Abstract
We investigated a Klebsiella oxytoca isolate demonstrating resistance to imipenem, meropenem, extended-spectrum cephalosporins, and aztreonam. The MICs of both imipenem and meropenem were 32 μg/ml. The β-lactamase activity against imipenem and meropenem was inhibited in the presence of clavulanic acid. Isoelectric focusing studies demonstrated five β-lactamases with pIs of 8.2 (SHV-46), 6.7 (KPC-2), 6.5 (unknown), 6.4 (probable OXY-2), and 5.4 (TEM-1). The presence of the blaSHV and blaTEM genes was confirmed by specific PCR assays and DNA sequence analysis. Transformation and conjugation studies with Escherichia coli showed that the β-lactamase with a pI of 6.7, Klebsiella pneumoniae carbapenemase-2 (KPC-2), was encoded on an approximately 70-kb conjugative plasmid that also carried SHV-46, TEM-1, and the β-lactamase with a pI of 6.5. The blaKPC-2 determinant was cloned in E. coli and conferred resistance to imipenem, meropenem, extended-spectrum cephalosporins, and aztreonam. The amino acid sequence of KPC-2 showed a single amino acid difference, S174G, when compared with KPC-1, another carbapenem-hydrolyzing β-lactamase from K. pneumoniae 1534. Hydrolysis studies showed that purified KPC-2 hydrolyzed not only carbapenems but also penicillins, cephalosporins, and aztreonam. KPC-2 had the highest affinity for meropenem. The kinetic studies revealed that KPC-2 was inhibited by clavulanic acid and tazobactam. An examination of the outer membrane proteins of the parent K. oxytoca strain demonstrated that it expressed detectable levels of OmpK36 (the homolog of OmpC) and a higher-molecular-weight OmpK35 (the homolog of OmpF). Thus, carbapenem resistance in K. oxytoca 3127 is due to production of the Bush group 2f, class A, carbapenem-hydrolyzing β-lactamase KPC-2. This β-lactamase is likely located on a transposon that is part of a conjugative plasmid and thus has a very high potential for dissemination.
The carbapenems, such as imipenem and meropenem, are used with increasing frequency in the United States and elsewhere for the treatment of multiresistant gram-negative nosocomial pathogens (20, 26). Resistance to carbapenems, while uncommon in enteric organisms, can be mediated by three unique mechanisms. The first mechanism of carbapenem resistance entails the production of large quantities of a chromosomal AmpC cephalosporinase combined with decreased drug permeability through the bacterial outer membrane due to loss or alteration of porins. This has been reported for Enterobacter cloacae (25, 39), Enterobacter aerogenes (7, 10), Proteus rettgeri (39), Citrobacter freundii (28), Escherichia coli (8, 45), and Klebsiella pneumoniae (2, 4). The second mechanism is the production of a β-lactamase that is capable of hydrolyzing carbapenems (5, 41, 47). The third mechanism of resistance, although uncommon, involves changes in the affinity of the target enzymes, the penicillin-binding proteins, for carbapenems (14).
K. pneumoniae carbapenemase-1 (KPC-1) is a class A β-lactamase that is capable of hydrolyzing carbapenems (47). KPC-2 is a closely related enzyme that also hydrolyzes carbapenems and has been reported from isolates of Salmonella enterica serotype Cubana (31) and K. pneumoniae (32). The amino acid sequence of KPC-2 showed a single amino acid difference, S174G, when compared with KPC-1, a carbapenem-hydrolyzing β-lactamase from K. pneumoniae 1534. In this study, a Klebsiella oxytoca strain manifesting carbapenem resistance was identified through project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) (1, 19) and analyzed for its mechanism(s) of carbapenem resistance. The results of our study suggest that the carbapenem resistance phenotype of the strain was caused solely by the production of KPC-2 and was not attributableto modifications of the organism's porins. In addition, we provide evidence that the gene encoding this carbapenemase is located on a mobile element related to IS21.
MATERIALS AND METHODS
Bacterial strains.
The carbapenem-resistant strain K. oxytoca 3127 was collected as part of project ICARE (1, 19) from the urine of a hospitalized patient in New York in 1998. Identification of the isolate was confirmed by using standard biochemical tests (16). E. coli HB101 [F− supE44 lacY1 ara-14 galK2 xyl-5 mtl-1 leuB6 Δ(mcrC-mrr) recA13 rpsL20 thi-1 Δ(gpt-proA)62 hsdSB20 λ−] (42) was used for electroporation of plasmid DNA isolated from strain 3127 and as a recipient in conjugal mating experiments (42). E. coli DH5α (supE44 ΔlacU169 [φ80 lacZΔM15] hsdR17 recA1 gyrA96 thi-1 relA1) (maA-10) was used for cloning the β-lactamase and plasmid DNA preparation of the clone for DNA sequence analysis (42). K. pneumoniae ATCC 13883 (the type strain) was used as a control for porin profiles.
Antimicrobial susceptibility testing.
