Abstract
Five major lineages of methicillin-resistant Staphylococcus aureus (MRSA) have evolved since the introduction of methicillin for the treatment of infections caused by penicillin-resistant S. aureus in 1959. The clones of these lineages are responsible for the vast majority of hospital-acquired MRSA disease globally. We have constructed high-resolution evolutionary models for each lineage using a parsimony approach with 15 partial gene sequences from 147 geographically diverse isolates. On the basis of these models, we infer that MRSA has emerged at least 20 times upon acquisition of the methicillin resistance determinant, which is carried on a mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec). The acquisition of SCCmec by sensitive clones was four times more common than the replacement of one SCCmec with another. Notably, SCCmec type IV was found in twice as many clones as any other SCCmec type, and it is this SCCmec type which is commonly found in clones from patients with community-acquired MRSA disease. Our findings suggest that most clones of MRSA arise by the acquisition of SCCmec type IV by methicillin-sensitive isolates.
Staphylococcus aureus has a proven ability to adapt to the selective pressure of antibiotics. The emergence of strains with resistance to penicillin and methicillin was reported in 1948 and 1961, respectively (2, 17). In both cases, resistance developed within a few years of the introduction of the antibiotics into clinical medicine. At present, methicillin-resistant S. aureus (MRSA) strains with resistance to vancomycin are emerging (4, 13). This trend is a cause of great public health concern, as vancomycin is the antibiotic of last resort for the treatment of MRSA infections. There are indications that the epidemiology of MRSA may also be expanding, from being a major cause of hospital-acquired infections to becoming a cause of community-acquired infections (3, 14). Consequently, there has been keen interest in understanding how natural populations of MRSA have evolved over the past half century.
Our understanding of the evolution of MRSA has benefited from the development of molecular tools that allow characterization of both the strain phylogeny and the methicillin resistance determinant. Strain phylogeny can be resolved by multilocus sequence typing, which identifies a strain unambiguously on the basis of its sequence at seven housekeeping genes (8). The product of the mecA gene confers methicillin resistance and is carried on a mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec).Four main types of SCCmec, which differ in size and composition, have been described for S. aureus (16, 22). The application of multilocus sequence typing and SCCmec typing to international collections of MRSA and methicillin-susceptible S. aureus (MSSA) isolates has revealed that (i) methicillin resistance has emerged in five phylogenetically distinct lineages, (ii) methicillin resistance has emerged on multiple occasions within a given phylogenetic lineage, and (iii) most MRSA disease is caused by a relatively small number of pandemic clones (9).
The frequency with which SCCmec is acquired in nature is unknown. Are new clones of MRSA sporadically emerging by a frequent acquisition of SCCmec, perhaps on a daily basis in the hospital setting, or is SCCmec a relatively stable locus that has been acquired on rare historical occasions? To address this question, we developed a high-resolution multilocus typing method that could detect SCCmec acquisitions among closely related isolates of S. aureus. The data allowed us to construct detailed evolutionary models upon which inferences regarding the acquisition of SCCmec could be based.
MATERIALS AND METHODS
Bacterial isolates.
A previous study of 912 MRSA and MSSA isolates found that methicillin resistance had emerged in five phylogenetically distinct lineages of S. aureus (9). We selected 136 of these isolates, which represented all hospital-acquired MRSA clones with an international distribution, plus related MRSA and MSSA isolates. We also included four MSSA isolates from Cuba and seven historically early MSSA isolates (5). Multiple isolates of the same clone were selected to be geographically diverse. A total of 147 isolates from 18 countries were included in this study, of which 92 were MRSA and 55 were MSSA. All isolates were stored at −80°C and grown overnight on blood agar plates at 37°C.
Sequence typing.
We selected seven S. aureus surface protein (sas) genes that encode the LPXTG cell wall attachment motif. The genes chosen were among the least well-characterized sas genes and have been named sasA, sasB, sasD, sasE, sasF, sasH, and sasI by Mazmanian et al. (24). sasB has homology with fmtB, a gene required for methicillin resistance (20). sasE has also been named sai-1, which encodes a 29-kDa surface protein (GenBank accession number AB042826), and isdA, an iron-responsive surface determinant (25). sasH has homology with a gene that encodes a 5′ nucleotidase. On the basis of the COL genome sequence (http://www.tigr.org), sasA and sasF were located ∼11 kb from each other, and all other sas loci were >50 kb from each other.
