Abstract
We report the discovery and characterization of a novel ribosome inhibitor (NRI) class that exhibits selective and broad-spectrum antibacterial activity. Compounds in this class inhibit growth of many gram-positive and gram-negative bacteria, including the common respiratory pathogens Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, and Moraxella catarrhalis, and are nontoxic to human cell lines. The first NRI was discovered in a high-throughput screen designed to identify inhibitors of cell-free translation in extracts from S. pneumoniae. The chemical structure of the NRI class is related to antibacterial quinolones, but, interestingly, the differences in structure are sufficient to completely alter the biochemical and intracellular mechanisms of action. Expression array studies and analysis of NRI-resistant mutants confirm this difference in intracellular mechanism and provide evidence that the NRIs inhibit bacterial protein synthesis by inhibiting ribosomes. Furthermore, compounds in the NRI series appear to inhibit bacterial ribosomes by a new mechanism, because NRI-resistant strains are not cross-resistant to other ribosome inhibitors, such as macrolides, chloramphenicol, tetracycline, aminoglycosides, or oxazolidinones. The NRIs are a promising new antibacterial class with activity against all major drug-resistant respiratory pathogens.
Respiratory tract infections are the number 1 killer worldwide, responsible for over 50 million deaths each year. Although antibacterial therapy has successfully stemmed the tide against infection since the middle of the last century, antibacterial resistance of Streptococcus pneumoniae, the most commonly identified pathogen associated with community-acquired pneumonia, is on the rise (2, 4, 8, 22). A recent worldwide study documents that a significant fraction of S. pneumoniae isolates have reduced susceptibility to penicillin (36%) and macrolides (31%) (5). Although overall rates of resistance to fluoroquinolones are low, these rates were found to be increasing rapidly in Canada (1). In general, pathogenic bacteria continuously evolve mechanisms of resistance to currently used antibacterial agents. The discovery of novel antibacterial classes would be the most powerful way to generate new therapy against these resistant pathogens. Unfortunately, novel antibacterial classes have been difficult to discover, with the oxazolidinones, identified in 1980, being the last example to successfully reach the clinic.
The bacterial ribosome is a proven target for antibacterial chemotherapy (6, 17, 20, 21). Since the 1940s, small-molecule ribosome inhibitors such as chloramphenicol, tetracyclines, macrolides, aminoglycosides, and, more recently, oxazolidinones have been used to combat bacterial infections in humans (11). These diverse chemical classes of ribosome inhibitors each bind to a different site on the ribosome, which is not surprising given its large size and complexity. Accordingly, drug resistance to each class generally develops separately, such that resistance to one class does not confer resistance to another. Therefore, from a drug discovery standpoint, the ribosome is actually a large collection of validated, broad-spectrum targets (9). A novel class of ribosome inhibitor binding to yet another ribosomal binding site should not be affected by any existing resistance mechanisms.
We attempted to discover a novel class of ribosome inhibitors by directly screening a library of compounds for this activity. This screen resulted in the identification of a novel class of antibacterial agents (novel ribosome inhibitors [NRIs]) that is unaffected by existing drug resistance mechanisms.
MATERIALS AND METHODS
Reagents.
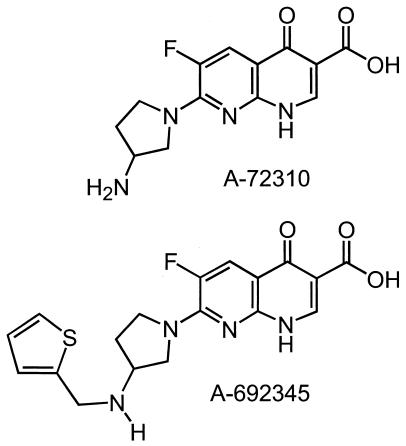
RNase-free water was used throughout. Bacterial culture media and yeast extract were purchased from Difco Laboratories, Sparks, Md. Chloramphenicol, tetracycline, minocycline, puromycin, streptomycin, cycloheximide, erythromycin, lincomycin, thiostrepton, phenylmethylsulfonyl fluoride, diisopropyl fluorophosphate, o-nitrophenyl-β-d-galactopyranoside (ONPG), coenzyme A, dimethyl sulfoxide (DMSO), PerfectHyp hybridization solution, and spermidine were purchased from Sigma, St. Louis, Mo. Recombinant RNaseOUT RNase inhibitor, RPMI 1640 culture medium, fetal calf serum, l-glutamine, and penicillin-streptomycin (Pen/Strep) were purchased from Gibco BRL, Rockville, Md. HindIII restriction enzyme, RNase-free DNase I, SOC medium, and Superscript II reverse transcriptase were purchased from Invitrogen Corp., Carlsbad, Calif. SUPERase-In RNase inhibitor was purchased from Ambion, Inc., Austin, Tex. Slide-A-Lyzer dialysis cassettes were purchased from Pierce Biotechnology, Rockford, Ill. Plasmid pSP-luc+, S30 PreMix, complete amino acid mixture, beetle luciferin, luciferase control RNA, the RiboMAX SP6 in vitro transcription system (used for all in vitro transcription reactions), and the nuclease-treated rabbit reticulocyte lysate system were purchased from Promega Corp., Madison, Wis. The EcoPro T7 Escherichia coli extract system was purchased from Novagen, Madison, Wis. E. coli gyrase was purchased from TopoGEN, Inc., Columbus, Ohio. Supercoiled ColE1 DNA was isolated by a published procedure (19). Costar microtiter plates were purchased from Corning, Inc., Acton, Mass. Millipore MultiScreen-FB filter plates and Montage PCR96 plates were purchased from Millipore Corp., Billerica, Mass. Laemmli sample buffer was purchased from Bio-Rad, Hercules, Calif. [35S]methionine, [3H]UTP, [α-33P]dCTP, and Sephadex G-25 spin columns were purchased from Amersham Pharmacia, Piscataway, N.J. Supermix liquid scintillation cocktail for microplates was purchased from Perkin-Elmer Life Sciences Inc., Boston, Mass. RNeasy columns and Plasmid Midi kits were purchased from Qiagen, Inc., Valencia, Calif. TaKaRa Ex Taq polymerase was purchased from Fisher Scientific, Pittsburgh, Pa. Bacillus subtilis cDNA labeling primers and Panorama gene arrays containing sequences from B. subtilis strain 168 were purchased from Sigma-Genosys, The Woodlands, Tex. Alamar blue reagent solution was purchased from BioSource International, Camarillo, Calif. Bacterial strains and the Jijoye human lymphoma B-cell line were obtained from the Abbott Laboratories collection or from the American Type Culture Collection, Manassas, Va. Rifampin was purchased from Calbiochem, San Diego, Calif. Oxytetracycline was purchased from Fluka, Buchs, Switzerland. Neamine was purchased from ICN Pharmaceuticals, Costa Mesa, Calif. Linezolid, ciprofloxacin, levofloxacin, norfloxacin, A-72310, and A-692345 were synthesized at Abbott Laboratories. The structures of A-72310 and A-692345 are shown in Fig. 1.