Organisms were tested by broth microdilution using Mueller-Hinton broth (BD Biosciences, Sparks, Md.) and the National Committee for Clinical Laboratory Standards (NCCLS) reference method (33) and by disk diffusion using Mueller-Hinton agar (Difco Laboratories, Detroit, Mich.) as described previously by NCCLS (34). Antimicrobial agent powders were obtained from the following sources. Amikacin, amoxicillin, ampicillin, cefotaxime, ceftriaxone, chloramphenicol, gentamicin, piperacillin, tetracycline, and trimethoprim-sulfamethoxazole were from Sigma Chemical Co. (St. Louis, Mo.); aztreonam was from Bristol-Myers Squibb (Princeton, N.J.); ceftazidime and tobramycin were from Eli Lilly (Indianapolis, Ind.); cefoxitin was from Merck (Rahway, N.J.); cefpodoxime was from Pharmacia-Upjohn (Kalamazoo, Mich.); clavulanic acid was from SmithKline Beecham (King of Prussia, Pa.); and tazobactam was from Lederle (Pearl River, N.Y.). All antimicrobial agent-containing disks were obtained from Fisher Scientific (Pittsburgh, Pa.). E. coli ATCC 25922, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 27853 (33-35), E. coli HB101, and E. coli DH5α were used for quality control.
Isoelectric focusing of β-lactamases.
Crude cell lysates were prepared by a previously described freeze-thaw procedure (46). Isoelectric focusing (IEF) was performed as described by Matthew and Harris (30). Cell extracts were analyzed by using commercially prepared polyacrylamide gel plates (pH 3.5 to 9.5; Amersham-Pharmacia, Piscataway, N.J.) and electrophoresed to equilibrium using an LKB Multiphor II apparatus (Pharmacia LKB, Piscataway, N.J.). β-Lactamases were visualized by staining the IEF gel with a 0.05% (0.96 mM) solution of nitrocefin (BD Biosciences). The isoelectric points of SHV-46 (8.2), TEM-1 (5.4), OXY-2 (6.4), and KPC-2 (6.7) were estimated by comparison to those of TEM-1 (5.4), SHV-5 (8.2), TEM-3 (6.3), and MIR-1 (8.6).
Examination of porin genes and porin expression.
PCR amplifications were performed in a Thermoline Amplitron 1 thermal cycler by using Taq polymerase (Pharmacia) with 30 cycles of amplification (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C). The primers used to amplify porin genes were U681 and L1316, which anneal to conserved sequences in porin genes located 215 and 850 bp downstream of the ompK36 start codon (12), respectively.
Outer membrane proteins (OMPs) were isolated by Sarkosyl extraction of total membrane preparations as described previously (22). Protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) as described by the manufacturer. The proteins were examined on either 8 to 15% sodium dodecyl sulfate-polyacrylamide linear gradient gels or 4 to 12% NuPAGE gels with morpholinepropanesulfonic acid (MOPS) buffer (Invitrogen, Carlsbad, Calif.). For OmpK37 analysis, electrophoresis of OMPs was performed on 11% acrylamide-0.2% bisacrylamide-0.1% sodium dodecyl sulfate gels (12). Samples were boiled for 5 min in Laemmli's sample buffer before electrophoresis. Gels were visualized by staining with Coomassie blue R250.
Western blotting of OmpK35, OmpK36 and OmpK37 was performed as described previously (12, 22). Filters were blocked in 1% bovine serum albumin in phosphate-buffered saline (PBS). After being washed, the filters were incubated with 1:100-diluted anti-OmpK35 or anti-OmpK36 or anti-OmpK37 antibody (12, 22) and then with alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G (Sigma; 1:5,000). The filters were developed as previously described (12, 22). All the incubations were carried out at room temperature for 1 h in 1% bovine serum albumin-0.05% Tween 20-PBS and, after incubation with the antiserum, the filters were washed with 0.05% Tween 20-PBS.
Plasmid profile analysis.
Plasmid DNA from K. oxytoca 3127 was isolated by using the method described by Portnoy et al. (36). Supercoiled plasmid DNAs of pDK9 (165 kb) and R1 (97.6 kb) and the plasmids in E. coli V517 (56.7, 5.8, 4.09, 3.15, 2.83, and 2.2 kb) were used as size standards.
Carbapenem inactivation assay.
In order to determine whether resistance to imipenem and meropenem was caused by production of a β-lactamase, a disk diffusion bioassay using E. coli DH5α was performed as previously described (47). Negative controls for carbapenemase production were E. coli HB101 and K. pneumoniae ATCC 13883. The positive control was K. pneumoniae 1534 (47).
Filter mating.
Filter mating studies were performed at both 30 and 37°C (42). E. coli HB101 was used as the recipient. The transconjugants were selected on Luria-Bertani (LB) agar containing 30 μg of tetracycline per ml, 2 μg of imipenem per ml, and 120 μg of streptomycin per ml.
Transformation.
Plasmid DNA prepared from K. oxytoca 3127 via Qiagen plasmid midiprep kit (Qiagen, Chatsworth, Calif.) was electroporated into E. coli HB101 as described previously (42). Transformants were selected on LB agar containing 120 μg of streptomycin per ml and 1.5 μg of imipenem per ml.
Cloning of blaKPC-2.
The cloning and sequencing of KPC-2 were carried out as described by Yigit et al. (47).
blaSHV-, blaTEM-, and blaKPC-2-specific PCR and DNA sequence analysis.
The primers and the PCR conditions used for amplification of blaSHV and blaTEM were those described by Rasheed et al. (40). The blaKPC-2 determinant was amplified from the parent strain, K. oxytoca 3127, by using the protocol and primers for blaKPC-1 described by Yigit et al. (47).
DNA sequencing data were analyzed by using DNASIS for Windows (Hitachi Software Genetic Systems, San Francisco, Calif.). The DNA and protein sequences of the other β-lactamases were obtained from the EMBL and the Swiss-Prot data banks. BLAST and BLASTX programs from the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST) were used to identify blaKPC-2.
β-Lactamase purification.