Polymorphic regions of the sas genes were identified by using the publicly available S. aureus genomic sequences, and primers were designed to amplify ∼450-bp fragments (Table 1). The primers of Enright et al. (8) were used for multilocus sequencing of the seven housekeeping genes, arcC, aroE, glpF, gmk, pta, tpi, and yqiL. Primers were also designed to amplify the short sequence repeats of the spa gene (Table 1). Chromosomal DNA was isolated with a DNeasy kit (Qiagen). The PCR conditions were identical to those of Enright et al. (8), with a few modifications. The PCR mixtures used 10 pmol of the sas- and housekeeping gene-specific primers per μl and 100 pmol of the spa-specific primers per μl. PCR annealing temperatures were 45°C for the sas genes and 55°C for the housekeeping and spa genes. Sequencing of the DNA of both strands was performed as described by Enright et al. (8) with an ABI 3700 automated sequencer (PE Applied Biosystems).
TABLE 1.
PCR primers
| Primer pair | Sequence (5′-3′) |
|---|---|
| Primary sas-specific primers | |
| sasAF | TCAACATCCTCAAAGAATACTACA |
| sasAR | ATGCGTTACTTAAGCCACCAATAC |
| sasBF | GTTGCAGCGCTTGTGACT |
| sasBR | ATTTTTGAGATTTCTTCGTTTTTA |
| sasDF | GGCGGAGTAGTACCACAAGGAA |
| sasDR | AATGCTAAGAATAACCCAGATACT |
| sasEF | TTACAATGCAAACAATCAAGA |
| sasER | GTTTAGGCGTTTCGTTATGTTTT |
| sasFF | GGATAGCAAAGACAATAAAAGTTC |
| sasFR | TGATATGTGTAATGTTGCGTTGAG |
| sasHF | CGCACCAACTAACAAACCAACTAC |
| sasHR | TACGCCAATAATTCCATAACGA |
| sasIF | ATACTATCACTTTTTCAGCATCAA |
| sasIR | TCATTCGTTTTATCGTTAGTATTA |
| Alternative sas-specific primers | |
| sasDF4 | ATTTTGTTGCATTTCTTT |
| sasDR4 | TCACACGATTTTTCTATTAT |
| sasEF2 | ACCCTGGTAAAGTGATT |
| sasER2 | CTAAAAGGGCAAGTGTT |
| sasIF3 | CAAACACTGCGAAAAACTATCCT |
| sasIR3 | TTTCACCTTTATCATTTTTCATTT |
| spa-specific primersspa1095u | AGACGATCCTTCGGTGA |
| spa1517d | CAGCAGTAGTGCCGTTTG |
| mec-specific primersmec | |
| 3490u | ATGATTCAATGCCTAAACCTAATCG (common) |
| 4110Ad | GAATTATAACTGGGAATATTTTAAATCCCA (class B mec) |
| 4110Bd | CTTTTTGTTTCAAAGTCATACTATTTTCAAC (class A mec) |
For the sas and housekeeping genes, unique sequences defined alleles, and the unique series of alleles at each locus defined a sequence type (ST). For the sas genes, a database of alleles was compiled and is available upon request. For the housekeeping genes, the alleles were identified by using the MLST database, available at http://www.mlst.net. The spa repeats were characterized as described by Shopsin et al. (31). The resolution obtained by different typing methods was compared by using Simpson's index of diversity (D) and was calculated as 1 − {[1/N(N − 1)]Σni(ni − 1)}, where n is the number of isolates belonging to the ith ST and N is the total number of isolates in the sample (12, 15).
SCCmec typing.
SCCmec types were assigned by PCR analysis of the ccr and mec genes, as described by Okuma et al. (27). The primers of Ito et al. (16) were used for PCR of the ccr genes, and new primers (Table 1) were used for PCR of the mec genes. Structural variants of SCCmec were detected by the multiplex PCR analysis of Oliveira and de Lencastre (28).
Nomenclature.
This study used a newly proposed nomenclature for MRSA clones that was based on a combination of the STs at seven housekeeping genes and the SCCmec type (9). For example, an MRSA clone of ST250 and SCCmec type I is referred to as ST250-MRSA-I, and an MSSA clone of ST8 is referred to as ST8-MSSA.
Phylogenetic analyses.