FIG. 1.
Structure of NRI compounds A-72310 and A-692345.
Construction of luciferase reporter plasmid pAS10rbs3.
The pA promoter region from pEVP3 (3) was subcloned as a KpnI-BglII fragment into pSP-luc+ to generate the plasmid pAS10. A new Shine-Dalgarno site (SD) composed of a consensus sequence from S. pneumoniae genes (10) was added with optimized spacing between the SD and the methionine start codon to yield the plasmid pAS10rbs3.
Preparation of S. pneumoniae S30 extract.
The procedure used to prepare S. pneumoniae S30 extract was adapted from those described by Mackie et al. (13) and LeGault et al. (12). Five 1-liter medium bottles containing water (800 ml), Todd-Hewitt broth (30,000 mg/liter), and yeast extract (5,000 mg/liter) were each inoculated with 48 ml of S. pneumoniae ATCC 49619 (overnight cultures) and grown without shaking at 37°C to an optical density at 600 nm of 0.4 to 0.5. Cultures were chilled in an ethanol-ice bath for 30 min, and cells were pelleted at 6,000 × g. Cell pellets were pooled and washed twice with 500 ml of cold high-salt buffer (10 mM Tris HCl [pH 7.5], 1 M NH4Cl, 15 mM Mg acetate, 50 mM KCl, 6 mM 2-mercaptoethanol) and once with 500 ml of low-salt buffer (same as high-salt buffer, except for 60 mM NH4Cl). Cell pellets were stored at −80°C. Frozen cells were thawed for 30 min on ice water and suspended in 50 ml of low-salt buffer. Phenylmethylsulfonyl fluoride and diisopropyl fluorophosphate were added to final concentrations of 1 and 0.1 mM, respectively, along with 1,500 U of RNaseOUT RNase inhibitor. The cells were lysed in a French press at 12,500 lb/in2, followed by centrifugation at 4°C for 10 min at 14,000 × g. The supernatant was centrifuged at 4°C for 40 min at 30,000 × g, and the resulting extract was dialyzed in Slide-A-Lyzer cassettes (10,000 molecular weight cutoff) at 4°C against three changes (1 liter each) of low-salt buffer (twice for 1 h each and then once overnight). The resulting extract was divided into aliquots, frozen in a dry ice-ethanol bath, and stored at −80°C. Each batch of extract was tested against a panel of different transcription and translation inhibitors and several negative controls to ensure batch-to-batch consistency.
S. pneumoniae transcription/translation assay (luciferase readout).
Experiments were carried out in 96-well half-area black plates (Costar no. 3694) in a final volume of 16.5 μl. Test compounds were dispensed into plates as DMSO solutions and dried in a vacuum centrifuge at 42°C for 15 min. Diluted S. pneumoniae S30 extract (8.5 μl), composed of 2.33 μl of S30 extract, 1.17 μl of buffer A [10 mM Tris-HCl (pH 7.5), 60 mM NH4Cl, 15 mM Mg(AcO)2, 50 mM KCl, 6 mM 2-mercaptoethanol], and 5 μl of H2O, was added to each well. Plates were incubated with shaking at 25°C for 10 min, followed by addition of 7.5 μl of plasmid solution containing 5 μl of PreMix, pAS10rbs3 (0.5 μl of a 1-μg/μl solution in water), 1.25 μl of 1 mM complete amino acid mix, and 1.25 μl of H2O. Plates were incubated at 25°C with shaking for 2 h. Reactions were stopped by adding 20 μl of kanamycin solution (20 μg of kanamycin per ml in water) to each well. Plates were analyzed on a Wallac Victor2 plate reader by automatically injecting 50 μl of luciferin reagent (20 mM Tricine, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM dithiothreitol [DTT], 270 μM coenzyme A, 470 μΜ beetle luciferin, 530 μΜ ATP, 1.07 mM MgCO3, adjusted to pH 7.8 with NaOH) one well at a time and immediately reading luminescence after each addition.
S. pneumoniae transcription/translation assay (electrophoretic readout).
The electrophoretic assay was identical to the coupled transcription/translation luciferase assay, with the following exceptions. Reactions involving mixtures containing 14 μl of S30, 7 μl of buffer A, 8.6 μl of H2O, 3.5 μl of pAS10rbs3 (1 μg/μl in water), 20 μl of PreMix, 5 μl of amino acid mix (no methionine), and 24 μCi of [35S]methionine (1,000 Ci/mmol) were carried out in duplicate in 1.5-ml polypropylene tubes. The contents of duplicate tubes were pooled and split: half of the volume was analyzed for luminescence as in the standard luciferase assay, and the other half was treated with 260 μl of cold acetone, incubated on ice for 15 min, and centrifuged at 21,000 × g for 5 min. The supernatant was aspirated, and the pellets containing precipitated protein were dried in a vacuum centrifuge. Pellets were dissolved in 80 μl of Laemmli sample buffer and electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [10% polyacrylamide]) gel. Gel visualization and band quantitation were performed on a Molecular Dynamics Storm 860 PhosphorImager with the supplied software.
S. pneumoniae translation assay (luciferase readout).
The mRNA-driven translation assay was identical to the coupled transcription/translation luciferase assay, with the following exceptions. mRNA encoding the luciferase gene was synthesized from linearized (EcoRI or EcoRV) pAS10rbs3 plasmid and used instead of the pAS10rbs3 plasmid. Each batch of mRNA was assayed for its ability to generate luminescence signal in the assay, and the assay concentration was adjusted accordingly prior to routine use; about 500 ng of mRNA per well was typical.
E. coli translation assay (β-galactosidase readout).
Reagents and methods from the Novagen EcoPro T7 extract system were used with modifications. mRNA was synthesized from a linearized (HindIII) plasmid containing a β-galactosidase gene. Reactions were performed in 96-well, clear, flat-bottom plastic plates (Costar no. 3596). EcoPro T7 extract (14 μl) was added to each well, and plates were agitated at room temperature on a platform shaker. After 10 min, 16 μl of mRNA solution containing β-galactosidase mRNA (2.5 μg in water) and SUPERase-In (0.7 μl, 14 U) was added to each well. The plates were covered with 96-well plate lids (Costar no. 3080) and agitated at room temperature on a platform shaker. After 30 min, 20 μl of kanamycin solution and 50 μl of β-galactosidase substrate buffer (200 mM sodium phosphate [pH 7.3] containing 2 mM MgCl2, 100 mM 2-mercaptoethanol, and 4.41 mM ONPG) were added, and the plates were incubated at 37°C. After 20 min, 150 μl of 1 M Na2CO3 was added to stop β-galactosidase activity, and plates were read for A420 on a Wallac Victor2 plate reader. Control wells containing no mRNA generated a background signal that was subtracted from test well values prior to calculation of the 50% inhibitory concentration (IC50).