The KPC-2 β-lactamase was purified from the E. coli DH5α strain containing cloned blaKPC-2 (E. coli DH5α pBR322-catI-blaKPC-2) for kinetic analysis studies. Four 1-liter cultures of tryptic soy broth supplemented with 100 μg of ampicillin per ml were grown overnight at 37°C. Bacteria were harvested by centrifugation and washed with 50 mM phosphate buffer (pH 7.0). The pellets were resuspended in 10 ml of 0.2 M sodium acetate (pH 5.5) and subjected to five freeze-thaw cycles. The lysate was centrifuged at 20,000 × g, and the β-lactamase activity of the supernatant was enriched by chromatography through Sephadex G-100 in 50 mM phosphate buffer (pH 7.0). Protein in peak fractions containing nitrocefin-hydrolyzing activity was precipitated with 90% ammonium sulfate; pellets were resuspended in 20 mM morpholineethanesulfonic acid (MES) buffer (pH 6.0)-10% glycerol, and dialyzed in 2 liters of the same buffer at 4°C. The β-lactamase was desalted through a HighTrap desalting column (Amersham-Pharmacia) and eluted from a HiTrap-S cation exchange column in 20 mM MES (pH 6.0)-10% glycerol by a 0 to 0.5 M NaCl gradient. The protein concentration of the HiTrap-S fractions was determined with the Micro Coomassie Plus protein assay (Pierce). The purity of the KPC-2 fractions was determined by scanning densitometry of a Colloidal Blue-stained NuPAGE 10% Bis-Tris gel. Purity of the fractions used for kinetic analysis was >90%.
Kinetic studies.
Initial hydrolysis rates were measured on a Shimadzu UV-1601 spectrophotometer at 25°C in 50 mM phosphate buffer (pH 7.0). Km and Vmax values were obtained by averaging results from Eadie-Hofstee, Hanes-Woolf, and direct linear plot analyses. Spectrophotometric measurements were determined on several different days, with cephaloridine assayed as a reference each day. The standard error for calculated kinetic parameters was ≤15%. Inhibition of hydrolysis was measured after a 5-min preincubation of enzyme with inhibitor in phosphate buffer (pH 7.0). Nitrocefin at a concentration of 100 μM was the substrate used for the inhibition studies. Ki values were determined by the method of Dixon (11). For comparison, a preparation of KPC-1 purified under the same conditions as KPC-2 was used to determine Ki values.
Gene dosage assays.
The gene dosage assays were performed as described by Heritage et al. (21). The plasmid pBR322-catI (47) was used for this assay. Transposition of a blaKPC-2-containing element onto pBR322-catI should increase the copy number for blaKPC-2, which, in turn, should increase the imipenem MIC because of the gene dosage effect. The overnight culture of E. coli DH5α carrying both the 70-kb plasmid and pBR322-catI was plated on the LB agar containing 128 μg of imipenem per ml and 40 μg of chloramphenicol per ml. The overnight cultures of E. coli DH5α transformant containing only the 70-kb plasmid were used as background controls and were serially diluted and plated on the LB agar containing 128 μg of imipenem per ml.
Nucleotide sequence accession numbers.
The nucleotide sequence of blaKPC-2 reported in this study will appear under the GenBank accession number AY210886, and that of blaSHV-46 will appear under accession number AY210887.
RESULTS
Antimicrobial susceptibility patterns of K. oxytoca 3127.
The MICs of a variety of antimicrobial agents tested against K. oxytoca 3127 are shown in Table 1. The isolate was resistant to imipenem and meropenem, with MICs of 32 μg/ml for each drug. The isolate was also resistant to extended-spectrum cephalosporins and aztreonam. The MICs of both imipenem and meropenem decreased from 32 to 4 μg/ml when tested in the presence of clavulanic acid (4 μg/ml) (Table 1). In the E. coli transformants, the imipenem and meropenem MICs decreased by five doubling dilutions from 16 to 0.5 μg/ml and 8 to ≤0.25 μg/ml, respectively, when the carbapenems were tested in combination with clavulanic acid. The MICs of ceftazidime, ceftriaxone, and cefotaxime also decreased by two to four doubling dilutions in the presence of clavulanic acid in the parent strain (data not shown).
TABLE 1.
Antimicrobial susceptibility patterns of K. oxytoca 3127, E. coli DH5α clone, and E. coli HB101 transformanta
| Antimicrobial agents | K. oxytoca 3127 (parent) | E. coli DH5α | E. coli DH5α (pBR322-catI-blaKPC-2) | E. coli HB101 transformant containing blaKPC-2 | E. coli HB101 |
|---|---|---|---|---|---|
| Imipenem | 32 | ≤0.25 | 16 | 15 | ≤0.25 |
| Imipenem-clavulanic acidb | 4 | ≤0.25 | 0.5 | 0.5 | ≤0.25 |
| Meropenem | 32 | ≤0.25 | 8 | 8 | ≤0.25 |
| Meropenem-clavulanic acidb | 4 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Ampicillin | >64 | 2 | >64 | >64 | 4 |
| Amoxicillin-clavulanic acid | >32/16 | 2/1 | >32/16 | >32/16 | 2/1 |
| Piperacillin-tazobactam | >128/4 | ≤1/4 | >128/4 | >128/4 | ≤1/4 |
| Aztreonam | >64 | ≤1 | >64 | >64 | ≤1 |
| Ceftazidime | >64 | ≤2 | 32 | 64 | ≤2 |
| Cefoxitin | >32 | 2 | 32 | 32 | 4 |
| Cefpodoxime | >16 | 0.5 | >16 | >16 | ≤0.25 |
| Cefepime | >32 | ≤1 | 16 | >32 | ≤1 |
| Cefotaxime | >64 | ≤1 | 8 | 64 | ≤1 |
| Ceftriaxone | >64 | ≤1 | 32 | 64 | ≤1 |
| Chloramphenicol | 8 | 4 | >32 | 4 | 4 |
| Gentamicin | 8 | ≤0.25 | ≤0.25 | 8 | ≤0.25 |
| Tobramycin | 16 | ≤0.25 | ≤0.25 | 16 | ≤0.25 |
| Trimethoprim-sulfamethoxazole | >8 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 |
Values are MICs and are given in micrograms per milliliter.