The alleles of the sas and the housekeeping genes were aligned by using the CLUSTALW program (33) with default parameters, followed by manual inspection. Insertion and deletion polymorphisms were ignored during phylogenetic analyses. MEGA software (version 2.0) (21) was used to calculate ds/dn ratios by the modified Nei and Gojobori method. ds/dn is the ratio of the number of synonymous or silent nucleotide changes (ds) to the number of nonsynonymous or amino acid-replacing nucleotide changes (dn). The PAUP* program (version 4.0b10) (32) was used to construct neighbor-joining (NJ) and maximum-parsimony (MP) trees. NJ trees were constructed by using the absolute number of nucleotide differences between STs. MP trees of all STs were constructed with a heuristic search and random addition of taxa. MP trees of specific lineages were constructed with a branch-and-bound search to ensure that all most parsimonious trees were found. Bootstrapping was performed with 1,000 replicates.
Nucleotide sequence accession numbers.
The sas gene sequences reported in this study have been deposited in GenBank under accession numbers AY175407 to AY175464.
RESULTS
Development of a high-resolution multilocus typing method.
To develop a multilocus typing method that could detect SCCmec acquisitions among closely related isolates, we sequenced fragments of seven S. aureus surface protein (sas) genes, which we expected would accumulate variations more rapidly than the seven housekeeping genes. We sequenced a similar number of nucleotides (P = 0.710, Mann-Whitney U test) and observed a similar number of alleles (P = 0.165) for the sas and housekeeping genes (Table 2). However, the sas genes provided significantly more polymorphic sites (P = 0.011) and parsimony-informative sites (P = 0.026) than the housekeeping genes (Table 2). The variation found in sasD appeared to be similar to that found in the housekeeping genes. sasD and sasF both presented alleles with insertion or deletion polymorphisms.
TABLE 2.
Nucleotide sequence variation among alleles of sas and housekeeping loci
| Gene | Sequence length (bp) | No. of alleles | No. of polymorphic sitesa | No. of informative sitesa | ds/dn |
|---|---|---|---|---|---|
| sas genes | |||||
| sasA | 462 | 8 | 59 | 51 | 3.9 |
| sasB | 462 | 7 | 40 | 20 | 9.9 |
| sasD | 453 | 7 | 11 | 2 | |
| sasE | 450 | 7 | 41 | 28 | 2.0 |
| sasF | 439 | 14 | 34 | 24 | |
| sasH | 467 | 8 | 36 | 18 | 5.2 |
| sasI | 452 | 7 | 43 | 25 | 3.0 |
| Housekeeping genes | |||||
| arcC | 456 | 6 | 7 | 4 | 6.3 |
| aroE | 456 | 14 | 19 | 10 | 3.4 |
| glpF | 465 | 8 | 11 | 4 | 3.8 |
| gmk | 429 | 10 | 13 | 7 | 6.5 |
| pta | 474 | 9 | 11 | 4 | 5.3 |
| tpi | 402 | 11 | 13 | 6 | 4.0 |
| yqiL | 516 | 9 | 13 | 4 | 4.3 |
Excludes insertions and deletions at sasD and sasF.
There is reason to suspect that variations in genes encoding surface proteins may be influenced by the selective pressure of the host immune system, but we found no evidence of diversifying selection in the fragments sequenced. When all alleles were considered, the proportion of synonymous nucleotide changes was greater than the proportion of nonsynonymous nucleotide changes (see the ds/dn ratios in Table 2). There were no differences in the ds/dn ratios for the sas and housekeeping genes (P = 0.530).
The short sequence repeats of the spa gene, which encodes immunoglobulin G-binding protein A, provides a highly discriminatory sequence-based method for the typing of S. aureus (31). We compared the resolutions of the different typing methods using Simpson's index of diversity, which takes into account the observed numbers of STs and their frequencies of occurrence (12, 15). The diversity resolved by the spa repeats was significantly greater than the diversity resolved by either multilocus method (i.e., the 95% confidence intervals did not overlap), and the diversities resolved by the multilocus methods were similar to each other (Table 3). However, the multilocus methods in combination provided a significantly greater resolution than either multilocus method by itself and a resolution similar to that obtained with the spa repeats (Table 3).
TABLE 3.
