown in panel A probed with the biotin-labeled aac(6′)-aph(2") gene. Lanes: L, biotin-labeled HindIII-digested lambda phage DNA; 1 and 2, genomic and plasmid DNA from isolate 140 [aac(6′)-aph(2") gene on the chromosome]; 3 and 4, genomic and plasmid DNA from isolate 136 [aac(6′)-aph(2") gene on the chromosome]; 5 and 6, genomic and plasmid DNA from isolate 35 [aac(6′)-aph(2") gene on a plasmid]; 7 and 8, genomic and plasmid DNA from isolate 111 [aac(6′)-aph(2") gene on a plasmid].
TABLE 1.
Structures, locations, and transfer frequencies of the genetic elements conferring high-level gentamicin resistance in E. faecalis
| Isolate no./groupa | Size of amplicon of aac-aph-IS256 junction region (bp)
|
Size(s) of HindIII digest(s) hybridized with aac-aph gene probe (kb) | Locationb | Transfer frequency (transconjugants/recipient cell) | |
|---|---|---|---|---|---|
| Left | Right | ||||
| 140/G | 559 | 691 | 2.4 | c | <10−9 |
| 136/G | 559 | 691 | 2.4 | c | <10−9 |
| 203/C | 559 | 691 | 2.4 | c | <10−9 |
| 206/NG | 559 | 691 | 2.4 | c | <10−9 |
| 108/H | 559 | 691 | 2.4 | p | <10−9 |
| 11/C | 559 | 691 | 9.4, 2.4c | p, c | 3 × 10−7 |
| 324/NG | 559 | 691 | 6.6, 2.4c | p, c | 5 × 10−8 |
| 30/A | 559 | NAd | 9.4 | p | 1 × 10−2 |
| 35/A | 559 | NA | 9.4 | p | 6 × 10−3 |
| 110/A | 559 | NA | 9.4 | p | 2 × 10−3 |
| 111/A | 559 | NA | 9.4 | p | 2 × 10−3 |
| 101/B | 559 | NA | 9.4 | p | 1 × 10−2 |
| 102/B | 559 | NA | 9.4 | p | 1 × 10−2 |
| 275/B | 559 | NA | 9.4 | p | 2 × 10−3 |
| 268/NG | 559 | NA | 9.4 | p | 1 × 10−3 |
| 297/NG | 559 | NA | 9.4 | p | 1 × 10−3 |
| 345/NG | 559 | NA | 9.4 | p | 3 × 10−3 |
NG, nongroupable.
c, chromosome; p, plasmid.
Sizes apply to plasmid and chromosome DNA, respectively.
NA, not amplified.
The findings presented herein indicate that HLGR in our isolates was strongly associated with the aac(6′)-aph(2") bifunctional enzyme gene, which was carried on IS256-based elements with different structures, locations, and transfer characteristics.
The PCR experiments strongly suggest that some of these elements are similar or identical to transposon Tn4001 but that others are truncated, lacking the insertion sequence IS256 on the right. The structural differences noted by PCR mapping of Tn4001 paralleled the findings of restriction enzyme and hybridization analysis. Indeed, in all isolates which generated amplicons of the expected sizes from both left and right junction regions, the aac(6′)-aph(2") gene probe hybridized to a 2.4-kb HindIII fragment, equivalent in size to the corresponding HindIII fragment of Tn4001. In the isolates which yielded no amplicon corresponding to the right junction region, the aac(6′)-aph(2") gene probe hybridized to a 6.6- or 9.4-kb HindIII fragment, indicating that the bifunctional enzyme gene resides on variant forms of transposon Tn4001. It is possible that these variant forms have been generated by molecular rearrangements during the process of transposition from the chromosome to a plasmid or during in vivo conjugative transfer.
Interestingly, the frequencies of transfer of gentamicin resistance for the isolates containing truncated variants of Tn4001 differed significantly from those for isolates containing nontruncated variants; 24 of 30 isolates containing truncated variants transferred gentamicin resistance with frequencies higher than 10−4 transconjugants per recipient cell, whereas only 3 of 34 isolates containing nontruncated variants transferred gentamicin resistance with frequencies higher than 10−4 transconjugants per recipient cell (P < 0.001).