Clavulanic acid was tested at a fixed concentration of 4 μg/ml.
Imipenem and meropenem resistance involves production of β-lactamase.
IEF of crude extracts of strain 3127 revealed five β-lactamase bands with pIs of 8.2, 6.7, 6.5, 6.4, and 5.4 (Fig. 1A, lane 7). To determine whether resistance to carbapenems could be attributed to production of a β-lactamase, we performed a disk diffusion carbapenem inactivation assay (47). The assay results indicated that a β-lactamase was involved in hydrolysis of imipenem in 3127. The crude cell lysates prepared in the presence and absence of imipenem showed similar zone alterations (data not shown). The presence of EDTA did not inhibit the activity of the β-lactamase, nor did the addition of ZnCl2 enhance the β-lactamase activity against imipenem or meropenem (data not shown).
FIG. 1.
(A) Isoelectric focusing patterns of cell lysates from carbapenem-resistant strains. The gel was stained with nitrocefin. Lanes 1 to 4, cell lysates prepared from strains producing TEM-3 (pI 6.3), TEM-1 (pI 5.4), SHV-5 (pI 8.2), and MIR-1 (pI 8.4), respectively; lane 5, the imipenem-resistant E. coli HB101 transformant containing a 70-kb plasmid from 3127; lane 6, an imipenem-resistant E. coli HB101 transconjugant of 3127; lane 7, K. oxytoca 3127. (B) Isoelectric focusing patterns of cell lysates prepared from carbapenem-resistant clones of K. oxytoca 3127. Lane 1, strain producing SHV-46 (pI 8.2); lanes 2 to 3, clones of strain 3127; lane 4, an imipenem-resistant E. coli HB101 transconjugant of K. oxytoca 3127; lane 5, E. coli DH5α containing the blaKPC-1 clone. The pIs of the β-lactamases were calculated by using the known pIs of TEM-1 (5.4), TEM-3 (6. 3), SHV-5 (8.2), KPC-1 (6.7), and MIR-1 (8.4).
PCR and DNA sequence analysis of blaSHV and blaTEM.
IEF results for K. oxytoca 3127 suggested the presence of blaTEM (pI 5.4) and blaSHV (pI 8.2) determinants (Fig. 1A, lanes 2, 3, and 7). PCR analysis using blaSHV- and blaTEM-specific primers confirmed the presence of these genes in strain 3127 (data not shown). DNA sequence analysis identified the genes as blaTEM-1 and blaSHV-46. The amino acid sequence of SHV-46 showed three amino acid changes (T195N, G238S, and E240K) when compared to the sequence of SHV-1. The β-lactamase with a pI of 6.4 is presumed to be the chromosomal OXY-2, which has been reported to have a pI ranging from 5.2 to 6.8 (17, 18). Since the IEF of lysates prepared from transformants and transconjugants of strain 3127 did not produce this enzyme (Fig. 1A, lanes 5 and 6, respectively), it is presumed to be chromosomal in origin. The identity of the β-lactamase with a pI of 6.5 carried on the 70-kb plasmid, while consistent with OXA enzymes (9), remains elusive.
Cloning of the blaKPC-2 gene from the E. coli DH5α transformant.
The filter mating results between strain 3127 and E. coli HB101 showed that the carbapenem resistance in 3127 was associated with a 70-kb conjugative plasmid that encoded four β-lactamases with pIs of 8.2, 6.7, 6.5, and 5.4 (Fig. 1A, lane 6). Resistance to carbapenems, extended-spectrum cephalosporins, and aztreonam was encoded on the same plasmid (Table 1). The carbapenem MICs for the E. coli HB101 transconjugants containing the plasmid encoding blaSHV-46, blaTEM-1, blaKPC-2, and the fourth β-lactamase were similar to those for the parent isolate. The MIC results also suggested that the resistance determinant encoding gentamicin and tobramycin resistance was located on the 70-kb conjugative plasmid. The DNA isolated from a transconjugant was electroporated into E. coli HB101 and used for IEF. The IEF results showed that this plasmid encoded SHV-46, TEM-1, and a β-lactamase with a pI of 6.5, in addition to KPC-2 (Fig. 1A, lane 5). The presence of blaSHV-46 and blaTEM-1 was also confirmed in the transconjugants and transformants by PCR analysis.
To characterize the β-lactamase mediating carbapenem resistance, we cloned a 2.4-kb BamHI fragment encoding KPC-2 in DH5α using pBR322-catI as a cloning vector. E. coli (pBR322-catI-blaKPC-2) encoded a single β-lactamase with a pI of 6.7 as shown by IEF (Fig. 1B, lanes 2 and 3). The antibiogram of the E. coli DH5α blaKPC-2 clone is shown in Table 1. This demonstrates that blaKPC-2 is responsible for the resistance to carbapenems, extended-spectrum cephalosporins, and aztreonam.