Eukaryotic translation assay (luciferase readout).
The Promega rabbit reticulocyte lysate system was used as directed with minor modifications. Experiments were carried out in 96-well, half-area, black plates (Costar no. 3694) in a final volume of 16.5 μl. Test compounds were dispensed into plates as DMSO solutions and dried in a vacuum centrifuge at 42°C for 15 min. Rabbit reticulocyte lysate (11.5 μl) was added to each well, and plates were incubated at room temperature. After 10 min, 5 μl of a solution containing luciferase control RNA (0.033 μg; 0.033 μl of a 1-mg/ml solution in water), RNaseOUT (13.2 U; 0.33 μl of a 40-U/μl solution), complete amino acid mixture (0.33 μl), and water (4.3 μl) was added to each well, and the plates were incubated at 25°C for 2 h with shaking. Reactions were stopped by adding 20 μl of cycloheximide solution (100 μg/ml in water), and plates were read as described for the S. pneumoniae transcription/translation assay.
E. coli gyrase DNA cleavage assay.
For the E. coli gyrase DNA cleavage assay, reaction mixtures contained 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 1 mM ATP, 1.8 mM spermidine, 100 μg of bovine serum albumin per ml, 0.1 μg of supercoiled ColE1 DNA, 3.3 U of gyrase enzyme, 6.5% (wt/vol) glycerol, and various concentrations of the test compound. Otherwise, conditions were identical to those described by Saiki et al. (18).
Jijoye B-cell toxicity assay.
Log-phase Jijoye cells were suspended in fresh culture medium (RPMI 1640 containing 10% fetal calf serum, 1% l-glutamine, and 1% Pen/Strep) at a density of 106 cells/ml and deposited in 96-well plates (Costar no. 3596) at 200 μl per well. Compounds were dissolved in DMSO, serially diluted, and added to the cells over a final concentration range from 0.8 to 100 μg/ml, while maintaining a final DMSO concentration of 1%. The plates were incubated at 37°C in an atmosphere containing 5% CO2 for approximately 24 h, and the effect on cellular respiration was evaluated by using Alamar blue (16). Alamar blue reagent solution, prewarmed to 37°C, was added (10 μl/well), and incubation was continued for an additional 4 h. Plates were fluorometrically assayed (λex = 530 nm, λem = 572 nm) on a Wallac Victor2 plate reader. Vehicle controls (untreated cells containing 1% DMSO) and positive control cytotoxins were run in each experiment.
S. pneumoniae transcription assay.
Compounds were tested in V-bottom polypropylene 96-well plates (Costar no. 3363). Test compounds were dispensed into plates as DMSO solutions and dried in a vacuum centrifuge at 42°C for 15 min. S. pneumoniae S30 extract (2.3 μl per well) was dispensed, and the plates were incubated at room temperature for 10 min. Plasmid solution (7.5 μl) comprising 1 μg of pAS10rbs3 (1 μl of a 1-μg/μl solution in water), 3.5 μl of PreMix, 1 μl of [3H]UTP (1 mCi/ml), and 2 μl of water was added to each well, and plates were covered with plastic wrap and incubated at room temperature. After 1 h, the contents of the wells were transferred to the corresponding wells of a MultiScreen-FB filter plate, each containing 200 μl of 5% trichloroacetic acid (TCA), to precipitate total protein. Plates were refrigerated for at least 1 h and then vacuumed dry on a plate manifold. Wells were washed with 5% TCA (three times at 200 μl each) and 95% ethanol (once at 200 μl). Excess ethanol was removed by blotting, and plates were allowed to air dry. One hundred microliters of liquid scintillant was added to each well, and the plates were covered, agitated, and read on a Wallac Trilux 1450 Microbeta scintillation counter.
Antibacterial susceptibility determination.
Organisms were tested by the broth microdilution method as described by the National Committee for Clinical Laboratory Standards (14).
B. subtilis gene expression array experiments.
Ten milliliters of Luria-Bertani (LB) broth was inoculated with B. subtilis 2521 (frozen glycerol stock) and incubated at 37°C overnight. The cells were diluted 1:100 (vol/vol) in fresh LB medium and grown at 37°C to an optical density of 0.6 at 600 nm. The cell suspension was delivered in 2-ml aliquots to sterile polypropylene tubes containing DMSO solutions of individual test compounds. In all cases, including the vehicle control, the final DMSO concentration was 1.25%. After 90 min, the cells were harvested by centrifugation at 4°C for 2 min at 18,000 × g, and total RNA was immediately isolated with RNeasy columns, as directed by the manufacturer. The resulting RNA was treated with RNase-free DNase I for 15 min to completely eliminate chromosomal DNA, purified a second time with RNeasy columns, and quantitated spectrophotometrically. Radioactive cDNA probes were prepared by using the purified total RNA as a template. Reverse transcription reaction mixtures contained 1 μg of total RNA, [α-33P]dCTP (5 μl of a 10-μCi/μl stock), 200 U of Superscript II reverse transcriptase, 4 μl of B. subtilis cDNA labeling primers consisting of a mixture of open reading frame-specific oligonucleotides, and sufficient Superscript II buffer to bring the final volume to 60 μl. Reaction tubes were placed in a Perkin-Elmer GeneAmp (PCR System 9600) thermal cycler that had been preheated to 90°C. After 4 min, the temperature was ramped linearly to 42°C over 20 min and then held constant at 42°C for an additional 2 h. The radiolabeled cDNAs were separated from unincorporated nucleotides by using Sephadex G-25 spin columns. Prior to hybridization, B. subtilis Panorama gene array membranes were washed with 1× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) (twice at 350 ml each), blotted on filter paper to remove excess liquid, and placed individually into polyester bags (8 by 12 in.) containing 20 ml of PerfectHyp hybridization solution. Bags were immersed in a water bath at 64°C with constant shaking (120 rpm). After 2 to 3 h, the bags were drained, and a solution of radiolabeled cDNA (approximately 30 to 40 μCi) in 20 ml of fresh hybridization solution was added to each bag. Hybridization to the membrane was carried out for 2 to 3 days in the 64°C bath. The arrays were washed at room temperature with 0.1× SSPE containing 0.5% SDS (twice for 10 min each) and at 64°C (twice for 10 min each), blotted on filter paper to remove excess liquid, covered with plastic wrap, and exposed to phosphorimaging screens for 2 to 3 days. The screens were scanned at a 50-μm resolution on a Molecular Dynamics Storm 860 PhosphorImager with the supplied software. To graphically process the data, the inverse image of the array derived from drug-treated cells (light spots where radiolabeled mRNAs had hybridized) was made semitransparent (50% opacity in Adobe Photoshop) and overlaid on the array derived from untreated cells (dark spots where radiolabeled mRNAs had hybridized).