Resolution of sequence-based typing methods
| Typing method | No. of types | % Most frequent type | Index of diversity (95% CIa) |
|---|---|---|---|
| spa repeats | 48 | 11.0 | 0.959 (0.948-0.970) |
| sas genes | 33 | 14.4 | 0.929 (0.927-0.932) |
| Housekeeping genes | 45 | 15.6 | 0.927 (0.909-0.946) |
| Multilocus methods (sas + housekeeping) | 66 | 11.0 | 0.965 (0.953-0.977) |
| All methods (spa + sas + housekeeping) | 84 | 5.1 | 0.988 (0.983-0.993) |
CI, confidence interval.
Construction of evolutionary models.
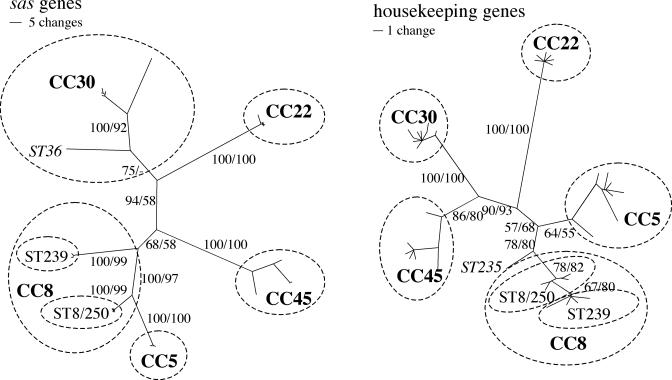
To compare the phylogenetic signals of the sas and housekeeping genes, we constructed NJ and MP trees based on the 3,185 nucleotides of the sas genes and the 3,198 nucleotides of the housekeeping genes. Previously, it was found that MRSA had emerged in five phylogenetically distinct lineages called clonal complexes (CCs) (9). The sas and housekeeping genes classified all but one isolate of ST36 into the same CCs on both NJ and MP trees with high bootstrap support (Fig. 1). The bootstrap support for the CCs was generally higher for the trees based on the sas genes, presumably because these genes provided the most polymorphisms (Table 2).
FIG. 1.
Phylogenetic trees of STs based on sas and housekeeping genes. The trees were constructed with the NJ algorithm and were based on the absolute number of nucleotide differences between STs. The scale shows the relative amount of change along branches. The trees were also constructed by MP analyses. The numbers on the branches refer to the NJ or MP bootstrap percentages. The two isolates of ST36 and ST235 that grouped anomalously on the trees are in italics. The five major CCs are circled.
The branching order between the CCs significantly differed between the sas and housekeeping genes, as witnessed by the high bootstrap support for conflicting arrangements of CCs (Fig. 1). Thus, no inferences regarding the relationships between CCs are made with these data. Of note, the housekeeping genes grouped the pandemic clones of ST8, ST250, and ST239 and their variants into a single lineage, called CC8 (Fig. 1). The sas genes grouped ST8 and ST250 into the same lineage but clearly showed that ST239 was a diverged branch of CC8. Moreover, the sas genes were identical for ST235 and some isolates of ST8, but these clones differed at five of seven housekeeping genes (Fig. 1).