Kinetic parameters.
The kinetic parameters for the KPC-2 β-lactamase are summarized in Table 2. The KPC-2 enzyme used in these studies was approximately 90% pure. KPC-2 hydrolyzed β-lactams from the penicillin, cephalosporin, carbapenem, and monobactam groups. The highest kcat values were obtained with cephaloridine, which demonstrated kcat values that were approximately seven times higher than those for cephalothin or nitrocefin and 2.5 times higher than that for ampicillin. The kcat values for penicillin G, cloxacillin, and aztreonam were similar, approximately 10 to 17 times lower than those for cephaloridine. KPC-2 showed hydrolytic activity against the carbapenems; hydrolysis of imipenem occurred at rates that were approximately 35 times slower than those for cephaloridine. Meropenem had kcat values four times lower than those for imipenem. Hydrolysis rates for cefotaxime and ceftazidime were 24 and 4,416 times lower than the values obtained for cephaloridine. Of the two extended-spectrum cephalosporins tested, cefotaxime had the highest kcat values, which were approximately 180 times higher than the kcat values for ceftazidime. Of the 12 substrates used in these experiments, cefoxitin and ceftazidime had the lowest hydrolysis rates.
TABLE 2.
Hydrolysis parameters of KPC-1 and KPC-2 β-lactamases
| Substrate or inhibitor |
kcat (s−1)
|
Relative kcat
|
Km (μM)
|
kcat/Km (μM−1 s−1)
|
Relative kcat/Km
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| KPC-1a | KPC-2 | KPC-1 | KPC-2 | KPC-1 | KPC-2 | KPC-1 | KPC-2 | KPC-1 | KPC-2 | |
| Substrates | ||||||||||
| Cephaloridine | 340 | 530 | 100 | 100 | 560 | 500 | 0.61 | 1.1 | 100 | 100 |
| Cephalothin | 75 | 69 | 22 | 13 | 53 | 82 | 1.4 | 0.84 | 230 | 76 |
| Cefotaxime | 14 | 22 | 4.2 | 4.2 | 160 | 220 | 0.09 | 0.10 | 14 | 9.1 |
| Cefoxitin | 0.26 | 0.31 | 0.08 | 0.06 | 120 | 180 | 0.002 | 0.002 | 0.36 | 0.16 |
| Ceftazidime | 0.10 | ≤0.12 | 0.03 | ≤0.02 | 94 | NDb | 0.001 | ND | 0.18 | ND |
| Benzylpenicillin | 32 | 51 | 9.6 | 9.6 | 23 | 27 | 1.4 | 1.9 | 230 | 170 |
| Ampicillin | 110 | 210 | 33 | 40 | 130 | 230 | 0.85 | 0.91 | 140 | 83 |
| Cloxacillin | 25 | 35 | 7.4 | 6.6 | 100 | 79 | 0.25 | 0.44 | 41 | 40 |
| Imipenem | 12 | 15 | 3.7 | 2.8 | 81 | 51 | 0.15 | 0.29 | 26 | 26 |
| Meropenem | 3.0 | 4.0 | 0.9 | 0.75 | 12 | 15 | 0.25 | 0.27 | 41 | 25 |
| Aztreonam | 20 | 30 | 5.9 | 5.7 | 310 | 360 | 0.07 | 0.08 | 11 | 7.6 |
| Inhibitors | ||||||||||
| Clavulanic acidc | 1.2 | 1.5 | ||||||||
| Tazobactamc | 0.23 | 0.18 | ||||||||
| EDTA | >5,000 | >5,000 | ||||||||
KPC-1 kcat and Km data are from reference 47.
ND, not determined.
Inhibitor constant (Ki) is micromolar.
KPC-2 had the highest affinity for meropenem, with a Km value of 15 μM. Other substrates with low Km values were nitrocefin, penicillin G, and imipenem, whose Km values ranged from 22 to 51 μM. Cephaloridine, at 500 μM, had the highest Km.
Hydrolytic efficiencies, measured by kcat/Km, revealed that penicillin G was hydrolyzed by KPC-2 approximately two times more efficiently than cephaloridine. Nitrocefin had the highest catalytic efficiency of the substrates tested, with a value 3.3 times that of cephaloridine. The hydrolytic efficiencies for imipenem and meropenem were <30% that of cephaloridine. Cefotaxime was the most efficiently hydrolyzed of the three extended-spectrum cephalosporins tested, with a kcat/Km value that was approximately 10% that of cephaloridine. The hydrolytic efficiency of cefoxitin was 600-fold lower than that for cephaloridine. The hydrolytic efficiency for ceftazidime could not be determined directly due to extremely slow hydrolysis.
The KPC-2 β-lactamase demonstrated Ki values of 1.5 μM for clavulanic acid and 0.18 μM for tazobactam. These were similar to the values obtained for KPC-1 under the same conditions (Table 2). No inhibition was observed when the enzyme was tested with 5 mM EDTA at pH 7.0.
blaKPC-2 may be located on a mobile element.