Spot intensities were also quantified in ImageQuant 5.0 (Molecular Dynamics [now Amersham Biosciences, Piscataway, N.J.]) Spots in the arrays corresponding to mRNAs of interest were manually selected, and the average pixel volume in each spot (raw value) was determined. Spot areas used for the quantitation of each mRNA were equal in both the inhibitor-treated and vehicle-treated controls. The average pixel intensity of a blank region in each array was subtracted from raw values for all of the spots in that image to yield background-corrected values. Values were then normalized to account for differences in total intensity between control and drug-treated arrays. The intensity value for each spot in the drug-treated array image divided by the intensity value for the corresponding spot in the untreated array image gave the fold change values listed in Table 4.
TABLE 4.
Quantitative analysis of expression array data for A-72310-, tetracycline-, clarithromycin-, and ciprofloxacin-treated B. subtilis
| Functional category and gene name | Protein | Fold changea
|
|||
|---|---|---|---|---|---|
| A-72310 | Tetracycline | Clarithromycin | Ciprofloxacin | ||
| Ribosomal proteins | |||||
| rplB | Ribosomal protein L2 (BL2) | 34.3 | 38.6 | 14.8 | 0.6 |
| rplC | Ribosomal protein L3 (BL3) | 6.4 | 7.1 | 11.8 | 0.5 |
| rplD | Ribosomal protein L4 | 2.9 | 2.0 | 4.7 | 1.0 |
| rplE | Ribosomal protein L5 (BL6) | 13.6 | 5.8 | 17.4 | 1.2 |
| rplF | Ribosomal protein L6 (BL8) | 7.7 | 7.1 | 10.8 | 1.4 |
| rplJ | Ribosomal protein L10 (BL5) | 1.8 | 1.4 | 6.1 | 1.1 |
| rplK | Ribosomal protein L11 (BL11) | 4.3 | 2.2 | 6.1 | 0.5 |
| rplL | Ribosomal protein L12 (BL9) | 7.9 | 6.7 | 16.2 | 0.7 |
| rplN | Ribosomal protein L14 | 24.8 | 16.8 | 17.2 | 0.6 |
| rplO | Ribosomal protein L15 | 1.8 | 1.6 | 4.1 | 1.0 |
| rplP | Ribosomal protein L16 | 5.6 | 4.9 | 18.3 | 0.5 |
| rplQ | Ribosomal protein L17 (BL15) | 36.6 | 27.5 | 10.4 | 0.9 |
| rplR | Ribosomal protein L18 | 46.5 | 42.1 | 19.0 | 0.8 |
| rplT | Ribosomal protein L20 | 10.9 | 25.3 | 11.8 | 0.6 |
| rplV | Ribosomal protein L22 (BL17) | 33.6 | 9.8 | 21.8 | 0.4 |
| rplW | Ribosomal protein L23 | 37.4 | 32.9 | 12.7 | 0.4 |
| rplX | Ribosomal protein L24 (BL23) (histone-like protein HPB12) | 10.1 | 7.7 | 15.5 | 0.8 |
| rpmA | Ribosomal protein L27 (BL24) | 4.0 | 4.1 | 4.1 | 0.9 |
| rpmC | Ribosomal protein L29 | 23.8 | 16.6 | 22.4 | 0.6 |
| rpmD | Ribosomal protein L30 (BL27) | 9.4 | 8.3 | 16.7 | 0.7 |
| rpmI | Ribosomal protein L35 | 2.5 | 4.6 | 8.4 | 0.7 |
| rpmJ | Ribosomal protein L36 (ribosomal protein B) | 6.0 | 4.5 | 4.5 | 1.0 |
| rpsC | Ribosomal protein S3 (BS3) | 24.5 | 11.0 | 22.1 | 0.7 |
| rpsD | Ribosomal protein S4 (BS4) | 0.6 | 1.1 | 4.4 | 0.4 |
| rpsE | Ribosomal protein S5 | 7.7 | 9.1 | 7.2 | 1.8 |
| rpsG | Ribosomal protein S7 (BS7) | 1.7 | 1.8 | 8.1 | 0.6 |
| rpsH | Ribosomal protein S8 (BS8) | 21.0 | 10.5 | 19.7 | 0.9 |
| rpsJ | Ribosomal protein S10 (BS13) | 8.6 | 7.8 | 8.5 | 0.3 |
| rpsL | Ribosomal protein S12 (BS12) | 6.9 | 3.0 | 5.8 | 0.4 |
| rpsM | Ribosomal protein S13 | 13.6 | 6.5 | 9.6 | 0.8 |
| rpsN | Ribosomal protein S14 | 4.1 | 4.6 | 11.0 | 1.0 |
| rpsQ | Ribosomal protein S17 (BS16) | 23.9 | 19.2 | 20.5 | 0.8 |
| rpsS | Ribosomal protein S19 (BS19) | 11.5 | 7.2 | 19.0 | 0.4 |
| ybxF | Similar to ribosomal protein L7AE family | 1.6 | 1.5 | 5.3 | 4.9 |
| Translation factors | |||||
| efp | Elongation factor P | 2.6 | 0.5 | 0.1 | 0.7 |
| frr | Ribosome recycling factor | 3.1 | 1.1 | 1.4 | 0.4 |
| fus | Elongation factor G | 3.4 | 1.4 | 9.9 | 1.3 |
| infA | Initiation factor IF-1 | 9.4 | 6.0 | 8.0 | 0.9 |
| infB | Initiation factor IF-2 | 3.0 | 1.9 | 1.1 | 1.0 |
| infC | Initiation factor IF-3 | 2.3 | 3.5 | 4.2 | 1.0 |
| prfA | Peptide chain release factor 1 | 0.9 | 0.9 | 1.0 | 0.8 |
| prfB | Peptide chain release factor 2 | 1.8 | 1.0 | 1.2 | 0.8 |
| tufA | Elongation factor Tu | 9.6 | 3.0 | 1.8 | 1.0 |
| ykrS | Similar to initiation factor elF-2B (alpha subunit) | 4.7 | 5.5 | 0.6 | 1.3 |
| ylaG | Similar to GTP-binding elongation factor | 3.2 | 2.1 | 0.4 | 0.2 |
| tRNA synthetases | |||||
| glyQ | Glycyl-tRNA synthetase (alpha subunit) | 1.7 | 1.8 | 1.0 | 0.9 |
| glyS | Glycyl-tRNA synthetase (beta subunit) | 3.5 | 1.3 | 0.2 | 0.7 |
| hisS | Hlstidyl-tRNA synthetase | 2.0 | 1.4 | 0.7 | 0.8 |
| ileS | Isoleucyl-tRNA synthetase | 3.9 | 0.2 | 0.5 | 0.1 |
| pheS | Phenylalanyl-tRNA synthetase (alpha subunit) | 2.8 | 1.1 | 0.6 | 1.1 |
| pheT | Phenylalanyl-tRNA synthetase (beta subunit) | 1.8 | 1.5 | 0.7 | 1.0 |
| serS | Seryl-tRNA synthetase | 1.2 | 1.1 | 0.8 | 1.2 |