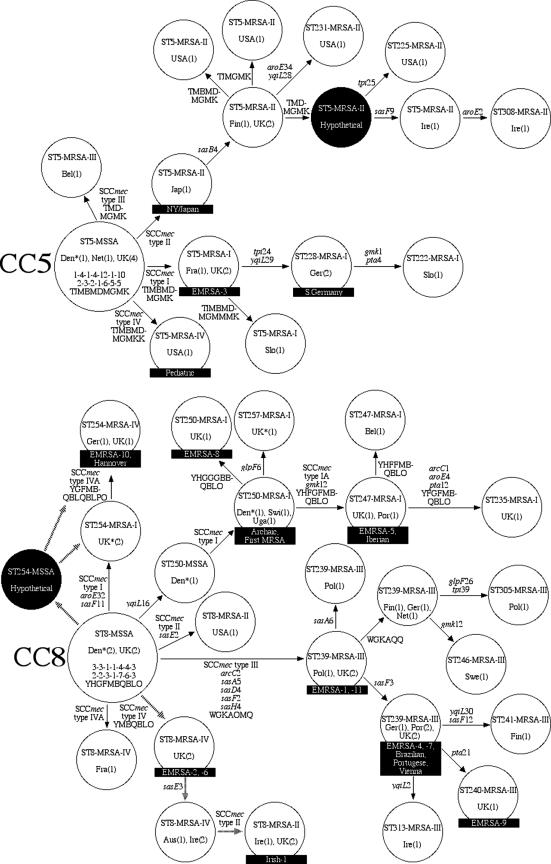
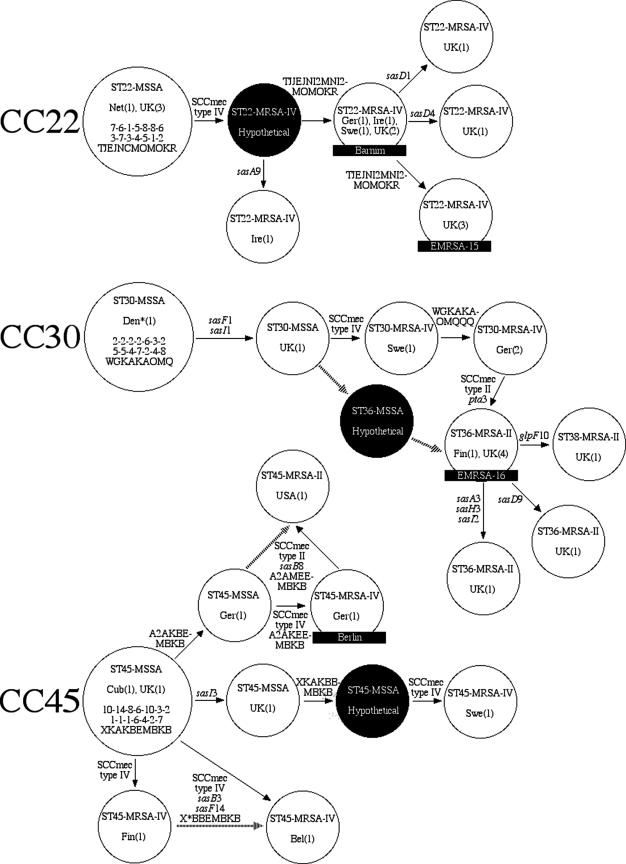
To investigate the branching order within CCs, we combined the nucleotide sequences of the 14 sas and housekeeping genes into a single data set with 6,383 nucleotides. This total evidence approach is justified, given that the data classified the isolates into the same CCs, with the two exceptions noted above. MP trees were constructed for each CC by using the combined data set. CC5, CC8 (consisting of separate trees for ST8-ST250 and ST239), and CC45 yielded single MP trees with perfect consistency. CC22 yielded two MP trees that differed in the placement of a single ST. CC30 yielded 17 MP trees. Evolutionary models for each CC were constructed by expanding the MP trees to include SCCmec type and spa type and assuming the fewest additional changes to the trees (Fig. 2).
FIG. 2.
Proposed evolutionary models for the emergence of MRSA in CC5 and CC8 (left panel) and CC22, CC30, and CC45 (right panel). The models were based on MP analyses of the combined sas and housekeeping genes. The SCCmec type and the spa type were added to the models by assuming the fewest additional changes to the trees. Only those branches on which MRSA isolates have emerged are shown. Large circles represent ancestral clones. Smaller circles represent descendant clones. Arrows indicate the directions and relative amounts of change between isolates. Names and countries of isolation are given for all clones. The numbers in parentheses after the country names represent the numbers of clones. Full genotypes are given for the ancestors with their housekeeping gene alleles at arcC-aroE-glpF-gmk-pta-tpi-yqiL (top line), sas gene alleles at sasA-sasB-sasD-sasE-sasF-sasH-sasI (middle line), and spa repeats (bottom line). Previously named clones are indicated in black boxes. Dotted lines represent alternative evolutionary hypotheses. Isolates from the 1950s and 1960s are indicated by asterisks next to the country of isolation; all other isolates are from the 1980s and 1990s. The country abbreviations are as follows: Aus, Australia; Bel, Belgium; Cub, Cuba; Den, Denmark; Fin, Finland; Fra, France; Ger, Germany; Ire, Ireland; Jap, Japan; Net, Netherlands; Pol, Poland; Por, Portugal; Slo, Slovenia; Swe, Sweden; UK, United Kingdom; USA, United States.