The 157-amino-acid partial sequence (GenBank accession number AAO53444.1) encoded in the 863-nucleotide sequence upstream of the blaKPC-2 coding region showed a high degree of similarity to several IstB-like proteins: 52% similarity to the IS21 putative ATP-binding protein (GenBank accession number P15026), 68% to the putative IS100 transposase from Yersinia pestis C092 (GenBank accession number NP_395401), 68% to the putative transposase from Y. pestis (GenBank accession number AAC44982), and 98% to the putative transposition helper protein from S. enterica subsp. enterica serotype Cubana (GenBank accession number AAM10642) (Fig. 2). IstB-like proteins are ATP-binding proteins that contain an ATP- or GTP-binding P-loop motif (http://www.ncbi.nlm.nih.gov). The IstB-like proteins are associated with the IS21 family of insertion sequences (6, 27). The functions of IstB-like proteins include stimulation of transposase and cointegrase-driven reactions (6). Thus, we investigated whether blaKPC-2 was located on an active transposable element.
FIG. 2.
Alignment of the partial sequence of the protein encoded upstream of KPC-2 (GenBank accession number AAO53444.1) to IstB-like proteins (6); IS21 putative ATP-binding protein (GenBank accession number P15026), putative IS100 transposase from Y. pestis C092 (GenBank accession number NP_395401), putative transposase from Y. pestis (GenBank accession number AAC44982), and putative transposition helper protein from S. enterica subsp. enterica serotype Cubana (GenBank accession number AAM10642).
The 70-kb plasmid from the E. coli HB101 transconjugant was electroporated into E. coli DH5α containing pBR322-catI. Since pBR322-catI is a multicopy cloning vector, we hypothesized that it would facilitate the detection of the transposition of blaKPC-2 from the 70-kb plasmid by a gene dosage assay (21). The frequency of blaKPC-2 transposition was 9.7 × 10−4, as indicated by the number of colonies with elevated imipenem MICs (i.e., those growing on 128-μg/ml versus 16-μg/ml imipenem). The background rate of colonies containing only the 70-kb plasmid growing on 128-μg/ml imipenem was 5.7 × 10−8.
Analysis of K. oxytoca 3127 OMPs.
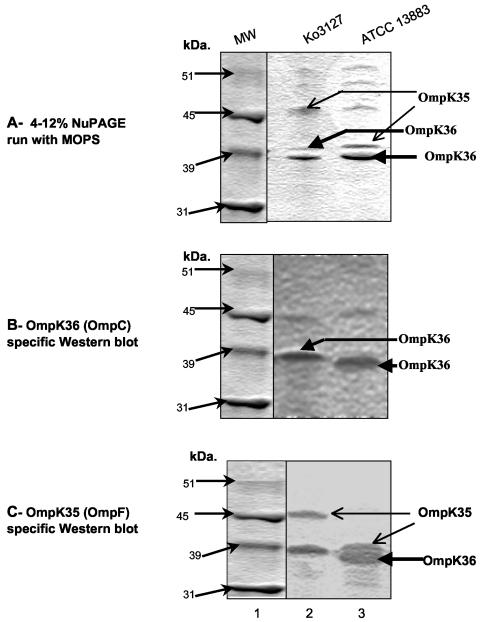
The MICs of meropenem, ceftazidime, and cefotaxime were lower for the E. coli HB101 transformants and E. coli DH5α(pBR322-catI-blaKPC-2) than for the parent K. oxytoca 3127. This may be due to species differences among the porins, which are known to increase the MICs of these drugs for K. pneumoniae isolates (2, 4, 29). PCR analysis showed that K. oxytoca 3127 and K. pneumoniae ATCC 13883 (the extended-spectrum cephalosporin-susceptible type strain) both carry all three porin genes, ompK35, ompK36, and ompK37 (data not shown). The porin profile of K. oxytoca 3127 was compared to that of K. pneumoniae ATCC 13883 (Fig. 3), and the expression of the porin genes was examined by Western blotting with polyclonal anti-OmpK35, anti-OmpK36, and anti-OmpK37 antisera. As reported by Hernandez-Alles et al. (22), the presence of OmpK35 and OmpK36 cannot be determined solely by their migration in gels, since in some strains OmpK35 migrates more slowly than OmpK36. This is the case (Fig. 3A) for K. pneumoniae ATCC 13883 (lane 3) and K. oxytoca 3127 (lane 2), where OmpK35 apparently migrates more slowly than OmpK36. A Western blot produced using anti-OmpK36-specific antisera (Fig. 3B) confirmed the identity of OmpK36 bands for ATCC 13883 (lane 3) and 3127 (lane 2). Anti-OmpK35 antisera are known to cross-react with OmpK36 (22) and reacted, as predicted, with two bands in ATCC 13883 (OmpK35 and OmpK36) (Fig. 3C, lane 3) and with two bands in 3127 (lane 2). These results suggest that OmpK35 was expressed in K. oxytoca 3127 but migrated more slowly than the OmpK35 in ATCC 13883. Neither of the two strains appeared to express OmpK37 when tested with OmpK37-specific antisera (data not shown).
FIG. 3.
NuPAGE gel and Western blot analysis of OMPs of K. oxytoca 3127 and a carbapenem-susceptible control strain, K. pneumoniae ATCC 13883. (A) NuPAGE gel analysis of OMPs. Lane 1, molecular mass markers; lane 2, OMPs prepared from K. oxytoca 3127; lane 3, OMPs prepared from K. pneumoniae ATCC 13883. (B) Western blot analysis of OMPs performed with anti-OMPK36 antisera. (C) Western blot analysis of OMPs performed with anti-OMPK35 antisera. Molecular mass is indicated in gels to the left of each panel.
DISCUSSION
Carbapenems, such as imipenem and meropenem, are antibacterial agents with activity against many gram-negative, gram-positive, and anaerobic microorganisms. Carbapenems are often used to treat infections caused by multidrug-resistant isolates, including strains producing extended-spectrum β-lactamases (ESBLs) (20, 26, 41, 43). However, the recent appearance of β-lactamases capable of hydrolyzing carbapenems, in addition to other mechanisms of carbapenem resistance, makes treating these infections more difficult (4, 20, 26, 41, 47).