| thrS | Threonyl-tRNA synthetase | 1.8 | 1.7 | 0.7 | 1.4 |
| tyrS | Tyrosyl-tRNA synthetase | 1.0 | 2.2 | 1.0 | 0.5 |
| valS | Valyl-tRNA synthetase | 0.8 | 0.8 | 0.4 | 1.0 |
| Amino acid biosynthesis | |||||
| ilvN | Acetolactate synthase (acetohydroxy-acid synthase) (small subunit) | 0.7 | 5.6 | 1.0 | 0.6/PICK> |
| leuD | 3-Isopropylmalate dehydratase (small subunit) | 1.7 | 8.4 | 5.8 | 1.0 |
| metK | S-Adenosylmethionine synthetase | 3.1 | 2.6 | 1.3 | 1.0 |
| trpA | Tryptophan synthase (alpha subunit) | 1.0 | 0.5 | 0.6 | 9.8 |
| Other categories | |||||
| adk | Adenylate kinase | 10.2 | 9.2 | 10.6 | 1.9 |
| lexA | Transcriptional regulator | 1.0 | 1.0 | 0.8 | 0.9 |
| map | Methionine aminopeptidase | 6.8 | 5.2 | 9.4 | 0.9 |
| recA | Multifunctional SOS repair regulator | 3.8 | 1.8 | 1.8 | 3.3 |
| rsbW | Switch protein/serine kinase and anti-sigma factor | 5.6 | 3.4 | 1.5 | 3.1 |
| rsbX | Serine phosphatase (dephosphorylation of RsbS) | 6.7 | 5.1 | 0.5 | 3.9 |
| ruvA | Holiday junction DNA helicase | 1.5 | 3.1 | 2.5 | 0.7 |
| ruvB | Holiday junction DNA helicase | 0.6 | 1.5 | 1.9 | 1.1 |
| secY | Preprotein translocase subunit | 10.4 | 9.2 | 8.4 | 1.1 |
| ssb | Single-strand DNA-binding protein | 17.9 | 9.6 | 15.6 | 0.6 |
| uvrA | Excinuclease ABC (subunit A) | 3.1 | 2.1 | 1.1 | 2.4 |
| ybyB | Unknown | 2.5 | 2.6 | 0.1 | 6.1 |
| yqgZ | Unknown | 0.8 | 0.9 | 0.5 | 3.9 |
| ywzA | Unknown | 3.0 | 1.2 | 1.1 | 3.2 |
Fold change relative to vehicle-treated control.
RESULTS AND DISCUSSION
Discovery and characterization of biochemical properties of A-72310.
In an effort to identify novel inhibitors of bacterial translation, we executed a cell-free transcription/translation high-throughput screen using extract from the gram-positive pathogen S. pneumoniae. Although these experiments would have been easier and less expensive to conduct with extracts from E. coli, we opted for the relevant system to maximize the probability of identifying a lead series with specific utility against this important pathogen.
S. pneumoniae S30 extract was used to catalyze coupled transcription/translation of a plasmid-borne luciferase gene, encoding an mRNA with optimized ribosome binding site and spacing. Activity of the resulting luciferase protein was quantified in a luminescence assay. As expected, compounds inhibiting transcription, translation, or the luminescence reaction itself reduced the luminescence output relative to the uninhibited control. A battery of control compounds, including the transcription inhibitor, rifampin, and translation inhibitors, chloramphenicol, linezolid, tetracycline, erythromycin, and streptomycin, was used to validate the assay (Table 1). The goal of the screen and subsequent characterization of hits was to identify compounds capable of inhibiting bacterial, but not eukaryotic RNA or protein synthesis. Ideally, these would therefore also exhibit selective antibacterial cell activity. Out of a screening library of∼300,000 small molecules, only one compound, A-72310, had this desirable profile of selectively inhibiting the bacterial coupled assay (IC50 = 10 μM).
TABLE 1.
In vitro data for NRI compounds and control antibacterials
| Antibacterial compound | IC50 (μM)
|
MIC (μg/ml)
|
Jijoye B-cell cytotoxicity LD50 (μg/ml)c | ||||
|---|---|---|---|---|---|---|---|
| S. pneumoniae transcription/translation | S. pneumoniae translation | E. coli translation | Eukaryotic translation | S. pneumoniae ATCC 6303a | S. pneumoniae ATCC 7257b | ||
| A-72310 | 10 | 5 | 5 | >100 | >64 | 32 | >100 |
| A-692345 | 14 | 19 | >100 | 32 | 8 | >100 | |
| Chloramphenicol | 10 | 3.4 | 0.5 | >100 | 2 | 1 | >100 |
| Linezolid | 1.6 | 2.9 | 1.2 | >100 | 2 | 0.5 | >100 |
| Minocycline | 1.2 | 3.5 | >0.06 | >0.06 | 20 | ||
| Oxytetracycline | 5.4 | 30 | 0.5 | 0.125 | >100 | ||
| Tetracycline | 0.35 | 0.55 | 2.6 | >100 | 0.5 | 0.125 | 50 |
| Erythromycin | 0.3 | 0.4 | 0.25 | >100 | ≤0.06 | ≤0.06 | >100 |
| Lincomycin | 0.2 | 10.9 | 0.125 | ≤0.06 | >100 | ||
| Neamine | 0.65 | 4 | 32 | 16 | >100 | ||
| Puromycin | 0.1 | 0.2 | 1 | 1 | <0.8 | ||
| Streptomycin | <0.02 | 0.03 | 0.04 | >100 | >64 | 16 | >100 |
| Thiostrepton | 0.2 | 0.5 | ≤0.06 | ≤0.06 | >50 | ||
| Rifampicin | 0.1 | >100 | >100 | ≤0.06 | ≤0.06 | >100 | |
| Cycloheximide | >100 | >100 | >100 | 0.3 | >64 | >64 | 1.5 |
| Ciprofloxacin | >100 | >100 | >100 | >100 | 0.5 | 8 | 70 |
| Levofloxacin | >100 | >100 | 1 | 16 | >100 | ||
| Norfloxacin | >100 | >100 | 1 | 32 | 110 | ||
Quinolone susceptible.
Quinolone resistant.
LD50, 50% lethal dose.