Of the 59 steps depicted in our evolutionary models (Fig. 2), the housekeeping genes and SCCmec type resolved 21 steps, the spa repeats and sas genes resolved 24 steps, and a combination of these two categories of typing methods resolved 14 steps. While the spa repeats and sas genes provided a modest but significant increase in typing resolution of 6.1% (98.8% diversity − 92.7% diversity) over that of the housekeeping genes (Table 3), these markers provided a large increase in phylogenetic resolution of 40.7% (24/59 steps, respectively) over that provided by the housekeeping genes and SCCmec type.
Evolution within CC5 and CC8.
CC5 and CC8 represented the most diversified lineages of MRSA and contained the most pandemic clones (Fig. 2, left panel). The putative ancestors of these lineages were the STs that had the largest number of single-step variants (i.e., single-locus variants and single-nucleotide variants) and were represented by historically early MSSA isolates (5). Within CC5, we propose that all four SCCmec types were acquired once by ST5-MSSA ancestors (Fig. 2, left panel). Within CC8, we propose that three SCCmec types were acquired on multiple occasions (Fig. 2, left panel).
Of note, we propose that ST254-MRSA-I, represented by two isolates from the United Kingdom from 1962 and 1965, respectively, arose independently of the archaic clone (30) in the early days of MRSA emergence. It is simpler to assume that ST254-MRSA-I evolved from ST8-MSSA by acquiring SCCmec type I and point mutations in aroE and sasF rather than to assume that ST254-MRSA-I evolved from ST250-MRSA-I, which would require a back-mutation at yqiL and the point mutations in aroE and sasF. To our knowledge, there are no known isolates of ST254-MSSA, but this possibility is indicated by dashed lines in Fig. 2 (left panel).
The integration of pUB110 into the SCCmec locus (28) gave rise to the variant SCCmec types IA and IVA (Fig. 2, left panel). The lineage leading to the Irish-1 clone is marked by several uncharacterized modifications in the SCCmec locus. ST8-MRSA-IV from Australia and Ireland have the ccr and mec genes characteristic of SCCmec type IV (27) but amplify only mecA in a multiplex PCR (28). Likewise, ST8-MRSA-II from the United Kingdom and Ireland, the Irish-1 clone, have the ccr and mec genes characteristic of SCCmec type II but either lack the kdp gene characteristic of SCCmec type II or have a multiplex PCR pattern identical to that of SCCmec type IV. Thus, the evolutionary pathways of this lineage remain unclear, as indicated by the dashed lines in Fig. 2 (left panel).
A hypothesis for the origin of ST239.
It was noted that ST239 represents a distinct branch within CC8 (Fig. 1). This lineage includes numerous clones such as the epidemic methicillin-resistant S. aureus type 1 (EMRSA-1); EMRSA-4; EMRSA-7; EMRSA-9; EMRSA-11; and the Brazilian, Portuguese, and Vienna clones (7, 19, 23, 34). The proposed path leading from ST8 to ST239 involved the acquisition of SCCmec type III and alleles at arcC, sasA, sasD, sasF, sasH, and spa (Fig. 2A). Surprisingly, with the exception of sasD4, the derived alleles were exclusive to ST239 and ST30 and their descendants; sasD4 also occurred in two isolates from CC22. These data suggest either that parallel evolution of multiple genes had occurred in unrelated lineages of S. aureus or that recombination had occurred. We favor the latter hypothesis for two reasons. First, the sasD4 allele had two characteristic deletions of 18 and 42 nucleotides, respectively, which are unlikely to arise independently in different lineages. Second, the genes arcC, sasF, sasA, sasH, spa, and sasD are contiguous on the COL genome sequence and are centered around the origin of replication. We propose that ST239 arose from a single recombination event that involved the exchange of >200 kb of contiguous DNA between ST30 and ST8. The donor of SCCmectype III remains unknown; the only clone to carry this element besides the clones in the ST239 branch was a single isolate from ST5 (Fig. 2, left panel).
Evolution within CC22, CC30, and CC45.
CC22, CC30, and CC45 represent less diversified lineages of MRSA strains mostly isolated from Europe (Fig. 2, right panel). The putative ancestors for CC22 and CC45 were the STs that had the largest number of single-step variants. No historically early MSSA isolates were available for these lineages. The putative ancestor for CC30 was more difficult to assign. ST30 had one less single-step variant than ST36 but was represented by an historically early MSSA isolate (5) and shared sas alleles with ST39, presumably an old clone within CC30 that has also evolved several single-step variants (10). Thus, our evolutionary model for CC30 is based on the assumption that ST30 should be considered ancestral.