Here we describe the appearance of the class A β-lactamase, KPC-2, in a strain of K. oxytoca. KPC-2 is closely related to the KPC-1 β-lactamase from K. pneumoniae 1534. KPC-2 was initially isolated from S. enterica serotype Cubana (31) (GenBank accession number AF481906). KPC-2 differs from KPC-1 by a single amino acid substitution, namely S174G. Our kinetic data show that the S174G substitution did not cause changes in the hydrolytic profile of the enzyme, as the kcat, Km, and Ki values for β-lactam substrates were similar for KPC-1 and KPC-2 (47).
During the last decade, many hospital outbreaks caused by ESBL-producing Enterobacteriaceae spp. have been reported. Most of the ESBL-producing strains carried derivatives of blaTEM-1, blaTEM-2, or blaSHV-1 (13, 23, 40). These β-lactamases are typically encoded on large conjugative plasmids, as KPC-2 is (3, 15, 24, 38, 44). More recent reports have highlighted the emergence of ESBL-producing strains that are multiply resistant to amikacin, gentamicin, sulfonamides, streptomycin, and trimethoprim (37, 38). The blaKPC-2 determinant of K. oxytoca 3127 was located on a 70-kb conjugative plasmid that also encodes SHV-46, TEM-1, and a fourth unidentified β-lactamase. This plasmid also encodes resistance to gentamicin and tobramycin. Although the exact genetic structure has not been determined, the blaKPC-2 determinant is presumably located on a transposable element, as suggested by gene dosage assays. The amino acid sequences inferred from DNA sequencing of the upstream region suggests that the KPC-2-encoding plasmid is 98% identical to the plasmid identified in the Salmonella serotype Cubana strain (GenBank accession number AF481906). Thus, the KPC-2 β-lactamase is likely being disseminated among species of Enterobacteriaceae through both conjugal plasmid transfer and transposition. Further studies are required to determine the identity and structure of this mobile element and its other resistance determinants.
We searched for alterations in one or more of the three porin proteins described for K. pneumoniae strains that are associated with increased MICs for extended-spectrum cephalosporins and carbapenems (12, 22, 29). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of K. oxytoca 3127 porin profiles, in combination with the Western blot analysis with OmpK35-, OmpK36-, and OmpK37-specific antibodies, showed that strain 3127 expresses OmpK36 (the OmpC homolog) and a slower-migrating OmpK35 (the OmpF homolog) (Fig. 3B and C). This finding is consistent with the observations of Hernandez-Alles et al., who reported that ESBL-producing K. pneumoniae strains can have OmpK35 porins that migrate more slowly than OmpK36 porins (22). Studies by Domenech-Sanchez et al. suggest that the newly identified porin, OmpK37, might be used by carbapenems to gain access to the cell (12); however, this porin is strongly down-regulated under standard laboratory conditions and is often seen only in the absence of OmpK35 and OmpK36 expression. Thus, its contribution to resistance in 3127 remains unclear.
In conclusion, we have isolated and characterized the class A carbapenemase determinant, KPC-2, from a clinical isolate of K. oxytoca. The data presented here show that KPC-2 is responsible for the carbapenem resistance of this strain, which, unlike the K. pneumoniae strain harboring KPC-1, has no detectable porin alterations.
Acknowledgments
H. Yigit is a recipient of an American Society for Microbiology-National Center for Infectious Diseases postdoctoral fellowship.
We offer special thanks to A. Karls and W. S. Reznikoff for their assistance with the gene dosage assay. We also thank J. Swenson for her help in obtaining the required media and antimicrobial agents for this study, and we thank C. Gownley for help with the KPC-2 purification. R. Clonno and J. Fung-Tomc are also thanked for their support.
The use of trade names is for identification purposes only and does not constitute endorsement by the Department of Health and Human Servicesor the U.S. Public Health Service.