To dissect the mechanism of action of A-72310, we used an uncoupled cell-free S. pneumoniae translation assay, initiated by the addition of mRNA encoding luciferase. In contrast to the coupled transcription/translation system, this assay is sensitive only to compounds that inhibit translation and is not affected by those that block transcription. A-72310 showed potent inhibitory activity (IC50 = 5 μM), as did all of the positive control ribosome inhibitors (Table 1). This inhibitory activity is within the experimental error of that observed in the coupled assay and is sufficient to account for the activity observed in the coupled assay.
To prove A-72310 did not also inhibit RNA synthesis, we tested it in a cell-free S. pneumoniae transcription assay, monitoring incorporation of radiolabeled uridine into RNA. In contrast to the transcription inhibitors rifampin and streptolydigin (IC50 = 0.1 and 1 μM, respectively), control translation inhibitors and A-72310 were inactive. (Table 1)
We conducted experiments to eliminate the possibility that A-72310 disrupts the luminescent assay readout by directly inhibiting the luciferase enzymatic activity or quenching the luminescence reaction. In particular, we ran uninhibited translation reactions to generate luciferase in situ, stopped translation with kanamycin, and then added A-72310. A-72310 did not inhibit light output in these tests, showing that it does not inhibit luciferase enzymatic activity or quench luminescence. Furthermore, when aliquots of coupled transcription/translation reaction mixtures containing A-72310 and [35S]methionine were electrophoresed, the inhibition of luciferase production measured on the gel was the same as that observed when using the luminescence output (data not shown).
Because of our interest in broad-spectrum agents, we tested A-72310 in a bacterial translation assay using cell extracts from E. coli. For this purpose, we used a commercially available E. coli extract system from Novagen charged with mRNA encoding β-galactosidase. The β-galactosidase concentration was quantified by its enzymatic action on a chromogenic substrate. A-72310 showed potent inhibitory activity (IC50 = 5 μM), as did all the positive control ribosome inhibitors (Table 1). These data all confirm that A-72310 is a bona fide inhibitor of bacterial translation.
A-72310 was inactive in a eukaryotic translation assay (IC50, >100 μM). The assay was validated with cycloheximide, a potent eukaryotic translation inhibitor. The observed biochemical selectivity of A-72310 for bacterial protein synthesis was its most unique characteristic compared to all other high-throughput screening hits.
Given its close structural similarity to antibacterial quinolones that inhibit DNA gyrase and topoisomerase IV, we were surprised that A-72310 inhibited bacterial translation. Indeed, none of the control quinolones we tested inhibited translation (Table 1). Furthermore, although thousands of related quinolone analogs were tested in the course of screening, none of them were detected as assay hits. These data support the conclusion that the structural similarity to quinolones is irrelevant to the biochemical properties of A-72310.
The vast majority of antibacterial quinolones, including thousands of other quinolones in our screening library, have an aryl or alkyl substituent at the N-1 position. Before the discovery of the NRI class, A-72310 would have been considered only as a synthetic intermediate for the corresponding N-1-substituted naphthyridine. Placement of typical antibacterial quinolone substituents at the N-1 position results in a substantial loss of translation inhibitory activity. For example, esafloxacin, the N-1 ethyl derivative of A-72310, inhibits coupled S. pneumoniae transcription/translation too weakly (IC50 = 100 μM) to have been detected in the screen. Conversely, addition of the N-1 ethyl substituent drastically enhances the compound’s ability to form cleavable DNA-gyrase complexes, a biochemical property common to marketed quinolones (19). The propensity of a compound to induce formation of cleavable complexes can be expressed as the concentration of compound required to induce 50% of maximal DNA linearization (CC50). Whereas esafloxacin is a potent inhibitor with CC50 = 0.4 μM, A-72310 exhibits an E. coli gyrase CC50 > 100 μM.
A-72310 has been the subject of a medicinal chemistry effort resulting in an extensive series of ribosome inhibitors in the NRI class. Details of the structure-activity relationships (SAR) for the NRI series will be the subject of other articles, but it is worth noting that the SAR for the NRI series diverges significantly from quinolone SAR, not only at N-1, but also at many other positions.
Biological characterization of NRIs.
In addition to their cell-free biochemical activity, NRIs also inhibit the growth of gram-positive and gram-negative bacteria in vitro. During the course of chemical optimization, we prepared analogs such as A-692345 that exhibit a biochemical profile similar to A-72310 and possess improved antibacterial activity (Table 1). The spectrum of antibacterial activity for A-692345, which is representative of the series, includes the key pathogens associated with respiratory tract infections (Staphylococcus aureus, S. pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis), but not E. coli or Pseudomonas aeruginosa (Table 2). In addition to inhibiting susceptible strains, NRIs inhibit the growth of clinical isolates of S. aureus and S. pneumoniae with all major mechanisms of drug resistance (Table 3). MICs for antibacterial-resistant strains, including quinolone-resistant S. pneumoniae 7257 and S. aureus 3405, 7702, and 7681 (all of which have point mutations in the genes encoding DNA gyrase and topoisomerase IV) were the same as or lower than those for susceptible strains. These quinolone-resistant strains exhibit at least 16-fold reduced susceptibility to ciprofloxacin and other quinolones (data not shown). Major mechanisms of antibacterial resistance do not affect the ability of A-692435 to inhibit bacterial growth, because the NRI inhibits the ribosome via a novel mechanism.
TABLE 2.
Antibacterial spectrum of NRI compounds
| NRI | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S. pneumoniae 6303 | S. pneumoniae 7257 | H. influenzae GYR 1435 | S. aureus 6538P | M. catarrhalis 2604 | E. faecalis PIU 1967 | E. coli 5358 | P. aeruginosa 5007 | |
| A-72310 | >64 | 32 | 4 | >64 | 4 | >64 | >64 | >64 |
| A-692345 | 16 | 4 | 1 | 16 | 0.5 | 16 | >64 | >64 |
| Levofloxacin | 1 | 16 | 0.015 | 0.12 | ≤0.12 | 1 | NDa | 0.5 |
ND, not determined.
TABLE 3.