Within CC22, we propose that SCCmec type IV was acquired once (Fig. 2, right panel). Within CC30, we propose that both SCCmec types II and IV were acquired once (Fig. 2, right panel). To our knowledge, there are no known isolates of ST36-MSSA, but this possibility is indicated by dashed lines in Fig. 2 (right panel). Within CC45, we propose that SCCmec type IV was acquired four times and that SCCmec type II was acquired once (Fig. 2, right panel). Alternative evolutionary hypotheses within CC45 are indicated by dashed lines in Fig. 2 (right panel).
Patterns of acquisition and variation of SCCmec.
Our evolutionary models indicate that a minimum of 20 acquisitions of SCCmec have occurred in S. aureus. There were 16 acquisitions of SCCmec by an MSSA clone and 4 putative reacquisitions of SCCmec by an MRSA clone (Table 4). There were 10 acquisitions of SCCmec type IV and 10 acquisitions of the other SCCmec types (Table 4). Nearly half (9 of 20) of all acquisitions involved an MSSA clone that acquired SCCmec type IV.
TABLE 4.
Characteristics of SCCmec
| SCCmec type | Size of element (kb) | No. of inferred acquisitions by:
|
No. of variants | |
|---|---|---|---|---|
| MSSA clones | MRSA clones | |||
| I | 34 | 3 | 2 | |
| II | 52 | 2 | 3 | 5 |
| III | 67 | 2 | 7 | |
| IV | 21-24 | 9 | 1 | 3 |
SCCmec variants IA and IVA have been characterized by others (28) and were included in our evolutionary models. We note that there were two conflicts in the classification of SCCmec types based on the ccr and mec gene (16, 27) and multiplex PCR (28) analyses. These cases involved isolates of SCCmec types II and IV and types II and III, which differed in the ccr and mec analyses but which had identical multiplex patterns (data not shown). We observed a total of 17 variants of SCCmec (Table 4). The largest SCCmec types, types II and III, presented the most STs. As further work is necessary to characterize the new variants, we have not included them in our evolutionary models.
DISCUSSION
For a decade it has been recognized that methicillin resistance has emerged multiple times within S. aureus (11, 26), but the frequency of resistance acquisition and any underlying historical patterns have remained unknown. Our previous work (9) has shown that MRSA has arisen on multiple occasions within successful MSSA lineages, but we were unable to resolve the full evolutionary history of MRSA since multiple acquisitions of the same SCCmec type by a single ST could not be discerned. In this work, we augmented the standard multilocus sequence typing scheme with an additional set of highly variable sas genes which allowed us to resolve a more complete evolutionary history of MRSA.
We propose several novel hypotheses for the emergence of MRSA. First, we propose that two MRSA clones (ST247, ST257) emerged from the first MRSA clone (ST250) by stepwise evolution and that one MRSA clone (ST254) emerged independently within 4 to 5 years of the first reported isolation of MRSA. These data make a case for multiple emergences of MRSA early in its history. Second, we propose that ST239 was created by the exchange of >200 kb of contiguous DNA between ST30 and ST8. This hypothesis is supported by partial sequencing of an additional 26 genes (D. A. Robinson and M. C. Enright, submitted for publication). ST239 and its descendants represent a phylogenetically distinct and clinically important branch within CC8. de Lencastre and colleagues (29) have reported that clones of ST239 are among the most prevalent in Portuguese hospitals and were recently found to be highly prevalent in hospitals in China and Taiwan (6). Third, we propose that CC22, CC30, and CC45 represent major lineages of MRSA that account for 8 of 20 SCCmec acquisitions. CC45, in particular, has not been widely recognized as a major lineage of MRSA, but we found that isolates of CC45 were geographically distributed in both the Eastern and the Western Hemispheres and that the lineage has acquired SCCmec four or five times.