REFERENCES
- 1.Archibald, L., L. Phillips, D. Monnet, J. E. McGowan, Jr., F. C. Tenover, and R. Gaynes. 1997. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: increasing importance of the intensive care unit. Clin. Infect. Dis. 24:211-215. [DOI] [PubMed] [Google Scholar]
- 2.Ardanuy, C., J. Linares, M. A. Dominguez, S. Hernandez-Alles, V. J. Benedi, and L. Martinez-Martinez. 1998. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 42:1636-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen, E. H., P. Desjardins, G. Arlet, F. Bourgeois, P. Mariani-Kurkdjian, N. Y. Lambert-Zechovsky, E. Denamur, A. Philippon, and J. Elion. 1993. Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J. Clin. Microbiol. 31:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 328-330. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 7.Chow, J. W., and D. M. Shlaes. 1991. Imipenem resistance associated with the loss of a 40-kDa outer membrane protein in Enterobacter aerogenes. J. Antimicrob. Chemother. 28:499-504. [DOI] [PubMed] [Google Scholar]
- 8.Cornaglia, G., L. Guan, R. Fontana, and G. Satta. 1992. Diffusion of meropenem and imipenem through the outer membrane of Escherichia coli K-12 and correlation with their antibacterial activities. Antimicrob. Agents Chemother. 36:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couture, F., J. Lachapelle, and R. C. Levesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 10.De Champs, C., C. Henquell, D. Guelon, D. Sirot, N. Gazuy, and J. Sirot. 1993. Clinical and bacteriological study of nosocomial infections due to Enterobacter aerogenes resistant to imipenem. J. Clin. Microbiol. 31:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon, M. 1972. The graphical determination of Km and Ki. Biochem. J. 129:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domenech-Sanchez, A., S. Hernandez-Alles, L. Martinez-Martinez, V. J. Benedi, and S. Alberti. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Bois, S. K., M. S. Marriott, and S. G. B. Amyes. 1995. TEM- and SHV-derived extended-spectrum β-lactamases: relationship between selection, structure and function. J. Antimicrob. Chemother. 35:7-22. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, R., and D. Greenwood. 1996. Mechanisms responsible for reduced susceptibility to imipenem in Bacteroides fragilis. J. Antimicrob. Chemother. 38:941-951. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, D., E. G. Russell, M. Tymms, E. J. Roper, M. L. Grayson, and J. Turnidge. 1995. Random amplified polymorphic DNA and plasmid analyses used in investigation of an outbreak of multiresistant Klebsiella pneumoniae. J. Clin. Microbiol. 33:713-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer, J. J. 1999. Enterobacteriaceae: introduction and identification, p. 442-458. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 17.Fournier, B., A. Gravel, D. C. Hooper, and P. H. Roy. 1999. Strength and regulation of the different promoters for chromosomal β-lactamases of Klebsiella oxytoca. Antimicrob. Agents Chemother. 43:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier, B., P. H. Roy, P. H. Lagrange, and A. Philippon. 1996. Chromosomal β-lactamase genes of Klebsiella oxytoca are divided into two main groups, blaOXY-1 and blaOXY-2. Antimicrob. Agents Chemother. 40:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridkin, S. K., C. D. Steward, J. R. Edwards, E. R. Pryor, J. E. McGowan, Jr., L. K. Archibald, R. P. Gaynes, and F. C. Tenover. 1999. Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: project ICARE phase 2. Project Intensive Care Antimicrobial Resistance Epidemiology (ICARE) hospitals. Clin. Infect. Dis. 29:245-252. [DOI] [PubMed] [Google Scholar]
- 20.Hawkey, P. M. 1997. Resistance to carbapenems. J. Med. Microbiol. 46:451-454. [PubMed] [Google Scholar]
- 21.Heritage, J., P. M. Hawkey, N. Todd, and I. J. Lewis. 1992. Transposition of the gene encoding a TEM-12 extended-spectrum β-lactamase. Antimicrob. Agents Chemother. 36:1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Alles, S., S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas, and V. J. Benedi. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby, G. A. 1997. Extended-spectrum β-lactamases and other enzymes providing resistance to oxymino-β-lactams. Infect. Dis. Clin. N. Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 24.Jacoby, G. A., and L. Sutton. 1991. Properties of plasmids responsible for production of extended-spectrum β-lactamase. Antimicrob. Agents Chemother. 35:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, E. H., M. H. Nicolas, M. D. Kitzis, G. Pialoux, E. Collatz, and L. Gutmann. 1991. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob. Agents Chemother. 35:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livermore, D. M. 1995. Bacterial resistance to carbapenems. Adv. Exp. Med. Biol. 390:25-47. [DOI] [PubMed] [Google Scholar]
- 27.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mainardi, J. L., P. Mugnier, A. Coutrot, A. Buu-Hoi, E. Collatz, and L. Gutmann. 1997. Carbapenem resistance in a clinical isolate of Citrobacter freundii. Antimicrob. Agents Chemother. 41:2352-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Martinez, L., A. Pascual, S. Hernandez-Alles, D. Alvarez-Diaz, A. I. Suarez, J. Tran, V. J. Benedi, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthew, M., and A. M. Harris. 1976. Identification of β-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J. Gen. Microbiol. 94:55-67. [DOI] [PubMed] [Google Scholar]
- 31.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moland, S. E., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 34.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A7, 7th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 35.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 36.Portnoy, D. A., H. Wolf-Watz, I. Bolin, A. B. Beeder, and S. Falkow. 1984. Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect. Immun. 43:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston, K. E., M. A. Kacica, R. J. Limberger, W. A. Archinal, and R. A. Venezia. 1997. The resistance and integrase genes of pACM1, a conjugative multiple-resistance plasmid, from Klebsiella oxytoca. Plasmid 37:105-118. [DOI] [PubMed] [Google Scholar]
- 38.Prodinger, W. M., M. Fille, A. Bauernfeind, I. Stemplinger, S. Amann, B. Pfausler, C. Lass-Florl, and M. P. Dierich. 1996. Molecular epidemiology of Klebsiella pneumoniae producing SHV-5 β-lactamase: parallel outbreaks due to multiple plasmid transfer. J. Clin. Microbiol. 34:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raimondi, A., A. Traverso, and H. Nikaido. 1991. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob. Agents Chemother. 35:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shannon, K., P. Stapleton, X. Xiang, A. Johnson, H. Beattie, F. El Bakri, B. Coohson, and G. French. 1998. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains causing nosocomial outbreaks of infection in the United Kingdom. J. Clin. Microbiol. 36:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirot, D. L. 1995. Extended-spectrum plasmid-mediated β-lactamases. J. Antimicrob. Chemother. 36(Suppl. A):19-34. [DOI] [PubMed] [Google Scholar]
- 45.Stapleton, P. D., K. P. Shannon, and G. L. French. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sykes, R. B., D. P. Bonner, K. Bush, and N. H. Georgopapadakou. 1982. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob. Agents Chemother. 21:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]