Antibacterial activity of A-692345 against clinically resistant S. pneumoniae and S. aureus
| Organism | Resistancea | MIC (μg/ml)
|
|
|---|---|---|---|
| A-692345 | Levofloxacin | ||
| S. pneumoniae | |||
| ATCC 6303 | 32 | 0.5 | |
| ATCC 49619 | PEN | 16 | 1 |
| ATCC 5957 | PEN | 16 | 1 |
| ATCC 7257 | LVX | 4 | 8 |
| ATCC 6396 | ERYb, CLX, TET | 16 | 1 |
| ATCC 5649 | PEN, ERYc | 8 | 4 |
| S. aureus | |||
| ATCC 29213 | 32 | 0.25 | |
| ATCC 1350 | PEN, ERY, CLX | 32 | 0.12 |
| ATCC 7667 | PEN, OXA | 32 | 0.25 |
| ATCC 7702 | PEN, OXA, CHL, TET, GEN, CIP | 32 | 16 |
| ATCC 2175 | PEN, OXA, ERY, CLX, TET, GEN, CHL | 32 | 0.25 |
| ATCC 7681 | PEN, OXA, ERY, CLX, CHL, GEN, CIP | 32 | 8 |
| ATCC 3405 | PEN, OXA, ERY, CLX, TET, GEN, CIP | 32 | 16 |
Resistance phenotype determined by NCCLS criteria (data not shown). Tested antibiotics included penicillin (PEN), erythromycin (ERY), clindamycin (CLX), tetracycline (TET), chloramphenicol (CHL), and levofloxacin (LVX) for S. pneumoniae and penicillin, oxacillin (OXA), erythromycin, clindamycin, chloramphenicol, tetracycline, gentamicin (GEN), and ciprofloxacin (CIP) for S. aureus.
erm (B)
mef (A)
Consistent with the lack of eukaryotic translation inhibition activity, the NRIs were inactive in a Jijoye B-cell assay (Table 1).
To prove that members of the NRI series inhibit translation in bacteria, gene expression array and mutational studies were undertaken. When administered to bacterial cells, ribosome inhibitors induce an increase in production of mRNAs encoding ribosomal protein genes (15). We used commercially available gene expression arrays to monitor global changes in mRNA levels in response to A-72310 and several control antibacterials. We made initial attempts to observe changes in gene expression by treating cells with a concentration of inhibitor equal to the MIC for B. subtilis 2521, measured under standard conditions. In some cases, we observed that the inhibitor concentration needed to be significantly higher in order to observe reproducible changes in gene expression. Specifically, the concentrations required to observe a reproducible pattern were as follows: clarithromycin, 10 μg/ml (MIC = 0.03 μg/ml); ciprofloxacin, 0.1 μg/ml (MIC = 0.01 μg/ml); tetracycline, 0.1 μg/ml (MIC = 0.06 μg/ml); and A-72310, 128 μg/ml (MIC = 128 μg/ml). Notably high concentrations of clarithromycin and ciprofloxacin relative to their standard MICs were required to induce reproducible gene expression changes in our experiments, while the concentration of A-72310 required to induce this effect was 128 μg/ml, equal to the MIC. At lower concentrations, A-72310 did not induce reproducible changes in gene expression, demonstrating that an especially high drug concentration relative to the MIC is not required to induce a regulatory response in the cell.
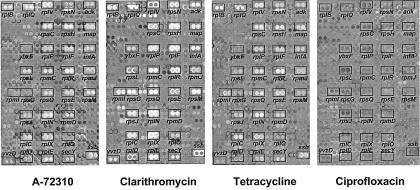
Composite gene expression array images that graphically display the relative changes in concentration of individual B. subtilis mRNAs induced by antibacterial treatment are shown in Fig. 2. In the processed images, upregulated mRNAs appear as white spots, downregulated mRNAs appear as dark spots, and mRNAs whose level is unaffected by antibacterial treatment are gray and of similar intensity to the neutral gray background; each gene is arrayed in duplicate. The translation inhibitors clarithromycin, tetracycline, and A-72310 all caused the induction of ribosomal protein genes in particular, consistent with their common mechanism of action. In sharp contrast, ciprofloxacin caused no significant change in the levels of the same mRNAs. Although semiquantitative, this graphical approach is a fast and effective way to visualize patterns of mRNA regulation across the genome and to match response patterns of compounds of unknown mechanism to those of control compounds.
FIG. 2.
Composite B. subtilis gene expression array images for A-72310 (128 μg/ml), clarithromycin (10 μg/ml), tetracycline (0.1 μg/ml), and ciprofloxacin (0.1 μg/ml). Drug-induced changes in mRNA concentration are indicated by either light (upregulated), dark (downregulated), or neutral gray (unchanged) spots (in duplicate).
We also quantitatively analyzed changes in B. subtilis mRNA expression that occurred in response to treatment with A-72310, clarithromycin, tetracycline, and ciprofloxacin. These mRNAs encode a range of proteins, including ribosomal proteins, tRNA synthetases, translation factors, proteins involved in amino acid biosynthesis, proteins in the pur gene cluster, and an additional seven mRNAs from other classifications whose expression level was significantly altered (greater-than-threefold increase or decrease in mRNA level relative to the level of the vehicle-treated control) by treatment with at least one of the compounds. The relative changes in mRNA expression for all of these genes are listed in Table 4. Spots in Fig. 2 are annotated with the corresponding gene name so that spot intensity in the figure can be correlated with the fold change in Table 4. Importantly, the spots in the array can be visually rank ordered, consistent with the quantitative data. For instance, the brightest spots in Fig. 2 (A-72310) occur for rplR mRNA, the most highly induced message for this inhibitor (37-fold increase). Similarly, spots for infA and rpsM in Fig. 2 (A-72310) are roughly equivalent and have medium intensity (9- and 14-fold increases, respectively), while spots corresponding to ybxF appear unchanged, consistent with data in Table 4.
All three translation inhibitors induced changes in mRNA levels that are quantitatively different from the changes induced by ciprofloxacin. Levels of mRNA encoding 26 of the ribosomal proteins are increased by more than a factor of 3 in the A-72310-, tetracycline-, and clarithromycin-treated samples. Ten of these messengers increase by greater than an order of magnitude. These levels of upregulation are considerably higher than those observed in S. pneumoniae (15) but agree qualitatively. In contrast, the levels of mRNA encoding these same genes did not change significantly in the ciprofloxacin-treated cells. All of the translation inhibitors also induced mRNAs for translation initiation factor IF-1 (infA) and translation elongation factor G (fus) by more than threefold.
Contrary to the observations in S. pneumoniae (15), levels of mRNAs encoding genes in the pur cluster (pbuX, purB, purE, purF, purH, purK, purL, purM, purN, and xpt) were not significantly upregulated in these experiments (data not shown), nor did we observe any significant downregulation of any aminoacyl-tRNA synthetase mRNAs with drug treatment, although levels of a few of these mRNAs (ileS and glyS) actually increased in the presence of A-72310 (Table 4). Messenger encoding most of the amino acid biosynthesis genes we analyzed was not significantly changed by any of the inhibitors. These unaffected species included glyA, asd, dapA, ilvD, trpC, ilvC, aspB, and metB (data not shown).
Messenger for genes from additional classes was also upregulated uniformly by the three translation inhibitors and not by ciprofloxacin. These included mRNA encoding a single-stranded DNA-binding protein (ssb), adenylate kinase (adk), methionine aminopeptidase (map), a preprotein translocase subunit (secY), and a gene of unknown function (yvzd).