We provide the first estimate of the number of times that SCCmec has been acquired by S. aureus in nature. In most cases, SCCmec is retained as the lineages evolve. Our data suggest that the spontaneous excision of SCCmec does not occur frequently from an evolutionary perspective. We propose that on four occasions SCCmec was reacquired by an MRSA clone. The proposed evolutionary events leading to the Irish-1 clone within CC8 are unclear. However, the proposed evolutionary events leading to the Hannover clone within CC8 and the EMRSA-16 clone within CC30 are clearer, and there are no known MSSA intermediates to support an alternative evolutionary path. The proposed acquisition of SCCmec type II by an SCCmec type IV clone within CC45 can be interpreted differently and requires testing. A mechanism for reacquisition of SCCmec could involve the loss of one SCCmec type followed by acquisition of another SCCmec type within a short time period. Although experiments have shown that the recombinase genes encoded by SCCmec, ccrA, and ccrB are sufficient to transfer the element from a multicopy plasmid into the chromosome in a site-specific and orientation-specific manner (18), the precise mechanism by which SCCmec is transferred in nature is not known. We note that our estimate provides a lower bound of the actual number of acquisitions that have occurred in nature. The estimate will undoubtedly increase as more isolates from patients with community-acquired disease and more isolates from a wider geographic area are studied.
The observation that the largest SCCmec elements, types II and III, had the most structural variants, as detected by PCR, might be explained by the fact that these elements carry more copies of transposable elements, such as IS431 and Tn554 (16). The activities of these transposable elements might play a role in remodeling the structure of SCCmec and, thus, lead to a greater number of structural variants. The observation that the smallest SCCmec element, type IV, had been acquired most often might be explained by a size dependence on the efficiency of DNA transfer. Smaller SCCmec elements may simply transfer more efficiently than larger SCCmec elements.
Alternatively, SCCmec type IV may have been acquired and subsequently retained in S. aureus most often because it has been the selectively favored element of methicillin resistance. Hiramatsu and colleagues (22, 27) suggest that SCCmec type IV may have a lower cost on fitness because it carries only the structural and regulatory genes for methicillin resistance and the recombinase genes for movement of the element. In contrast, SCCmec types I to III can carry additional genes, such as those encoding resistance to non-β-lactam antibiotics and heavy metals (16). The combination of smaller size and lower cost on fitness may make SCCmec type IV the selectively favored element for transfer among all S. aureus isolates.
SCCmec type IV was first discovered in recent studies that examined isolates of community-acquired MRSA (1, 22). Several new clones that carry SCCmec type IV have also been identified from samples from patients with community-acquired MRSA (27). Our results, based on inferences from evolutionary models, show that SCCmec type IV is also the most frequently acquired element within the five major lineages responsible for most hospital-acquired MRSA infections. While the prevalence of disease caused by clones that carry SCCmec types I to III at present may be higher than that caused by clones that carry SCCmec type IV, the more frequent acquisition of SCCmec type IV has markedly increased the genetic diversity of MRSA and suggests that the prevalence of disease caused by clones that carry this element will increase.
Acknowledgments
We thank Brian Spratt and Ed Feil for comments on the manuscript and Paul Wilkinson for technical assistance.
M.C.E. is a Royal Society University Research Fellow. This work was supported by the Wellcome Trust.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 2.Barber, M., and M. Rozwadowska-Dowzenko. 1948. Infection by penicillin-resistant staphylococci. Lancet ii:641-644. [DOI] [PubMed]
- 3.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 5.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Sousa, M. A., M. I. Crisostomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa, M. A., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., K. Okuma, X. X. Ma, M. Yamamoto, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 15.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jevons, M. P. 1961. Celbenin-resistant staphylococci. Br. Med. J. i:124-125.
- 18.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr, S., G. E. Kerr, C. A. Mackintosh, and R. R. Marples. 1990. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J. Hosp. Infect. 16:35-48. [DOI] [PubMed] [Google Scholar]
- 20.Komatsuzawa, H., K. Ohta, M. Sugai, T. Fujiwara, P. Glanzmann, B. Berger Bachi, and H. Suginaka. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 45:421-431. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marples, R. R., and E. M. Cooke. 1985. Workshop on methicillin-resistant Staphylococcus aureus held at the headquarters of the Public Health Laboratory Service on 8 January 1985. J. Hosp. Infect. 6:342-348. [PubMed] [Google Scholar]
- 24.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 25.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 30:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 30.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony and other methods, version 4. Sinauer, Sunderland, Mass.
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witte, W. 1999. Antibiotic resistance in gram-positive bacteria: epidemiological aspects. J. Antimicrob. Chemother. 44(Suppl. A):1-9. [DOI] [PubMed] [Google Scholar]