In agreement with a prior report (3), the truncated variants of Tn4001 were located only on plasmids, whereas the nontruncated elements were carried on the chromosome in six isolates and on a plasmid in one (isolate 108). The location and structure relationship of Tn4001 were demonstrated better in isolates 11 and 324, which contained two copies of the elements: one copy on a plasmid, in a truncated form, and another copy on the chromosome, in a nontruncated form. Interestingly, conjugative transfer of gentamicin resistance was not possible when the aac(6′)-aph(2") gene was carried on nontruncated forms, regardless of their location on a plasmid or on the chromosome.
In the present study it was not elucidated whether the structure and location of Tn4001-like elements had a cause-and-effect relationship with element transfer frequencies. Our findings, however, indicate that the nontruncated elements are more stable, whereas the truncated variants are mobile and transfer gentamicin resistance more efficiently.
REFERENCES
- 1.Byrne, M. E., M. T. Gillespie, and R. A. Skurray. 1990. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob. Agents Chemother. 34:2106-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 3.Casetta, A., A. B. Hoi, G. D. Cespedes, and T. Horaud. 1998. Diversity of structures carrying the high-level gentamicin resistance gene (aac6-aph2) in Enterococcus faecalis strains isolated in France. Antimicrob. Agents Chemother. 42:2889-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clewell, D. B., P. K. Tomich, C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotranferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes, B. A., and D. R. Schaberg. 1983. Transfer of resistance plasmids from Staphylococcus epidermidis to Staphylococcus aureus: evidence for conjugative exchange of resistance. J. Bacteriol. 153:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodel-Christian, S. L., and B. E. Murray. 1991. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob. Agents Chemother. 35:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodel-Christian, S. L., and B. E. Murray. 1992. Comparison of the gentamicin resistance transposon Tn5281 with regions encoding gentamicin resistance in Enterococcus faecalis isolates from diverse geographic locations. Antimicrob. Agents Chemother. 36:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann, S. A., and R. C. Moellering. 1987. The Enterococcus: “putting the bug in our ears.” Ann. Intern. Med. 106:757-761. [DOI] [PubMed] [Google Scholar]
- 10.Lyon, B. R., M. T. Gillespie, M. E. Byrne, J. W. May, and R. A. Skurray. 1987. Plasmid-mediated resistance to gentamicin in Staphylococcus aureus: the involvement of a transposon. J. Med. Microbiol. 23:101-110. [DOI] [PubMed] [Google Scholar]
- 11.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1999. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Patterson, J. E., and M. J. Zervos. 1990. High-level gentamicin resistance in Enterococcus: microbiology, genetic basis, and epidemiology. Rev. Infect. Dis. 12:644-652. [DOI] [PubMed] [Google Scholar]
- 15.Rice L. B., and L. L. Carias. 1998. Transfer of Tn5385, a composite, multiresistance chromosomal element from Enterococcus faecalis. J. Bacteriol. 180:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice, L. B., L. L. Carias, and S. H. Marshall. 1995. Tn5384, a composite enterococcal mobile element conferring resistance to erythromycin and gentamicin whose ends are directly repeated copies of IS256. Antimicrob. Agents Chemother. 39:1147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice, L. B., and A. S. Thorisdottir. 1994. The prevalence of sequences homologous to IS256 in clinical enterococcal isolates. Plasmid 32:344-349. [DOI] [PubMed] [Google Scholar]
- 18.Rouch, D. A., M. E. Byrne, Y. C. Kong, and R. A. Skurray. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J. Gen. Microbiol. 133:3039-3052. [DOI] [PubMed] [Google Scholar]
- 19.Straut, M., G. de Cespedes, F. Delbos, and T. Horaud. 1997. Molecular typing of Enterococcus faecalis strains resistant to high levels of gentamicin isolates in Romania. J. Antimicrob. Chemother. 39:483-491. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thal, L. A., J. W. Chow, D. B. Clewell, and M. J. Zervos. 1994. Tn924, a chromosome-borne transposon encoding high-level gentamicin resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thal, L. A., J. W. Chow, J. E. Patterson, M. B. Perri, S. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Molecular characterization of highly gentamicin-resistant Enterococcus faecalis isolates lacking high-level streptomycin resistance. Antimicrob. Agents Chemother. 37:134-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]