In B. subtilis, ciprofloxacin treatment significantly induced mRNA encoding the alpha subunit of tryptophan synthase (trpA), an unknown protein similar to the ribosomal protein L7AE family (ybxF), and two unknown genes, ybyB and yqgZ. Levels of these mRNAs were not affected by any the translation inhibitors. Two messengers were also downregulated by ciprofloxacin: isoleucyl-tRNA synthetase (ileS) and a protein similar to GTP-binding elongation factor (ylaG). These were unchanged by tetracycline and clarithromycin, whereas the levels were actually increased in the case of A-72310—the exact opposite of ciprofloxacin. In H. infuenzae, ciprofloxacin was previously shown to induce genes encoding proteins involved in SOS repair (recA, uvrA, and lexA), DNA helicases (ruvA, ruvB), and the excinuclease ABC subunit A (uvrA) (7). Of these genes in B. subtilis, ciprofloxacin only caused a significant, but modest increase for recA, which was also apparent in the A-72310-treated samples. Taken together, these data demonstrate that genes upregulated by ciprofloxacin in B. subtilis are not generally upregulated by the NRI or other translation inhibitors and support the conclusion that the NRI acts by a non-quinolone mechanism in the cell.
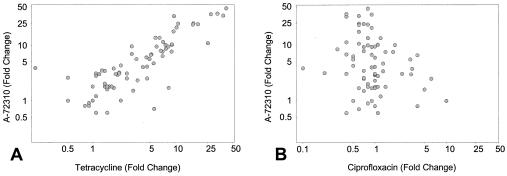
Scatter plots of the fold change in mRNA levels induced by A-72310 and tetracycline further demonstrate that the patterns of gene expression induced by these two inhibitors very closely resemble each other (Fig. 3A). Conversely, there is no correlation in the data for A-72310 and ciprofloxacin (Fig. 3B). The results support the conclusion that A-72310 inhibits translation in B. subtilis and does not induce a cellular response similar to that of ciprofloxacin
FIG. 3.
Scatter plots of the fold change in B. subtilis mRNA levels induced by A-72310 and tetracycline (A) and A-72310 and ciprofloxacin (B).
To further understand the NRI mechanism, we carried out experiments to isolate and characterize NRI-resistant S. pneumoniae point mutants. S. pneumoniae strains resistant to compounds in the NRI series could be selected in the laboratory, albeit at low frequency (≤1 × 10−8). Resistance arose from genetically transferable point mutations in the 16S rRNA gene and the S3 protein. These mutants, while two- to fivefold resistant to multiple compounds in the NRI series, are not cross-resistant to chloramphenicol, linezolid, erythromycin, tetracycline, oxytetracycline, spectinomycin, streptomycin, puromycin, or lincomycin—indicating that NRI class is likely to interact with the ribosome at a new site (A. M. Nilius and R. K. Hickman, personal communication).
The NRI class is a novel antibacterial class that inhibits translation in cell extracts from gram-positive and gram-negative organisms and is biochemically selective versus eukaryotic translation. Compounds in this class inhibit translation in bacterial cells, as evidenced by expression array and mutational experiments, and, despite some chemical similarity, are mechanistically distinct from quinolones, since drug-resistant strains for each class are not cross-resistant with each other. In summary, the NRI is a promising novel antibacterial class, and further chemical optimization could yield a new antibacterial class for treatment of respiratory and other infections.
REFERENCES
- 1.Chen, D. K., A. McGeer, J. C. de Azavedo, and D. E. Low. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 2.Chenoweth, C. E., S. Saint, F. Martinez, J. P. Lynch III, and A. M. Fendrick. 2000. Antimicrobial resistance in Streptococcus pneumoniae: implications for patients with community-acquired pneumonia. Mayo Clin. Proc. 75:1161-1168. [DOI] [PubMed] [Google Scholar]
- 3.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 4.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50(Suppl. 1):25-37. [DOI] [PubMed] [Google Scholar]
- 6.Gale, E. F., E. Cundliffe, P. E. Reynolds, M. H. Richmond, and M. J. Waring. 1981. Molecular basis of antibiotic action. John Wiley and Sons, London, United Kingdom.
- 7.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure to Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibilities to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles, D. J. C., N. Foloppe, N. B. Matassova, and A. I. Murchie. 2002. The bacterial ribosome, a promising focus for structure-based drug design. Curr. Opin. Pharmacol. 5:501-506. [DOI] [PubMed] [Google Scholar]
- 10.Lacks, S. A., B. Greenberg, and A. G. Sabelnikov. 1995. Possible regulation of DNA methyltransferase expression by RNA processing in Streptococcus pneumoniae. Gene 157:209-212. [DOI] [PubMed] [Google Scholar]
- 11.Lafontaine, D. L. J., and D. Tollervey. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2:514-520. [DOI] [PubMed] [Google Scholar]
- 12.Legault-Demare, L., and G. H. Chambliss. 1974. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J. Bacteriol. 120:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackie, G. A., C. Donly, and P. C. Wong. 1990. Coupled transcription/translation of ribosomal proteins, p. 191-211. In G. Spedding (ed.), Ribosomes and protein synthesis: a practical approach. Oxford University Press, New York, N.Y.
- 14.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Document no. M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagé, B., M. Pagé, and C. Noël. 1993. A fluorometric assay for cytotoxicity measurements in vitro. Int. J. Oncol. 3:473-476. [PubMed] [Google Scholar]
- 17.Pestka, S. 1971. Inhibitors of ribosome functions. Annu. Rev. Microbiol. 25:487-562. [DOI] [PubMed] [Google Scholar]
- 18.Saiki, A. Y. C., L. L. Shen, C.-M. Chen, J. Baranowski, and C. G. Lerner. 1999. DNA cleavage activities of Staphylococcus aureus gyrase and topoisomerase IV stimulated by quinolones and 2-pyridones. Antimicrob. Agents Chemother. 43:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen, L. L., W. E. Kohlbrenner, D. Weigl, and J. Baranowski. 1989. Mechanism of quinolone inhibition of DNA gyrase: appearance of unique norfloxacin binding sites in enzyme-DNA complexes. J. Biol. Chem. 264:2973-2978. [PubMed] [Google Scholar]
- 20.Spahn, C. M., and C. D. Prescott. 1996. Throwing a spanner in the works: antibiotics and the translation apparatus. J. Mol. Med. 74:423-439. [DOI] [PubMed] [Google Scholar]
- 21.Vázquez, D. 1979. Inhibitors of protein sythesis. Springer-Verlag, New York, N.Y.
- 22.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, C. Lexau, A. Reingold, L. Lefkowitz, P. R. Cieslak, M. Cetron, E. R. Zell, J. H. Jorgensen, A. Schuchat, R. R. Facklam, and N. M. Bennett. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917-1924. [DOI] [PubMed] [Google Scholar]